ABSTRACT
Although virulence of Streptococcus pneumoniae is associated with its capsule, some pathogenic S. pneumoniae isolates lack capsules and are serologically nontypeable (NT). We obtained 64 isolates that were identified as NT “pneumococci” (i.e., bacteria satisfying the conventional definition but without the multilocus sequence typing [MLST]-based definition of S. pneumoniae) by the traditional criteria. All 64 were optochin sensitive and had lytA, and 63 had ply. Twelve isolates had cpsA, suggesting the presence of a conventional but defective capsular polysaccharide synthesis (cps) locus. The 52 cpsA-negative isolates could be divided into three null capsule clades (NCC) based on aliC (aliB-like ORF1), aliD (aliB-like ORF2), and our newly discovered gene, pspK, in their cps loci. pspK encodes a protein with a long alpha-helical region containing an LPxTG motif and a YPT motif known to bind human pIgR. There were nine isolates in NCC1 (pspK+ but negative for aliC and aliD), 32 isolates in NCC2 (aliC+ aliD+ but negative for pspK), and 11 in NCC3 (aliD+ but negative for aliC and pspK). Among 52 cpsA-negative isolates, 41 were identified as S. pneumoniae by MLST analysis. All NCC1 and most NCC2 isolates were S. pneumoniae, whereas all nine NCC3 and two NCC2 isolates were not S. pneumoniae. Several NCC1 and NCC2 isolates from multiple individuals had identical MLST and cps regions, showing that unencapsulated S. pneumoniae can be infectious among humans. Furthermore, NCC1 and NCC2 S. pneumoniae isolates could colonize mice as well as encapsulated S. pneumoniae, although S. pneumoniae with an artificially disrupted cps locus did not. Moreover, an NCC1 isolate with pspK deletion did not colonize mice, suggesting that pspK is critical for colonization. Thus, PspK may provide pneumococci a means of surviving in the nasopharynx without capsule.
IMPORTANCE
The presence of a capsule is critical for many pathogenic bacteria, including pneumococci. Reflecting the pathogenic importance of the pneumococcal capsule, pneumococcal vaccines are designed to elicit anticapsule antibodies. Additional evidence for the pathogenic importance of the pneumococcal capsule is the fact that in pneumococci all the genes necessary for capsule production are together in one genetic locus, which is called the cps locus. However, there are occasional pathogenic pneumococci without capsules, and how they survive in the host without the capsule is unknown. Here, we show that in these acapsular pneumococci, the cps loci have been replaced with various novel genes and they can colonize mouse nasopharynges as well as capsulated pneumococci. Since the genes that replace the cps loci are likely to be important in host survival, they may show new and/or alternative capsule-independent survival mechanisms used by pneumococci.
Introduction
Streptococcus pneumoniae is an important human pathogen and is well known for expressing diverse polysaccharide (PS) capsules (1) that are considered to be essential for its pathogenicity (2, 3). Extensive studies have identified more than 90 serologically distinct capsules (1, 4). The capsular polysaccharide synthesis (cps) locus, which contains the genes necessary for capsule production and is flanked by dexB and aliA genes, has been characterized for all serotypes (1, 5, 6). Despite such a close association between pneumococcal pathogenesis and the capsule, some pneumococcal isolates show no serological evidence for capsule expression. These nontypeable (NT) “pneumococcal” isolates (i.e., bacteria satisfying the conventional definition but without the multilocus sequence typing [MLST]-based definition of S. pneumoniae) have been associated with infectious conjunctivitis (7), occasional invasive pneumococcal diseases (IPD) (8–10), otitis media (11, 12), and nasopharyngeal (NP) carriage (8). Also, the prevalence of NT pneumococcal isolates appears to be increasing (from 1.5% in 2001 to 5.1% in 2006) following the use of a very effective conjugate vaccine (13).
Despite its increase in prevalence (13), little is known about NT “pneumococci.” A major issue is that some of them may not even be S. pneumoniae. Traditionally, S. pneumoniae has been identified as alpha-hemolytic colonies that are optochin sensitive and have genes such as lytA and ply. However, these traditional chemical or genetic tests can fail to distinguish S. pneumoniae from other closely related species in the mitis group, which includes Streptococcus mitis, Streptococcus pseudopneumoniae, and Streptococcus oralis as well as S. pneumoniae (14–16). This difficulty in species identification can now be addressed, since it was recently found that the MLST analysis that examines the sequences of 7 different housekeeping gene loci can be used to reliably distinguish pneumococci from closely related species (17–19).
Using NT “pneumococci” that were typed only by traditional means, one study found two groups of cps among them: groups I and II (8). Group I NT isolates have the cps locus sequences of conventional capsule types (8), but their cps appears to be too disrupted (e.g., cpsE duplication) to produce capsular PS (20). Group II isolates lack all the genes usually found in the cps sequences of encapsulated S. pneumoniae isolates, but their cps contains an aliB-like ORF1 or aliB-like ORF2 (8), which appear to encode lipoproteins (8) and which are referred to here as aliC and aliD for simplicity. Since this study was performed before the use of MLST for species identification, many of these NT “pneumococci” may not be S. pneumoniae by MLST analysis (17, 21, 22). Moreover, the study reporting group I and group II isolates examined only European isolates. We were additionally interested in the possibility that the group II isolates from different regions might show additional diversity in their cps regions. We therefore studied 52 group II isolates from different continents for their MLST and cps diversity. Our study found a new subset of group II isolates containing a novel gene, pspK, which is necessary for nasopharyngeal carriage in mice.
RESULTS
Identification of group II NT “pneumococcal” isolates.
All 64 NT “pneumococcal” isolates from three continents were positive for lytA and ply except for one isolate (MNZ1052), which had lytA but not ply (Table 1). To exclude group I isolates, we used PCR to look for the presence of the cpsA gene. Twelve isolates had cpsA and were thus identified as group I isolates. The remaining 52 isolates were thus identified as group II isolates and chosen for additional studies (Table 1).
TABLE 1 .
