Abstract
OBJECTIVES
To develop a logistic regression-based model to predict prostate cancer biopsy at, and compare its performance to the risk calculator developed by the Prostate Cancer Prevention Trial (PCPT), which was based on age, race, prostate-specific antigen (PSA) level, a digital rectal examination (DRE), family history, and history of a previous negative biopsy, and to PSA level alone.
PATIENTS AND METHODS
We retrospectively analysed the data of 1280 men who had a biopsy while enrolled in a prospective, multicentre clinical trial. Of these, 1108 had all relevant clinical and pathological data available, and no previous diagnosis of prostate cancer. Using the PCPT risk calculator, we calculated the risks of prostate cancer and of high-grade disease (Gleason score ≥7) for each man. Receiver operating characteristic (ROC) curves for the risk calculator, PSA level and the novel regression-based model were compared.
RESULTS
Prostate cancer was detected in 394 (35.6%) men, and 155 (14.0%) had Gleason ≥7 disease. For cancer prediction, the area under the ROC curve (AUC) for the risk calculator was 66.7%, statistically greater than the AUC for PSA level of 61.9% (P < 0.001). For predicting high-grade disease, the AUCs were 74.1% and 70.7% for the risk calculator and PSA level, respectively (P = 0.024). The AUCs increased to 71.2% (P < 0.001) and 78.7% (P = 0.001) for detection and high-grade disease, respectively, with our novel regression-based models.
CONCLUSIONS
ROC analyses show that the PCPT risk calculator modestly improves the performance of PSA level alone in predicting an individual's risk of prostate cancer or high-grade disease on biopsy. This predictive tool might be enhanced by including percentage free PSA and the number of biopsy cores.
Keywords: prostatic neoplasms, needle biopsy, risk assessment, ROC curve
INTRODUCTION
With revitalised debate about PSA level thresholds and the declining frequency of palpable prostate lesions, physicians and men alike are often faced with a difficult decision of whether to prompt either an initial or a repeat prostate biopsy. There is no absolute PSA value that assures that a man does not have prostate cancer, as this risk lies on a continuum of PSA values [1]. In addition, changes in PSA level over time are more likely to be informative than any single value [2]. However, absolute PSA thresholds continue to be a central facet not only of PSA screening, but also for referral to a urologist and for taking a prostate biopsy.
Many men face the prospect of prostate biopsy with inadequate information about their probability of having prostate cancer. The decision to have a prostate biopsy might be aided by algorithms that estimate an individual's risk of having cancer detected. In the last decade, several nomograms and artificial neural networks have been developed to predict prostate cancer, either on initial or repeat biopsy [3–19]. In general, these models have been based on PSA values, a DRE and age, but have also used other variables, including race, family history, year of biopsy, prostate volume, number of needle cores, percentage free PSA (%fPSA), number of previous negative biopsies and PSA velocity. Although there has been significant enthusiasm for the development of models that integrate a few of these variables for individualized risk prediction, none has gained widespread acceptance.
Recently, investigators from the Prostate Cancer Prevention Trial [20] (PCPT) developed a risk calculator based on age, race, PSA, DRE, family history, and history of a previous negative prostate biopsy, using risk equations created from multivariable logistic regression models [21]. The risk of detecting prostate cancer was predicted using PSA level, DRE, family history and previous negative biopsy, whereas the risk of detecting high-grade disease (Gleason score ≥7) was predicted using age at biopsy, PSA, DRE, race and previous negative biopsy. More recently, the risk calculator was validated externally in a screened population using the San Antonio Center of Biomarkers of Risk of Prostate Cancer (SABOR) cohort of the Early Detection Research Network [22]. In the present study, we developed a novel logistic regression-based model that incorporates %fPSA and number of biopsy cores, and compared its performance to the PCPT risk calculator and to PSA level alone.
PATIENTS AND METHODS
We used the data from 1280 consecutive men scheduled for biopsy who had been previously enrolled in one of two prospective, multicentre clinical trials which studied complexed [23] and %fPSA [24], respectively (1995–2001). Serum PSA and %fPSA levels were measured in all men before a DRE and TRUS. Most men were referred and biopsied because of a suspicious DRE and/or abnormal PSA level. The studies were approved by the institutional review boards at all study sites, and all participants provided written informed consent. Of the 1280 men, 1108 (87%) men had all relevant clinical and pathological data available and no previous diagnosis of prostate cancer, and were included in the present study.
Clinical and pathological features were compared between the men diagnosed with cancer and those not diagnosed with cancer using the chi-square test for categorical variables and Wilcoxon Mann–Whitney test for continuous variables. A similar comparison was made between men diagnosed with high-grade disease (Gleason score ≥7) and those not diagnosed with cancer or with low-grade disease (Gleason ≤6). The %fPSA results are expressed as a percentage of total PSA calculated as ((fPSA/total PSA) × 100).
For predicting the prostate biopsy results, race was coded as 0 (not African-American) or 1 (African-American); DRE as 0 (negative or normal) or 1 (suspicious for cancer); family history as 0 (no) or 1 (yes); and previous negative prostate biopsy as 0 (no previous biopsy) or 1 (one or more previous biopsies, all negative for cancer) as described by Thompson et al. [21]. Age at prostate biopsy and the natural log of PSA (lnPSA) were treated as continuous variables. Logistic regression analysis was used to determine the odds ratio, 95% CI and P values using the six variables, as explained above, with %fPSA (<15 vs ≥15) and number of biopsy cores (≤9, 10–12 or >12).
Using the risk equations formulated from the inverse logistic functions previously published from the PCPT data [21], we calculated the risks of prostate cancer and of high-grade disease for each patient. To quantify the predictive accuracy of the risk calculator, receiver operating characteristic (ROC) curves for the risk calculator, PSA level alone and the multivariable model were generated and plotted as the false-positive rate (1 − specificity) vs sensitivity. The areas under the ROC curves (AUC) for the risk calculator, PSA level alone and the multivariable model were compared using the test for equality.
RESULTS
The clinical and pathological characteristics of the 1108 men are summarized in Table 1; of the 1108 men assessed, prostate cancer was detected in 394 (35.6%) and 155 (14.0%) had high-grade disease. Table 2 compares men with cancer to those without using the chi-square and Wilcoxon-Mann–Whitney tests. Men without cancer were significantly more likely to be young (P < 0.001) and not African-American (P = 0.002), and to have a lower PSA level (P < 0.001), a normal DRE (P < 0.001), no family history of prostate cancer (P = 0.006), a higher %fPSA (P < 0.001) and fewer needle biopsy cores (P < 0.001). The proportion with a previous negative prostate biopsy was similar among the two groups (P = 0.272). Men with high-grade prostate cancer, compared with those with no or low-grade disease, had statistically significant differences in age (P < 0.001), race (P = 0.008), PSA level (P < 0.001), DRE (P = 0.006), %fPSA (P < 0.001) and number of needle cores (P = 0.001); there were no differences in family history of prostate cancer (P = 0.079) or in previous negative prostate biopsy (P = 0.154; data not shown). The PCPT calculated risk was associated with an increasing risk of prostate cancer. The calculated risk was greater among those with prostate cancer (median 47.3%, 7.1–99.1%) than among those not diagnosed with prostate cancer on biopsy (median 39.9%, 6.2–85.4%; P < 0.001).
TABLE 1.
The clinical and pathological characteristics of the 1108 men
| Characteristic | Value |
|---|---|
| Age at biopsy, years | |
| Mean (SD) | 63.2 (8.4) |
| Median (range) | 64 (32–92) |
| PSA level, ng/mL | |
| Mean (SD) | 8.4 (26.4) |
| Median (range) | 5.4 (0.35–759.3) |
| %fPSA | |
| Mean (SD) | 14.5 (7.0) |
| Median (range) | 13.4 (2.7–61.1) |
| Race, n (%) | |
| White | 942 (85.0) |
| Black | 89 (8.0) |
| other | 77 (6.9) |
| DRE | |
| Normal/not suspicious | 915 (82.6) |
| Abnormal | 193 (17.4) |
| Family history of cancer | |
| No | 861 (77.7) |
| Yes | 247 (22.3) |
| Previous negative prostate biopsy | |
| No | 851 (76.8) |
| Yes | 257 (23.2) |
| Number of cores at prostate biopsy | |
| ≤9 | 168 (15.2) |
| 10–12 | 887 (80.1) |
| >12 | 53 (4.8) |
| Prostate biopsy result | |
| Benign | 714 (64.4) |
| Low-grade cancer (Gleason ≤6) | 239 (21.6) |
| High-grade cancer (Gleason ≥7) | 155 (14.0) |
| Clinical stage | |
| T1 | 214 (54.5) |
| T2a | 123 (31.3) |
| T2b | 42 (10.7) |
| T3 | 14 (3.6) |
TABLE 2.