Characterization of null-capsule (group II) “pneumococcal” isolatesa
| ID code | Source | Diagnosis | PCR result for: |
NC clade | STb | GenBank accession no. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| lytA | ply | aliC | aliD | Intergenic region (kb) | pspK | ||||||
| MNZ11 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 6151 | JF489996 |
| MNZ15 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 1106 | |
| MNZ24 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 6152 | |
| MNZ37 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 1106 | JF489997 |
| MNZ50 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 1106 | |
| MNZ66 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 1106 | |
| MNZ67 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 1464 | JF489998 |
| MNZ75 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 1106 | |
| MNZ731 | South Korea | CAR | + | + | − | − | ND | + | NCC1 | 1106 | |
| MNZ5 | South Korea | CAR | + | + | + | + | 1.5 | ND | NCC2a | 448 | |
| MNZ14 | South Korea | CAR | + | + | + | + | 1.5 | − | NCC2a | 448 | JF489999 |
| MNZ49 | South Korea | CAR | + | + | + | + | 1.5 | ND | NCC2a | 448 | |
| MNZ814 | United States | CONJ | + | + | + | + | 1.5 | ND | NCC2a | 448 | JF490003 |
| MNZ815 | United States | CONJ | + | + | + | + | ND | ND | NCC2a | 448 | |
| MNZ816 | United States | CONJ | + | + | + | + | ND | ND | NCC2a | 448 | |
| MNZ817 | United States | CONJ | + | + | + | + | ND | ND | NCC2a | 448 | |
| MNZ818 | United States | CONJ | + | + | + | + | 1.5 | ND | NCC2a | 448 | JF490004 |
| MNZ084 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ105 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ106 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ109 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ112 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ115 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ085 | Brazil | CAR | + | + | + | + | 1.5 | ND | NCC2a | 2315 | JF490000 |
| MNZ089 | Brazil | CAR | + | + | + | + | 1.5 | ND | NCC2a | 2315 | JF490001 |
| MNZ090 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ095 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ104 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ107 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ113 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ114 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ121 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ122 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ125 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ127 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ130 | Brazil | CAR | + | + | + | + | 1.5 | ND | NCC2a | 2315 | JF490002 |
| MNZ132 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ133 | Brazil | CAR | + | + | + | + | ND | ND | NCC2a | 2315 | |
| MNZ41 | South Korea | CAR | + | + | + | + | 1.0 | ND | NCC2b | 6153 | JF490005 |
| MNZ43 | South Korea | CAR | + | + | + | + | 1.0 | ND | NCC2b | 6153 | |
| MNZ45 | South Korea | CAR | + | + | + | + | 1.0 | ND | NCC2b | 6153 | |
| MNZ1053 | United States | HIV-CAR | + | + | + | + | 1.0 | ND | NCC2b | P1 | JF490006 |
| MNZ1065 | United States | HIV-CAR | + | + | + | + | 1.0 | ND | NCC2b | P2 | JF490007 |
| MNZ1050 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P3 | JF490008 |
| MNZ1049 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P4 | JF723379 |
| MNZ1054 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P5 | |
| MNZ1051 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P6 | |
| MNZ1056 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P7 | |
| MNZ1063 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P8 | JF723380 |
| MNZ1052 | United States | HIV-CAR | + | −c | − | + | ND | ND | NCC3 | Unknownd | |
| MNZ1055 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P9 | |
| MNZ1064 | United States | HIV-CAR | + | + | − | + | ND | ND | NCC3 | P10 | |
Bold type indicates isolates whose STs were used to create the neighbor-joining tree in Fig. 1. CAR, CONJ, and HIV-CAR, respectively, indicate that the isolates were obtained from the NP of healthy children, the eyes of patients with conjunctivitis, and the NP of HIV+ persons. ND, not done.
P1 to P10 are the provisional sequence types of HIV-CAR isolates. P2 and P5 are single-locus variants of P1 and P4, respectively.
PCR for ply was done twice for this sample.
The ST of MNZ1052 is unknown because its aroE PCR yielded no product.
MLST patterns of the 52 group II NT isolates.
Some streptococcal species related to S. pneumoniae can also meet the traditional genetic or biochemical definitions used for S. pneumoniae (14–16). To determine if the 52 group II isolates are truly S. pneumoniae, we performed MLST for 51 NT isolates (the ply-negative isolate was excluded). Thirty-six isolates (21 Brazilian, 5 U.S. conjunctivitis, and 10 Korean isolates) had pneumococcal sequence types (STs) that have already been included in the database at http://www.mlst.net (Table 1). All 21 isolates from Brazil belonged to ST2315, which shares four genetic loci with ST344. All 5 conjunctivitis isolates from the United States belonged to ST448, and 10 Korean isolates belonged to ST448 (3 isolates), ST1106 (6 isolates), or ST1464 (1 isolate). The remaining 5 Korean isolates belonged to new STs—ST6151 (1 isolate), ST6152 (1 isolate), and ST6153 (3 isolates)—but the new STs were single-locus variants of either ST1106 or ST344. However, all 10 isolates from HIV-positive patients (Table 1) that were analyzed by MLST have sequences that were 1 to 5% divergent from those known for any pneumococci, and they were identified as non-S. pneumoniae by the typing scheme at http://www.mlst.net. MLST analysis was not performed with one ply isolate (MNZ1052).
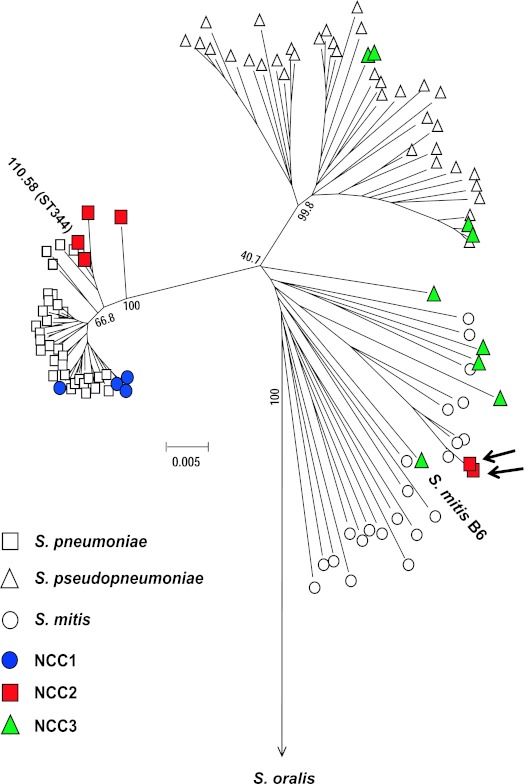
To visualize the genetic relationship of our group II NT isolates within the mitis group, we constructed a neighbor-joining tree with the concatenated sequences of six of the seven MLST loci (excluding ddl) of the 17 isolates (Table 1; Fig. 1). The 52 NT isolates expressed 17 different STs, and the 17 isolates were chosen to represent all the 17 STs. Figure 1 contains two additional data points (110.58 and S. mitis B6) that were based on published sequences (GenBank accession no. FN568063 for S. mitis and Hathaway et al. [8] for 110.58) and were added to the figure for comparison. Locations for S. pneumoniae, S. pseudopneumoniae, S. mitis, and S. oralis species were defined in the neighbor-joining tree using the MLST sequences described in other studies (17, 21, 23). The tree showed that the 10 U.S. isolates from HIV-positive patients that were analyzed by MLST cluster with the S. pseudopneumoniae or S. mitis species (Fig. 1). No isolates clustered with S. oralis. In contrast, 41 of the 52 isolates were clearly localized within S. pneumoniae. Due to duplicated MLST results, there were only 7 distinct STs among the 41 isolates clustering with S. pneumoniae (Fig. 1).