Comparison of risk factors in men with and without prostate cancer
| Variable | No cancer | Cancer | P |
|---|---|---|---|
| No. of patients | 714 | 394 | |
| n (%): | |||
| Age, years | <0.001* | ||
| ≤50 | 55 (7.7) | 15 (3.8) | |
| 51–60 | 239 (33.5) | 114 (28.9) | |
| 61–70 | 309 (43.3) | 168 (42.6) | |
| >70 | 111 (15.5) | 97 (24.6) | |
| Race | 0.002* | ||
| White | 606 (84.9) | 336 (85.3) | |
| Black | 47 (6.6) | 42 (10.7) | |
| Other | 61 (8.5) | 16 (4.1) | |
| PSA level, ng/mL | <0.001* | ||
| ≤2.5 | 125 (17.5) | 31 (7.9) | |
| 2.6–4 | 142 (19.9) | 48 (12.2) | |
| 4.1–10 | 341 (47.8) | 227 (57.6) | |
| >10 | 106 (14.8) | 88 (22.3) | |
| DRE | <0.001* | ||
| Normal | 617 (86.4) | 298 (75.6) | |
| Abnormal | 97 (13.6) | 96 (24.4) | |
| Family history of cancer | 0.006* | ||
| No | 573 (80.2) | 288 (73.1) | |
| Yes | 141 (19.8) | 106 (26.9) | |
| Previous negative prostate biopsy | 0.272* | ||
| No | 541 (75.8) | 310 (78.7) | |
| Yes | 173 (24.2) | 84 (21.3) | |
| Calculated risk of prostate cancer, % | <0.001† | ||
| Mean (SD) | 40.6 (14.1) | 50.0 (16.1) | |
| Median (range) | 39.9 (6.2–85.4) | 47.3 (7.1–99.1) | |
| Number of cores at prostate biopsy | <0.001* | ||
| ≤9 | 134 (18.8) | 34 (8.6) | |
| 10–12 | 549 (76.9) | 338 (85.8) | |
| >12 | 31 (4.3) | 22 (5.6) | |
| %fPSA | <0.001* | ||
| <15 | 388 (54.3) | 278 (70.6) | |
| ≥15 | 326 (45.7) | 116 (29.4) |
chi-squa re;
Wilcoxon-Mann-Whitney rank-sum test.
On multivariable logistic regression analyses, independent predictors of a biopsy positive for prostate cancer (and for high-grade disease) were an elevated ln(PSA), low %fPSA, advanced age, abnormal DRE findings, African-American race, positive family history, and more biopsy cores, whereas a previous negative biopsy was protective. Table 3 summarizes the odds ratios, 95% CI and P values for the multivariable logistic regressions combining these eight variables.
TABLE 3.
Multivariable logistic regression for predicting prostate cancer and high-grade disease
| Variable | OR (95% CI) | P |
|---|---|---|
| Prostate cancer | ||
| African-American race | 1.89 (1.17–3.06) | 0.009 |
| Age | 1.03 (1.01–1.05) | 0.001 |
| ln(PSA) | 1.69 (1.37–2.08) | <0.001 |
| Family history | 1.74 (1.28–2.38) | <0.001 |
| DRE | 2.42 (1.72–3.41) | <0.001 |
| Number of biopsy cores | 2.03 (1.45–2.84) | <0.001 |
| Previous negative biopsy | 0.56 (0.40–0.78) | 0.001 |
| %fPSA (<15 vs ≥15) | 1.95 (1.46–2.61) | <0.001 |
| High-grade disease | ||
| African-American race | 2.23 (1.22–4.07) | 0.009 |
| Age | 1.05 (1.02–1.07) | <0.001 |
| ln(PSA) | 2.40 (1.79–3.20) | <0.001 |
| Family history | 1.80 (1.17–2.75) | 0.007 |
| DRE | 1.95 (1.23–3.08) | 0.004 |
| Number of biopsy cores | 2.22 (1.33–3.70) | 0.002 |
| Previous negative biopsy | 0.40 (0.24–0.65) | <0.001 |
| %fPSA (<15 vs ≥15) | 2.84 (1.80–4.49) | <0.001 |
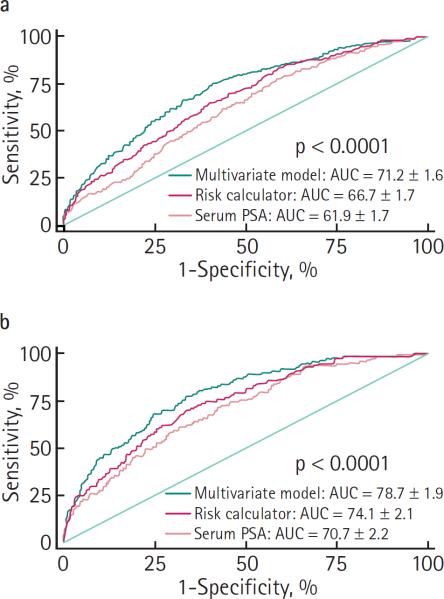
For predicting prostate cancer, the ROC analyses showed an AUC for the risk calculator of 66.7%, statistically greater than the AUC for PSA of 61.9% (P < 0.001; Fig. 1a). Adding the data on the number of biopsy cores and %fPSA, the AUC for the multivariable logistic regression model increased to 71.2% (P < 0.001). For predicting high-grade disease, the AUCs were 74.1% and 70.7% for the risk calculator and PSA, respectively (P = 0.024; Fig. 1b). This increased to 78.7% for the multivariable logistic regression model (P < 0.001).
FIG. 1.
ROC curves for PSA, the PCPT risk calculator and a novel logistic-regression based model for predicting: a, prostate cancer; and b, high-grade prostate cancer (Gleason score ≥7).
As the PCPT risk calculator was developed with older men, we tested its performance characteristics for men aged ≥50 years and found that its accuracy did not improve. When men aged <50 years were excluded, the AUCs for predicting prostate cancer using the risk calculator and PSA alone were 65.9% and 61.0%, respectively (P < 0.001). For predicting high-grade disease, the corresponding AUCs were 73.7 and 70.5 (P = 0.047).
DISCUSSION
Risk stratification is essential for comprehensive patient counselling and evidence-based decision-making. As the PSA level is limited by poor specificity and the DRE by poor sensitivity, algorithms that estimate an individual's risk of having prostate cancer detected might aid the decision to have a prostate biopsy. In the present study of 1108 men, we externally validated the PCPT risk calculator, comparing its performance to PSA alone and to a novel, logistic regression-based model which included %fPSA and number of biopsy cores. The risk calculator gave a modest improvement in the performance characteristics of PSA alone in predicting an individual's risk of prostate cancer or high-grade disease. Our novel regression-based model further improved the predictive accuracy, with AUCs of 71.2% and 78.7% for predicting prostate cancer and high-grade disease, respectively.
Not surprisingly, multivariable regression showed that increasing age, increasing PSA level, low %fPSA, abnormal DRE, African-American race, positive family history, increasing number of biopsy cores and no previous negative prostate biopsy indicate an increasing risk of detecting prostate cancer and high-grade disease on biopsy. It was also apparent that adding a more specific PSA variant (%fPSA) enhanced the AUC compared with the PCPT risk calculator, and that the probability of finding prostate cancer on biopsy depends on the extent of sampling. Our model was similar to previously reported regression-based nomograms and artificial neural networks, the AUCs for which are 69–88% [5,7–10,18,21].
The study by Thompson et al. [21] reported AUCs for predicting prostate cancer of 70.2% and 67.8% for the risk calculator and PSA alone, respectively; the AUC for the risk of high-grade disease was 69.8% [21]. They clearly stated `the independent risk factors of family history, DRE result, and previous prostate biopsy did not appreciably improve the sensitivity and specificity of PSA level.' In the external validation using the SABOR cohort [22], the AUC for the risk calculator was 65.5%, similar to that for PSA alone (64.0%). Despite a modest to no improvement over the predictive accuracy of PSA alone, both reports advocate the widespread use of this online calculator `incorporating the current best panel of risk factors' [21,22]. We show here that the PCPT risk calculator again performs with only a modest improvement over PSA alone, and that %fPSA and number of biopsy cores can be included in the panel of risk factors, as each improves on the predictive accuracy.