FIG 1 .
A neighbor-joining tree of the MLST sequences of various species in the mitis group in published studies (all open symbols and two solid symbols) and from our study (remaining solid symbols). The two solid symbols from published studies are labeled 110.58 (ST344) (8) and S. mitis B6 (GenBank accession no. FN568063). Data from published studied were used to provide reference points. The symbols indicate S. pneumoniae, S. pseudopneumoniae, S. mitis, NCC1, NCC2, and NCC3. The two short arrows indicate strains MNZ1053 and MNZ1065, which express NCC2b (Table 1). The 52 group II NT isolates contained only 17 unique STs (bold letters in Table 1), and the 17 unique STs are shown in the tree. The tree was constructed with the concatenated sequence (2,751 bp) of 6 of the 7 DNA loci used for MLST by using the method for minimum evolution. The bar indicates 0.005 substitution per site. Bootstrap values are shown for each branch.
Testing the 52 NT isolates with PCR for aliC and aliD genes and intergenic region.
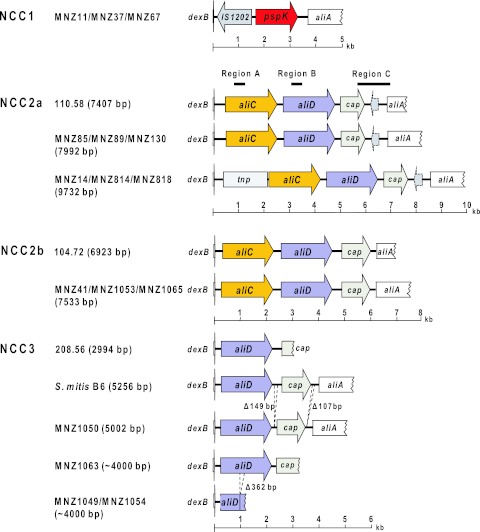
The cps sequences of four group II NT “pneumococcal” isolates have been reported (8). These four sequences are distinguished by the presence of aliC and aliD genes and also by the size of the intergenic region between the capN-like gene and aliA (8) (Fig. 2). To investigate heterogeneity among our 52 group II NT “pneumococci,” we developed PCR tests for aliC, aliD, and the intergenic region. Their amplicons are, respectively, designated regions A, B, and C (Fig. 2). Using the PCR tests, we found nine Korean isolates lacking aliC, aliD, and the intergenic region and assigned them to null capsule clade 1 (NCC1). The PCR tests showed that three other Korean isolates, five U.S. isolates from conjunctivitis patients, and all 21 Brazilian isolates had both aliC and aliD and long (1.5-kb) intergenic regions. This group was assigned to null capsule clade 2a (NCC2a). In contrast, three additional isolates from South Korea and two U.S. isolates from HIV patients had both aliC and aliD but had short (1.0-kb) intergenic regions. This group was assigned to the null capsule clade 2b (NCC2b). Finally, nine U.S. isolates obtained from the NP of HIV-positive patients had aliD but not aliC, and their intergenic regions were variable in size (ranging from 1 to 2 kb). They were assigned to the null capsule clade 3 (NCC3).
FIG 2 .
Diagrammatic representation of the cps regions of unencapsulated strains. aliC, aliD, and cap indicate aliB-like ORF1, aliB-like ORF2, and the capN-like region, respectively (8). tnp denotes a transposon-like insert. The cps regions were sequenced and analyzed for MNZ11, MNZ37, and MNZ67 in NCC1; MNZ14, MNZ41, MNZ85, MNZ89, MNZ130, MNZ814, MNZ818, MNZ1053, and MNZ1065 in NCC2; and MNZ1049, MNZ1050, and MNZ1063 in NCC3. Diagrams of the published sequences of 110.58 (NCC2a), 104.72 (NCC2b), 208.56 (NCC3), and S. mitis B6 (NCC3) (GenBank accession no. AY653209 to AY653212 and FN568063) are shown for comparison purpose. The numbers of bases in parentheses are the complete sequence sizes. MNZ1049 and MNZ1063 cps sequences are incomplete due to technical problems encountered during repeated sequencing attempts; their size estimates are based on electrophoresis of their PCR products.
Genetic heterogeneity among NCC2a, NCC2b, and NCC3 isolates.
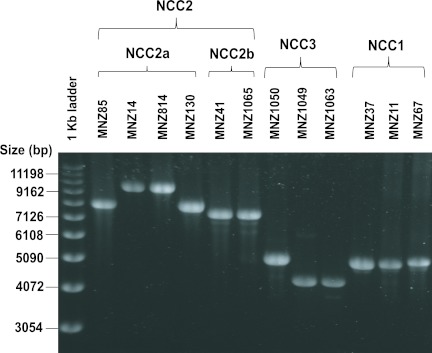
To better define the cps loci of the NCC2 and NCC3 isolates, we determined the DNA sequences of the PCR products from 12 representative isolates (Table 1). Figure 2 shows genetic diagrams of NCC2a, NCC2b, and NCC3 isolates along with those of published reference sequences. As expected, all NCC2 isolates have aliC and aliD as well as capN-like regions. Interestingly, the sequences of the three Brazilian NCC2a isolates (MNZ85, MNZ89, and MNZ130) are almost identical to the sequence of a European isolate (strain 110.58; accession no. AY653211). For example, our sequences of MNZ89, MNZ130, and MNZ85 have a 7,405-bp overlap with the published sequence of isolate 110.58. When the 7,405-bp sequences were compared, MNZ89, MNZ130, and MNZ85 differed from 110.58 by only 0, 2, and 5 nucleotides, respectively. The NCC2a isolates (MNZ14 from South Korea and MNZ814 and MNZ818 from the United States) with an ~10-kb PCR product (Fig. 3) have a 1.7-kb insert between dexB and aliC. Except for the insert region, the remainder of their cps loci (7,405 bp) was again almost identical (six nucleotide difference) to the corresponding cps region of 110.58 (Fig. 2). Thus, NCC2a isolates obtained from diverse geographic locations and in different years had very homologous cps sequences.
FIG 3 .
Electrophoresis patterns of the PCR products of the cps regions of group II unencapsulated strains. PCR was performed with primer sets 5419 and 3419 (Table 2).
The PCR products of the cps regions of three NCC2b isolates [one Korean isolate (MNZ41) and two U.S. isolates (MNZ1053 and MNZ1065)] were ~7.5 kb long. MNZ41 and MNZ1065 are shown in Fig. 3. Their sequences (6,935 bp) were ~97% identical to the published sequence of a European NCC2b isolate (strain 104.72; accession number AY653209) in all parts of the locus: 97% similar in aliC, 95% similar in aliD, and 94% similar in the 5′ region (835 bp) of the capN-like gene. When our three sequences were compared among themselves, the sequences for the two U.S. NCC2b cps regions were 99.8% identical to each other but were only 97.3% identical to the sequence for the Korean NCC2b cps region. Thus, while the general organizations of the cps regions of the NCC2b isolates are similar to those of NCC2a isolates, the NCC2b isolates have more sequence diversity.
As expected from their PCR test results, the sequencing studies with four NCC3 isolates showed them to have aliD and capN-like regions but not aliC regions. While the capN-like region was intact in one isolate (MNZ1050), sequencing of the region could not be completed for other isolates due to technical difficulties in sequencing (Fig. 2). Nevertheless, the sequences of our NCC3 isolates resemble those of isolate 208.56 from Europe and the S. mitis B6 genome (Fig. 2). When the cps locus of MNZ1050 is compared with the corresponding region of the S. mitis strain B6 genome (accession number NC013853), they are ~93% identical for bp 1 to 5060 except for 149-bp and 106-bp deletions at the ends of the capN-like gene (Fig. 2). Interestingly, genetic deletions are common in the NCC3 cps locus. For example, MNZ1049 and MNZ1054 have an incomplete aliD due to a 362-bp deletion. Thus, NCC3 isolates display much more cps sequence diversity than do NCC1 or NCC2 isolates.
Discovery of pspK among NCC1 isolates.
To begin investigating the cps loci of the nine NCC1 isolates from South Korea, we determined the size of their cps regions using PCR primers located in the dexB and aliA genes. We found that their PCR products measured ~5 kb. The PCR products from isolates MNZ37, MNZ11, and MNZ67 with different MLST types are shown in Fig. 3. In contrast, the PCR products of the NCC2 isolates were three different sizes (7.5, 8, and 10 kb), and those of the NCC3 isolates were two different sizes (4 and 5 kb). We then obtained DNA sequences of the PCR products of the three Korean NCC1 isolates (GenBank accession numbers JF489996 to JF489998). As seen in the genetic diagram in Fig. 2, the NCC1 cps region of MNZ37 has no typical genes involved in capsule biosynthesis (e.g., cpsA), but it contains a 1,410-bp insertion sequence and a 1,605-bp novel open reading frame (ORF) (Fig. 2). The insertion sequence is named IS1202, since its sequence is 98% identical to the IS1202 found in the cps locus of the S. pneumoniae strain 361/39 (serotype 35F; accession number CR931707). The novel ORF was found in all 9 NCC1 isolates, and the presence of the novel ORF was included in the definition of NCC1 here.
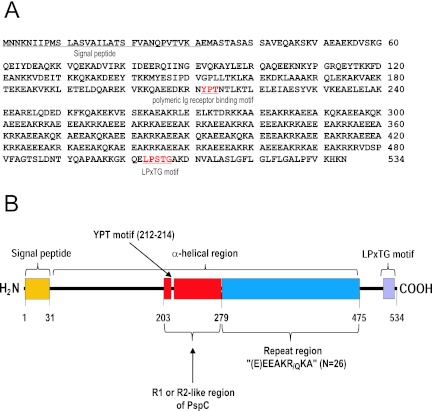
Analysis of the novel ORF showed that it likely encodes a 534-amino-acid (aa) polypeptide with a signal peptide at the N-terminal region, a long alpha-helix region in the middle (between residues 40 and 475), and an LPXTG motif at the C-terminal region (Fig. 4). This finding suggests that the novel ORF encodes a surface protein that is secreted and anchored to peptidoglycan. The ORF was named pneumococcal surface protein Korea (pspK). The alpha-helix region (residues 40 to 475) has two interesting subregions. One subregion (residues 203 to 280) is 67% and 76% identical, respectively, to the R1 (residues 197 to 274) and R2 (residues 350 to 427) domains of PspC/CbpA of S. pneumoniae TIGR4 (accession number NP_346601), which appear to be involved in pneumococcal adhesion to and invasion of human cells (24) and have YPT sequences. The YPT motif is known to be critical in binding human pIgR or secretory component (25) and is also found in the R1-like region of PspK. The other interesting subregion (residues 280 to 475) has 26 repeats containing 7 to 8 amino acids per repeat. The repeat sequence is typically (E)EEAKRKA or (E)EEAKQKA and generally has residues with opposite charges at the ith and (i + 4)th positions, which can stabilize the alpha-helix region (26, 27).
FIG 4 .
Predicted amino acid sequence of the ORF of PspK from MNZ37 (A) and schematic diagram of the PspK of MNZ37 (B). The ORF encodes a 534-aa polypeptide with a signal peptide (1 to 31), an alpha-helical region (39 to 475), and an LPXTG motif. The alpha-helical region from bases 113 to 279 resembles the R1 domain of pspC and has a YPT motif, which is associated with binding to the human secretory component. The repeat region between bases 279 and 475 has 26 repeats of (E)EEAK(R/Q)K.
Nasopharyngeal colonization by NCC1 and NCC2 isolates.
Previous studies using laboratory-derived strains of acapsular S. pneumoniae found that capsule expression is important or even essential in NP carriage in mice (28–30). In view of our finding that naturally acapsular S. pneumoniae isolates readily colonize NP of children (13, 31, 32), we tested the abilities of two naturally acapsular strains (MNZ11 of NCC1 and MNZ85 of NCC2) to colonize NP of mice using the previously established mouse model (28, 33–35). For comparison, we also did colonization studies with three strains (D39, AM1000, and R36A) that have been used in previous colonization studies. Similar results were obtained in two different experiments (Fig. 5), and the combined results were used for data analysis.