We also found that the PCPT risk calculator tended to overestimate the risk of high-grade disease, especially in older, African-American men. For example, an 85-year-old African-American man with a normal DRE, no family history, no previous biopsy and a serum PSA level of 6.0 ng/mL had calculated risks of prostate cancer and high-grade disease of 43% and 41%, respectively. This suggests that the risk of this man having Gleason ≤6 disease is only 2%. If that same man's PSA level was 8.0 ng/mL, his corresponding calculated risks would be 49% and 50%. Interestingly, in the present study cohort, there were 13 (1.2%) men whose calculated risk of high-grade disease was greater than their calculated risk of having any prostate cancer. This inaccuracy might stem from a lack of African-American men and men with elevated serum PSA levels in the PCPT cohort.
Limitations from our retrospective study include the potential for selection bias. Despite the many men assessed from several institutions, the cohort might not be representative of the general population of the USA undergoing screening for prostate biopsy. Men in the present cohort were referred for possible prostate biopsy and their characteristics might be different from those of a screening population. In addition, verification bias might also be present; as the present men were enrolled for clinical indications (unlike in the PCPT, in which end-of-study biopsies were taken), the operating characteristics of PSA were evaluated in a cohort over-represented by men with an elevated PSA level. Thus, our model is ideal for men identified at high risk and referred for possible prostate biopsy. As always, prostate biopsies are limited by the extent of sampling and do not perfectly reflect the true disease status, as up to 30% of clinically significant cancers might be missed on biopsy [25–27]. In the present study, family history was defined as positive or negative, with no information on the degree of relation to the individual whose risk was calculated. It might be possible that men enrolled in a prostate cancer study could report a higher incidence of a positive family history (potentially in a more distant relative); this effect would underestimate the effect of having a first-degree relative with prostate cancer. Information on the timing of the previous negative biopsy relative to the repeat biopsy was not available and was thus not considered.
However, the present cohort more closely resembles the target population (of men and of clinically significant tumours). The PCPT cohort used to create the risk calculator is likely to be enriched in small-volume, low-grade cancers, as it excluded men who had a PSA level of >3.0 ng/mL at enrolment. In fact, only 631 (11.4%) had a PSA level of >4.0 ng/mL at the time of biopsy. Another important distinction is in the biopsy regimen, as ≈80% of the men in the PCPT study had sextant biopsies, whereas in the present study only 10.5% had six or fewer cores and 84.8% had ≥10 cores taken. As the PCPT predictive model was based on sextant biopsy information, it must be updated according to the current standard of extended biopsy schemes. In our multivariable model, the number of biopsy cores was indeed found to be an independent predictor of detecting prostate cancer and high-grade disease, and thus is more appropriate for use in contemporary men.
Since the introduction of PSA screening, there has been a significant increase in the proportion of men newly diagnosed with clinically localized, low-risk prostate cancer. Although PSA remains the most commonly used serum biomarker for prostate cancer, it has clearly led to an increasing number of prostate biopsies, and the over-diagnosis and over-treatment in some cases of prostate cancer. As indicated by the results of the PCPT and others, a single PSA value cannot accurately identify men with and without prostate cancer. PSA is instead associated with a range of risk and there is no lower limit at which there is no risk of prostate cancer. Thus, the interpretation of an individual PSA value remains a distinct challenge. The importance of clinical recommendations using PSA levels is amplified by the potential impact on millions of men annually.
Certainly, the decision to take a prostate needle biopsy should integrate numerous clinicopathological features, in addition to the most recent PSA level, including the age, race, PSA kinetics, %fPSA, DRE results, family history, previous needle biopsy findings, and psychosocial factors such as the degree of anxiety and aversion to a cancer diagnosis, treatment or complications. Until molecular biomarkers with improved operating characteristics are developed, clinicians and patients will often be faced with a difficult decision due to flawed and limited prognostic information. This can be improved by multivariable, regression-based prediction tools, which provide a relatively bias-free estimate of an individual's risk with improved predictive accuracy, as compared to mental predictions even by expert clinicians [19,28].
We found that the PCPT risk calculator modestly improves the performance of PSA alone in predicting an individual's risk of prostate cancer or high-grade disease on biopsy. Our novel regression-based model had an improved predictive ability by incorporating %fPSA and number of biopsy cores.
ACKNOWLEDGEMENTS
Financial disclosures: Supported by the National Institutes of Health/National Cancer Institute SPORE Grant #P50CA058236 and Early Detection Research Network/National Cancer Institute/National Institutes of Health Grant U01-CA86323; Bayer Diagnostics, Tarrytown, NY and Dianon Systems Inc, Stratford, CT.
Abbreviations
- AUC
area under the curve
- ROC
receiver operating characteristic
- %fPSA
percentage free PSA
- PCPT
Prostate Cancer Prevention Trial
- SABOR
San Antonio Center of Biomarkers of Risk of Prostate Cancer
REFERENCES
- 1.Thompson IM, Ankerst DP, Chi C, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/ml or lower. JAMA. 2005;294:66–70. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 2.Carter HB, Pearson JD. PSA velocity for the diagnosis of early prostate cancer. A new concept. Urol Clin North Am. 1993;20:665–70. [PubMed] [Google Scholar]
- 3.Optenberg SA, Clark JY, Brawer MK, Thompson IM, Stein CR, Friedrichs P. Development of a decision-making tool to predict risk of prostate cancer: the Cancer of the Prostate Risk Index (CAPRI) test. Urology. 1997;50:665–72. doi: 10.1016/S0090-4295(97)00451-2. [DOI] [PubMed] [Google Scholar]
- 4.Carlson GD, Calvanese CB, Partin AW. An algorithm combining age, total prostate-specific antigen (PSA), and percent free PSA to predict prostate cancer: results on 4298 cases. Urology. 1998;52:455–61. doi: 10.1016/s0090-4295(98)00205-2. [DOI] [PubMed] [Google Scholar]
- 5.Eastham JA, May R, Robertson JL, Sartor O, Kattan MW. Development of a nomogram that predicts the probability of a positive prostate biopsy in men with an abnormal digital rectal examination and a prostate-specific antigen between 0 and 4 ng/mL. Urology. 1999;54:709–13. doi: 10.1016/s0090-4295(99)00213-7. [DOI] [PubMed] [Google Scholar]
- 6.Potter SR, Horniger W, Tinzl M, Bartsch G, Partin AW. Age, prostate-specific antigen, and digital rectal examination as determinants of the probability of having prostate cancer. Urology. 2001;57:1100–4. doi: 10.1016/s0090-4295(01)00980-3. [DOI] [PubMed] [Google Scholar]
- 7.Djavan B, Remzi M, Zlotta A, Seitz C, Snow P, Marberger M. Novel artificial neural network for early detection of prostate cancer. J Clin Oncol. 2002;20:921–9. doi: 10.1200/JCO.2002.20.4.921. [DOI] [PubMed] [Google Scholar]
- 8.Garzotto M, Hudson RG, Peters L, et al. Predictive modeling for the presence of prostate carcinoma using clinical, laboratory, and ultrasound parameters in patients with prostate specific antigen levels < or = 10 ng/mL. Cancer. 2003;98:1417–22. doi: 10.1002/cncr.11668. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Corona E, Ohori M, Scardino PT, Reuter VE, Gonen M, Kattan MW. A nomogram for predicting a positive repeat prostate biopsy in patients with a previous negative biopsy session. J Urol. 2003;170:1184–8. doi: 10.1097/01.ju.0000087451.64657.fa. [DOI] [PubMed] [Google Scholar]