FIG 5 .
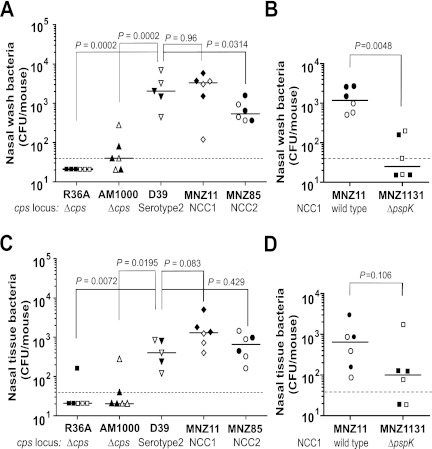
Nasopharyngeal carriage of S. pneumoniae in mice. C57BL/6 mice were inoculated with six S. pneumoniae strains (x axis), and pneumococci were recovered 5 days later from the nasal wash (A and B) and nasal tissue (C and D). Each group is identified with the names of the respective bacterial strains and descriptions of their cps loci. No mice died during the experiment, and each data point indicates an individual animal. Open symbols show results from the first experiment, and solid symbols show results from the second. The horizontal bars indicate the median for the group. The dotted lines indicate the detection limit, which is 40 CFU/mouse. Symbols below the dotted lines were arbitrarily placed to show individual data points. The Mann-Whitney test was used to obtain P values.
Consistent with the previous report (28), nasal washes contained many D39 colonies (median, 2,040 CFU/mouse) but very few colonies (median, 40 CFU/mouse or less) of AM1000 which is a recent unencapsulated variant of D39 (Fig. 5A), and the difference was statistically significant (P = 0.002). Also, R36A, another laboratory-derived acapsular variant of D39, was not detectable (<40 CFU/mouse) in nasal washes. In contrast, the two naturally acapsular strains, MNZ11 and MNZ85, were present in the nasal washes of mice (Fig. 5A), and their median numbers of CFU were 3,340 (P = 0.96 compared with D39) and 540 (P = 0.0314 compared with D39), respectively. The difference between MNZ85 and D39 carriages was significant (P = 0.0314). When the CFU in the nasal tissue were compared, similar relative patterns were observed, but this time there was no significant difference in the numbers of CFU of MNZ85 and D39 (Fig. 5C). Moreover, pneumococcal infection in all mice was limited to the NP, since no bacteria were found in the blood or lungs of infected mice of all groups (data not shown). This is consistent with the results of previous studies of carriage in the absence of induced aspiration, where the CFU are generally either not detectable or present in much lower numbers in the lungs than in the nasal wash or nasal tissue (35, 36).
To confirm the role of pspK in NP carriage, we prepared a new strain, MNZ1131, by deleting pspK from MNZ11 and examined both strains for colonizing mouse NP. Nasal washes of mice infected with the wild-type strain (MNZ11) contained many bacteria (median, 1,180 CFU/mouse), but very few bacteria (median, ≤40 CFU/mouse) were detected in the washes of MNZ1131-infected mice (P = 0.0048) (Fig. 5B). When the nasal tissues were compared, MNZ11 tended to colonize better than MNZ1131 (median, 640 versus 100 CFU/mouse) without achieving statistical significance (P = 0.106) (Fig. 5D). Taking these results together, we conclude that naturally acapsular pneumococci can colonize NP of mice and that pspK appears to be essential to the colonization.
DISCUSSION
Because of its pathogenic importance, the pneumococcal capsule has been extensively studied (37–39), and the identification of capsule types is considered important. In contrast, characterization of NT “pneumococcus” is usually limited to simple serologic tests, even though NT “pneumococcus” is known to cause infectious conjunctivitis (7, 12, 40) or even IPD (8). Such a simple serologic definition of NT “pneumococcus” is inadequate, since it may include (i) S. pneumoniae strains expressing rare, so-far-unidentified capsule types; (ii) S. pneumoniae strains with disrupted and nonfunctional cps loci (group I); (iii) S. pneumoniae strains with novel genes at their cps loci (group II); or (iv) streptococcal species closely related to but different from S. pneumoniae.
The last possibility is clearly illustrated with our NCC3 isolates and two NCC2b isolates from HIV patients attending the same clinic. Although they have the key features of S. pneumoniae (e.g., lytA or optochin sensitivity), they belong to the pneumococcus-like species S. pseudopneumoniae or S. mitis. Our finding is consistent with a report that states that NT “pneumococcal” isolates from HIV+ patients are not usually S. pneumoniae (i.e., 11 of 12 isolates in that study were not S. pneumoniae) (21), and healthy children can carry NCC3 isolates belonging to S. pneumoniae (41). In addition, our NT “pneumococcal” isolates from HIV+ patients have diverse MLST types and different cps sequences, suggesting that each isolate arose individually. Perhaps HIV+ patients provide a special environment where these pneumococcus-like isolates can arise from a vast pool of commensal species.
In contrast, NCC2a isolates were all revealed to be S. pneumoniae (Table 1) and also showed clear evidence of spreading. We found that many Brazilian children carry pneumococcal NCC2a isolates with an identical sequence type (ST2315) as well as cps loci. Indeed, NCC2a isolates have been associated with epidemic conjunctivitis in the past (7). Our finding almost identical cps sequences among many NCC2a isolates from diverse geographic locations, times, and diseases, as well as with different MLST types, suggests that isolates with NCC2a cps can readily spread among individuals. As NCC2a isolates account for the majority (29 of 52) of group II NT “pneumococcal” isolates and have been shown to cause IPD (8) as well, further studies are needed to understand their ability to spread and cause disease.
A surprising finding was the discovery of a new clade, NCC1, and its unique gene, pspK. MLST analysis clearly shows NCC1 isolates to be typical S. pneumoniae. While additional epidemiological studies are necessary to know their full pathogenic potential, all NCC1 isolates identified so far have been carriage isolates. The NCC1 isolates are clearly communicable, since NCC1 isolates expressing one MLST type are found among children living in different cities in South Korea. Several features of pspK suggest that it encodes a surface protein important in cell adhesion. PspK has an alpha helix with highly charged amino acids [(E)EEAKRKA], which may serve as a rigid stalk for the R1-like region of PspC. The R1-like region of PspC is known to be important in cell adhesion (24, 42). In addition to these molecular features, we have directly demonstrated that an NCC1 isolate can colonize NP of mice only when it has pspK. Thus, PspK may facilitate pneumococcal adhesion to host cells, as does PspC (24, 42).