- 10.Karakiewicz PI, Benayoun S, Kattan MW, et al. Development and validation of a nomogram predicting the outcome of prostate biopsy based on patient age, digital rectal examination and serum prostate specific antigen. J Urol. 2005;173:1930–4. doi: 10.1097/01.ju.0000158039.94467.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun FK, Briganti A, Graefen M, et al. Development and external validation of an extended 10-core biopsy nomogram. Eur Urol. 2007;52:436–44. doi: 10.1016/j.eururo.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 12.Chun FK, Graefen M, Briganti A, et al. Initial biopsy outcome prediction-head-to-head comparison of a logistic regression-based nomogram versus artificial neural network. Eur Urol. 2007;51:1236–40. doi: 10.1016/j.eururo.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 13.Chun FK, Briganti A, Graefen M, et al. Development and external validation of an extended repeat biopsy nomogram. J Urol. 2007;177:510–5. doi: 10.1016/j.juro.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H, Komiya A, Kamiya N, et al. Development of a nomogram to predict probability of positive initial prostate biopsy among Japanese patients. Urology. 2006;67:131–6. doi: 10.1016/j.urology.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 15.Yanke BV, Gonen M, Scardino PT, Kattan MW. Validation of a nomogram for predicting positive repeat biopsy for prostate cancer. J Urol. 2005;173:421–4. doi: 10.1097/01.ju.0000150522.82760.00. [DOI] [PubMed] [Google Scholar]
- 16.Yanke BV, Carver BS, Bianco FJ, Jr, et al. African-American race is a predictor of prostate cancer detection: incorporation into a pre-biopsy nomogram. BJU Int. 2006;98:783–7. doi: 10.1111/j.1464-410X.2006.06388.x. [DOI] [PubMed] [Google Scholar]
- 17.Nam RK, Toi A, Klotz LH, et al. Nomogram prediction for prostate cancer and aggressive prostate cancer at time of biopsy: utilizing all risk factors and tumor markers for prostate cancer. Can J Urol. 2006;13:2–10. [PubMed] [Google Scholar]
- 18.Stephan C, Cammann H, Semjonow A, et al. Multicenter evaluation of an artificial neural network to increase the prostate cancer detection rate and reduce unnecessary biopsies. Clin Chem. 2002;48:1279–87. [PubMed] [Google Scholar]
- 19.Chun FK, Karakiewicz PI, Briganti A, et al. Prostate cancer nomograms: an update. Eur Urol. 2006;50:914–26. doi: 10.1016/j.eururo.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 20.Thompson IM, Goodman PJ, Tangen CM, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003;349:215–24. doi: 10.1056/NEJMoa030660. [DOI] [PubMed] [Google Scholar]
- 21.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–34. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 22.Parekh DJ, Ankerst DP, Higgins BA, et al. External validation of the Prostate Cancer Prevention Trial risk calculator in a screened population. Urology. 2006;68:1152–5. doi: 10.1016/j.urology.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Partin AW, Brawer MK, Bartsch G, et al. Complexed prostate specific antigen improves specificity for prostate cancer detection: results of a prospective multicenter clinical trial. J Urol. 2003;170:1787–91. doi: 10.1097/01.ju.0000092695.55705.dd. [DOI] [PubMed] [Google Scholar]
- 24.Thiel RP, Oesterling JE, Wojno KJ, et al. Multicenter comparison of the diagnostic performance of free prostate-specific antigen. Urology. 1996;48:45–50. doi: 10.1016/s0090-4295(96)00609-7. [DOI] [PubMed] [Google Scholar]
- 25.Rabbani F, Stroumbakis N, Kava BR, Cookson MS, Fair WR. Incidence and clinical significance of false-negative sextant prostate biopsies. J Urol. 1998;159:1247–50. [PubMed] [Google Scholar]
- 26.Svetec D, McCabe K, Peretsman S, et al. Prostate rebiopsy is a poor surrogate of treatment efficacy in localized prostate cancer. J Urol. 1998;159:1606–8. doi: 10.1097/00005392-199805000-00052. [DOI] [PubMed] [Google Scholar]
- 27.Singh H, Canto EI, Shariat SF, et al. Predictors of prostate cancer after initial negative systematic 12 core biopsy. J Urol. 2004;171:1850–4. doi: 10.1097/01.ju.0000119667.86071.e7. [DOI] [PubMed] [Google Scholar]
- 28.Ross PL, Gerigk C, Gonen M, et al. Comparisons of nomograms and urologists' predictions in prostate cancer. Semin Urol Oncol. 2002;20:82–8. doi: 10.1053/suro.2002.32490. [DOI] [PubMed] [Google Scholar]