Mutations that block capsule expression in normally encapsulated strains have been reported to block nasal colonization (28–30), demonstrating that for these strains, the capsule is a critical colonization factor. In contrast, we show that NCC1 and NCC2a isolates spread and colonize the NP of many different children and that they can also colonize mice. Thus, expression of a PS capsule is not an absolute requirement for NP colonization of pneumococci, and naturally acapsular strains may have genes like pspK that provide critical survival advantages in the NP, compensating for the lack of a capsule. The survival advantages of such proteins may be novel and could lead to increased colonization by several different mechanisms, including increased hydrophilicity of the bacteria to prevent them from clumping and being swept out by mucus, increased adherence to epitopes on host cells, and/or more extensive biofilm formation. Studies of naturally acapsular S. pneumoniae strains may show us novel mechanisms of pneumococcal survival in the nasopharynx.
MATERIALS AND METHODS
Bacteria.
From three different countries (South Korea, Brazil, and the United States), we obtained 64 anonymized noninvasive isolates that had been identified as NT “pneumococci” in previous studies (31, 43, 44). Sixteen Korean isolates were obtained from the nasopharynges (NP) of healthy children (less than 5 years old) attending daycare centers (DCCs) in 2008 (43). Twenty-six Brazilian isolates were obtained from the NP of children (less than 5 years old) attending municipal DCCs in 2005 (31, 45). The 22 isolates from the United States included 5 isolates from patients with conjunctivitis (provided by Mary Marquart at the University of Mississippi Medical Center, Jackson, MS) and 17 isolates from the nasopharynges of HIV-positive outpatients (>18 years old) at a large academic referral clinic (44). All 64 isolates were identified as pneumococci by commonly used methods, such as colony morphology, alpha-hemolysis, optochin sensitivity, and bile solubility, and determined to be NT by the Quellung reaction. They tended to form smaller colonies than encapsulated pneumococci.
Strain R36A was obtained from D. Briles (University of Alabama at Birmingham, Birmingham, AL). Strains D39 and AM1000 were obtained from J. Yother (University of Alabama at Birmingham, Birmingham, AL). AM1000 was derived from D39 by disrupting its cps locus and does not express capsular PS (28). The three strains were previously used by the above laboratories in nasopharyngeal carriage experiments (28, 46). A pspK deletion mutant strain (MNZ1131) was created as follows. The 5′ and 3′ flanking regions of pspK were obtained by PCR using MNZ11 as the template and primer pairs 5419-3743 and 5743-3419, respectively (Table 2). A spectinomycin resistance cassette (aad9 gene) was obtained by PCR from plasmid vector pCLT1242 (47) using primers 5725 and 3725 (Table 2). The three DNA fragments were joined to an ~4.8-kb DNA fragment by an overlap extension PCR (48). MNZ1131 was obtained by transforming MNZ11 with the ~4.8-kb DNA and selecting a spectinomycin-resistant isolate. Correct insertion of aad9 and deletion of pspK were confirmed by sequencing the involved region of MNZ1131 DNA. All isolates were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) and stored at −80°C in 20% glycerol.
TABLE 2 .
Primers used in this study
| Primer | Genetic location | Sequence (5′ → 3′) | Reference |
|---|---|---|---|
| Forward primers | |||
| 5120 | dexB | TGTCCAATGAAGAGCAAGACTTGACAGTAG | 5 |
| 5419 | dexB | TCTTAGTTCCATGGGATGCTTTCTGTGTG | 8 |
| 5322 | lytA | CAACCGTACAGAATGAAGCGG | 53 |
| 5203 | ply | ATTTCTGTAACAGCTACCAACGA | 54 |
| 5202 | cpsA | GCAGTACAGCAGTTTGTTGGACTGACC | 55 |
| 5441 | Intergenic region between capN and aliA | CGTTCTGCGACTGACCTTCC | This study |
| 5433 | aliC | AACACTTGGAACGGAGAATG | 8 |
| 5434 | aliD | AGATGCCAAATGGTTCACGG | 8 |
| 5658 | pspK | GCAAATCAGCCAGTAACTGTGA | This study |
| 5725 | aad9 | TTTTCCTTGATTCGATTTTCGTTCGTGAATAC | This study |
| 5743 | pspK | TATTGCAGACAGCTTGTTAAACATAAA | This study |
| Reverse primers | |||
| 3126 | aliA | CAATAATGTCACGCCCGCAAGGGCAAGT | 5 |
| 3419 | aliA | CGCTGAACTTTTGTAGTTGCTGTCTGGTCAAC | 8 |
| 3322 | lytA | TTATTCGTGCAATACTCGTGCG | 53 |
| 3203 | ply | GAATTCCCTGTCTTTTCAAAGTC | 54 |
| 3202 | cpsA | GAATATTTTCATTATCAGTCCCAGTC | 55 |
| 3403 | Intergenic region between capN and aliA | GCTAAAACACCAGCTGCTAA | This study |
| 3433 | aliC | GCCCTTTGTTATACCTAGATGTTTC | 8 |
| 3434 | aliD | GAAATCTTTTAACAAATAAGGTCCG | 8 |
| 3658 | pspK | CAAGATAAGCTTTCTGCACCTCT | This study |
| 3725 | aad9 | GGGAAATATTCATTCTAATTGGTAATGCAGACAGCT | This study |
| 3743 | pspK | CGAATCAAGGAAAAATATTCTTATTATTCAT | This study |
Multilocus sequence typing (MLST).
PCR for MLST was performed as described elsewhere (49), and the DNA sequences of the PCR products were determined by the Genomics core facility at University of Alabama at Birmingham (UAB). All the DNA sequences were submitted to the pneumococcal MLST website (http://spneumoniae.mlst.net), which identified the sequences either as existing alleles or as new alleles with new allele numbers.
A neighbor-joining tree (Fig. 1) was created with concatenated DNA sequences (2,751 bp) of six of the seven genetic loci used by MLST (ddl was removed) for the 17 NT pneumococcal isolates using MEGA version 4 (50). The sequence types of these 17 isolates are in bold in Table 1. To include reference pneumococcal and nonpneumococcal species in the tree, the corresponding DNA sequences of 39 S. pneumoniae strains, 42 S. pseudopneumoniae strains, 25 S. mitis strains, and 8 S. oralis strains were retrieved from the GenBank or the MLST database. The sequences in these two databases were deposited by others (17, 21, 23). The statistical significance of a branching was determined by performing 1,000 bootstrap replications with the minimum evolution algorithm. The tree was drawn to scale, with branch lengths measured in the same units used for the evolutionary distances on the phylogenic tree.
PCR for pneumococcus-specific genes and sequencing of the cps.
To detect genes encoding pneumococcal autolysin (lytA), pneumolysin (ply), cpsA (wzg), aliC, aliD, pspK, and the intergenic region, PCR was performed as described elsewhere (51) with the primer sets listed in Table 2. The PCR products were visualized by agarose gel electrophoresis (51). To amplify the entire cps locus, a long-range PCR was carried out using the primers located at dexB and aliA (Table 2) with TaKaRa LA Taq (TaKaRa Bio United States, Madison, WI).
The DNA sequences of the PCR products were determined by the Genomics Core Facility at UAB. The DNA sequences were analyzed and aligned using DNASTAR Lasergene version 6.0. Insertional sequences and the open reading frames (ORFs) in the DNA sequences were identified using a BLAST search at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST), Softberry (http://www.softberry.com) free web-based software, and IS Finder (http://www-is.biotoul.fr).
Nasopharyngeal colonization by pneumococci in mice.
Pneumococcal carriage in mice was examined as previously described (28, 33, 35, 52). Eight-week-old C57BL/6 mice were obtained from Frederick Cancer Research UK (Baltimore, MD), and 10 µl of Ringer’s lactate solution containing bacteria was instilled into their nares without anesthesia in two separate experiments (Fig. 5A and C). Inoculating doses (CFU per mouse) were 1.5 × 107 for R36A, 9 × 106 for AM1000, 6 × 106 for D39, 3 × 107 for MNZ11, and 2 × 107 for MNZ85 in the first experiment. In the second experiment, they were 1.8 × 107 for R36A, 5 × 106 for AM1000, 5 × 106 for D39, 4.3 × 107 for MNZ11, and 2.7 × 107 for MNZ85. In a prior colonization study we demonstrated that the bacterial dose affected the frequency of mice with detectable colonization but had essentially no effect on the numbers of CFU in colonized mice (35). These doses are thus well within an acceptable range for comparison. After 5 days, the mice were sacrificed, and nasal wash samples were obtained by washing the nasal cavity with 1 ml of Ringer’s lactate solution. The nonwashable bacterial fraction was obtained by grinding the excised nasal tissue in 1 ml of Ringer’s lactate solution. The nasal washes and nonwashable fractions were serially diluted and plated on blood agar plates with 50 µg/ml of gentamicin. After overnight inoculation at 37°C with 5% CO2, the number of colonies in each plate was determined.
To study the involvement of pspK in NP carriage in mouse, additional experiments were performed exactly as described above with a ΔpspK strain (MNZ1131) and a wild-type strain (MNZ11) (Fig. 5B and D). The inoculating doses were 1.2 × 107 CFU of MNZ11 and 4.5 × 107 CFU of MNZ1131 in the first experiment and 3 × 107 CFU of MNZ11 and 1.8 × 107 CFU of MNZ1131 in the second experiment.
Statistical analysis.
CFU in different experimental groups were compared using the Mann-Whitney test and Prism 4 software (GraphPad Software, Inc., San Diego, CA).
Nucleotide sequence accession numbers.
The 12 DNA sequences determined here were registered in GenBank (accession numbers JF489999 to JF490008, JF723379, and JF723380).
ACKNOWLEDGMENTS
The work was supported by Public Health Service grants AI-31473 to M.H.N. and AI-21548 to D.E.B. from the National Institutes of Health, by the National Institute of Science and Technology for Health Technology Assessment/IATS to A.L.A., and by the Brazilian Council for Research and Development/CNPq grants 482646/2007-1 and 306096/2010-2 to A.L.A. D.E.B. is partially supported by the grant WCU R33-10045 of the Korea Science and Engineering Foundation (KOSEF).
We thank Mary Marquart for providing the conjunctivitis strains and Janet Yother for providing pneumococcal strains and for stimulating discussions.
ADDENDUM IN PROOF
Salter et al. (Microbiology, 8 March 2012, posting date, doi:10.1099/mic.0.056580-0) described the gene nspA among NT pneumococcal isolates from Thailand and the Netherlands. It is likely that nspA is identical to pspK.
Footnotes
Citation Park IH, et al. 2012. Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. mBio 3(3):e00035-12. doi:10.1128/mBio.00035-12.
REFERENCES
- 1. Henrichsen J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avery OT, Dubos R. 1931. The protective action of a specific enzyme against type III pneumococcus infection in mice. J. Exp. Med. 54:73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368–377 [DOI] [PubMed] [Google Scholar]
- 4. Park IH, et al. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bentley SD, et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mavroidi A, et al. 2007. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7841–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin M, et al. 2003. An outbreak of conjunctivitis due to atypical Streptococcus pneumoniae. N. Engl. J. Med. 348:1112–1121 [DOI] [PubMed] [Google Scholar]
- 8. Hathaway LJ, Stutzmann Meier P, Bättig P, Aebi S, Mühlemann K. 2004. A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae. J. Bacteriol. 186:3721–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lagos R, et al. 2008. Age- and serotype-specific pediatric invasive pneumococcal disease: insights from systematic surveillance in Santiago, Chile, 1994–2007. J. Infect. Dis. 198:1809–1817 [DOI] [PubMed] [Google Scholar]
- 10. Martin GS, Mannino DM, Eaton S, Moss M. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 348:1546–1554 [DOI] [PubMed] [Google Scholar]
- 11. Onwubiko C, Shires C, Quin LR, Swiatlo E, McDaniel LS. 2007. Characterization of Streptococcus pneumoniae isolated from children with otitis media. FEMS Immunol. Med. Microbiol. 50:119–125 [DOI] [PubMed] [Google Scholar]
- 12. Xu Q, et al. 2011. Nontypeable Streptococcus pneumoniae as an otopathogen. Diagn. Microbiol. Infect. Dis. 69:200–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sá-Leão R, et al. 2009. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin. Microbiol. Infect. 15:1002–1007 [DOI] [PubMed] [Google Scholar]
- 14. Messmer TO, et al. 2004. Comparison of four polymerase chain reaction assays for specificity in the identification of Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 49:249–254 [DOI] [PubMed] [Google Scholar]
- 15. Pease AA, Douglas CW, Spencer RC. 1986. Identifying non-capsulate strains of Streptococcus pneumoniae isolated from eyes. J. Clin. Pathol. 39:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shayegani M, Parsons LM, Gibbons WE, Jr, Campbell D. 1982. Characterization of nontypable Streptococcus pneumoniae-like organisms isolated from outbreaks of conjunctivitis. J. Clin. Microbiol. 16:8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanage WP, et al. 2005. Using multilocus sequence data to define the pneumococcus. J. Bacteriol. 187:6223–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kilian M, et al. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Whatmore AM, et al. 1999. Molecular characterization of equine isolates of Streptococcus pneumoniae: natural disruption of genes encoding the virulence factors pneumolysin and autolysin. Infect. Immun. 67:2776–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waite RD, Penfold DW, Struthers JK, Dowson CG. 2003. Spontaneous sequence duplications within capsule genes cap8E and tts control phase variation in Streptococcus pneumoniae serotypes 8 and 37. Microbiology 149:497–504 [DOI] [PubMed] [Google Scholar]
- 21. Leegaard TM, et al. 2010. Phenotypic and genomic characterization of pneumococcus-like streptococci isolated from HIV-seropositive patients. Microbiology 156:838–848 [DOI] [PubMed] [Google Scholar]
- 22. Simões AS, et al. 2010. Highly penicillin-resistant multidrug-resistant pneumococcus-like strains colonizing children in Oeiras, Portugal: genomic characteristics and implications for surveillance. J. Clin. Microbiol. 48:238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chi F, et al. 2007. New Streptococcus pneumoniae clones in deceased wild chimpanzees. J. Bacteriol. 189:6085–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo R, et al. 2005. Solution structure of choline binding protein A, the major adhesin of Streptococcus pneumoniae. EMBO J. 24:34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammerschmidt S. 2006. Adherence molecules of pathogenic pneumococci. Curr. Opin. Microbiol. 9:12–20 [DOI] [PubMed] [Google Scholar]
- 26. Spink BJ, Sivaramakrishnan S, Lipfert J, Doniach S, Spudich JA. 2008. Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI. Nat. Struct. Mol. Biol. 15:591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang E, Wang CL. 1996. (i, i + 4) Ion pairs stabilize helical peptides derived from smooth muscle caldesmon. Arch. Biochem. Biophys. 329:156–162 [DOI] [PubMed] [Google Scholar]
- 28. Magee AD, Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nelson AL, et al. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nelson AL, et al. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 75:83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andrade AL, et al. 2010. Non-typeable Streptococcus pneumoniae carriage isolates genetically similar to invasive and carriage isolates expressing capsular type 14 in Brazilian infants. J. Infect. 61:314–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee H, Nahm MH, Burton R, Kim KH. 2009. Immune response in infants to the heptavalent pneumococcal conjugate vaccine against vaccine-related serotypes 6A and 19A. Clin. Vaccine Immunol. 16:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Balachandran P, Brooks-Walter A, Virolainen-Julkunen A, Hollingshead SK, Briles DE. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect. Immun. 70:2526–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Briles DE, Novak L, Hotomi M, van Ginkel FW, King J. 2005. Nasal colonization with Streptococcus pneumoniae includes subpopulations of surface and invasive pneumococci. Infect. Immun. 73:6945–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu HY, et al. 1997. Establishment of a Streptococcus pneumoniae nasopharyngeal colonization model in adult mice. Micro. Pathog. 23:127–137 [DOI] [PubMed] [Google Scholar]
- 36. Briles DE, et al. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J. Infect. Dis. 188:339–348 [DOI] [PubMed] [Google Scholar]
- 37. Briles DE, Crain MJ, Gray BM, Forman C, Yother J. 1992. Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect. Immun. 60:111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hyams C, et al. 2010. Streptococcus pneumoniae resistance to complement-mediated immunity is dependent on the capsular serotype. Infect. Immun. 78:716–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Melin M, Trzcinski K, Meri S, Käyhty H, Väkeväinen M. 2010. The capsular serotype of Streptococcus pneumoniae is more important than the genetic background for resistance to complement. Infect. Immun. 78:5262–5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zegans ME, et al. 2009. Clinical features, outcomes, and costs of a conjunctivitis outbreak caused by the ST448 strain of Streptococcus pneumoniae. Cornea 28:503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hiller NL, et al. 2010. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 6:e1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quin LR, et al. 2007. Factor H binding to PspC of Streptococcus pneumoniae increases adherence to human cell lines in vitro and enhances invasion of mouse lungs in vivo. Infect. Immun. 75:4082–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim KH, et al. 2011. Nasopharyngeal pneumococcal carriage of children attending day care centers in Korea: comparison between children immunized with 7-valent pneumococcal conjugate vaccine and non-immunized. J. Korean Med. Sci. 26:184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onwubiko C, Swiatlo E, McDaniel LS. 2008. Cross-sectional study of nasopharyngeal carriage of Streptococcus pneumoniae in human immunodeficiency virus-infected adults in the conjugate vaccine era. J. Clin. Microbiol. 46:3621–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Franco CM, et al. 2010. Survey of nonsusceptible nasopharyngeal Streptococcus pneumoniae isolates in children attending day-care centers in Brazil. Pediatr. Infect. Dis. J. 29:77–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Slotved HC, Kaltoft M, Skovsted IC, Kerrn MB, Espersen F. 2004. Simple, rapid latex agglutination test for serotyping of pneumococci (pneumotest-latex). J. Clin. Microbiol. 42:2518–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dong S, et al. 2010. Characterization of the enzymes encoded by the anthrose biosynthetic operon of Bacillus anthracis. J. Bacteriol. 192:5053–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davis KM, Akinbi HT, Standish AJ, Weiser JN. 2008. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog. 4:e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144(Pt 11):3049–3060 [DOI] [PubMed] [Google Scholar]
- 50. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 51. Park IH, Park S, Hollingshead SK, Nahm MH. 2007. Genetic basis for the new pneumococcal serotype, 6C. Infect. Immun. 75:4482–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McDaniel LS, et al. 1987. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA). J. Exp. Med. 165:381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nagai K, et al. 2001. Evaluation of PCR primers to screen for Streptococcus pneumoniae isolates and beta-lactam resistance, and to detect common macrolide resistance determinants. J. Antimicrob. Chemother. 48:915–918 [DOI] [PubMed] [Google Scholar]
- 54. Salo P, Ortqvist A, Leinonen M. 1995. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J. Infect. Dis. 171:479–482 [DOI] [PubMed] [Google Scholar]
- 55. Pai R, Gertz RE, Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]