Abstract
Blood pressure (BP) is a heritable trait1 influenced by multiple biological pathways and is responsive to environmental stimuli. Over one billion people worldwide have hypertension (BP ≥140 mm Hg systolic [SBP] or ≥90 mm Hg diastolic [DBP])2. Even small increments in BP are associated with increased risk of cardiovascular events3. This genome-wide association study of SBP and DBP, which used a multi-stage design in 200,000 individuals of European descent, identified 16 novel loci: six of these loci contain genes previously known or suspected to regulate BP (GUCY1A3-GUCY1B3; NPR3-C5orf23; ADM; FURIN-FES; GOSR2; GNAS-EDN3); the other 10 provide new clues to BP physiology. A genetic risk score based on 29 genome-wide significant variants was associated with hypertension, left ventricular wall thickness, stroke, and coronary artery disease, but not kidney disease or kidney function. We also observed associations with BP in East Asian, South Asian, and African ancestry individuals. Our findings provide new insights into the genetics and biology of BP, and suggest novel potential therapeutic pathways for cardiovascular disease prevention.
Genetic approaches have advanced the understanding of biological pathways underlying inter-individual variation in BP. For example, studies of rare Mendelian BP disorders have identified multiple defects in renal sodium handling pathways4. More recently two genome-wide association studies (GWAS), each of >25,000 individuals of European-ancestry, identified 13 loci associated with SBP, DBP, and hypertension5,6. We now report results of a new meta-analysis of GWAS data that includes staged follow-up genotyping to identify additional BP loci.
Primary analyses evaluated associations between 2.5 million genotyped or imputed single nucleotide polymorphisms (SNPs) and SBP and DBP in 69,395 individuals of European ancestry from 29 studies (Supplementary Materials Sections 1–3, Supplementary Tables 1–2). Following GWAS meta-analysis, we conducted a three-stage validation experiment that made efficient use of available genotyping resources, to follow up top signals in up to 133,661 additional individuals of European descent (Supplementary Fig. 1 and Supplementary Materials Section 4). Twenty-nine independent SNPs at 28 loci were significantly associated with SBP, DBP, or both in the meta-analysis combining discovery and follow up data (Fig. 1, Table 1, Supplementary Figs 2–3, Supplementary Tables 3–5). All 29 SNPs attained association P <5×10−9, an order of magnitude beyond the standard genome-wide significance level for a single stage experiment (Table 1).
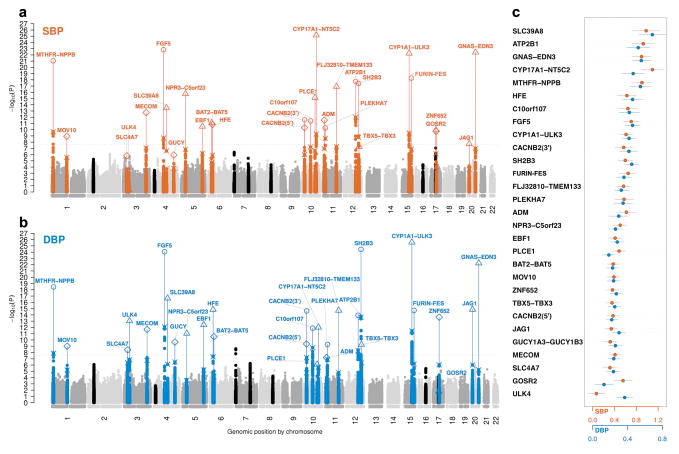
Fig. 1.
Genome-wide –log10 P-value plots and effects for significant loci.
Genome-wide –log10 P-value plots are shown for systolic (SBP: panel a) and diastolic (DBP: panel b). SNPs within loci reaching genome-wide significance are labeled in red for SBP and blue for DBP (±2.5Mb of lowest P-value) and lowest P-values in the initial genome-wide analysis as well as the results of analysis including validation data are labeled separately. The lowest P-values in the initial GWAS are denoted as an X. The range of different sample sizes in the final meta-analysis including the validation data are indicated as: circle (96–140k), triangle (>140–180k), and diamond (>180–220k). SNPs near unconfirmed loci are in black. The horizontal dotted line is P=2.5 × 10−8. Panel c shows the effect size estimates and 95% confidence bars per BP-increasing allele of the 29 significant variants for SBP (red) and DBP (blue). Effect sizes are expressed in mmHg/allele. GUCY = GUCY1A3-GUCY1B3.
Table 1. Summary association results for 29 BP SNPs.
Summary association statistics, based on combined discovery and follow-up data, for 29 independent SNPs in individuals of European ancestry are shown. New genome-wide significant findings (17 SNPs) are presented in the top half of the table, data on 12 previously published signals are presented in the lower half.
| Locus | Index SNP | Chr | Position | CA/NCA | CAF | nsSNP | eSNP | SBP
|
DBP
|
HTN
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | P-value | Effect in EA/SA/A | Beta | P-value | Effect in EA/SA/A | Beta | P-value | ||||||||
| MOV10 | rs2932538 | 1 | 113,018,066 | G/A | 0.75 | Y(p) | Y(p) | 0.388 | 1.2*10−9 | +/+/− | 0.24 | 9.9*10−10 | +/+*/− | 0.049 | 2.9*10−7 |
|
| |||||||||||||||
| SLC4A7 | rs13082711 | 3 | 27,512,913 | T/C | 0.78 | Y(p) | Y(p) | −0.315 | 1.5*10−6 | −/−/+ | −0.238 | 3.8*10−9 | −/−/+ | −0.035 | 3.6*10−4 |
|
| |||||||||||||||
| MECOM | rs419076 | 3 | 170,583,580 | T/C | 0.47 | - | - | 0.409 | 1.8*10−13 | +/+/+ | 0.241 | 2.1*10−12 | +/+/− | 0.031 | 3.1*10−4 |
|
| |||||||||||||||
| SLC39A8 | rs13107325 | 4 | 103,407,732 | T/C | 0.05 | Y | Y(+) | −0.981 | 3.3*10−14 | ?/+/+ | −0.684 | 2.3*10−17 | ?/+/+ | −0.105 | 4.9*10−7 |
|
| |||||||||||||||
| GUCY1A3-GUCY1B3 | rs13139571 | 4 | 156,864,963 | C/A | 0.76 | - | - | 0.321 | 1.2*10−6 | +/−/+ | 0.26 | 2.2*10−10 | +/−/+ | 0.042 | 2.5*10−5 |
|
| |||||||||||||||
| NPR3-C5orf23 | rs1173771 | 5 | 32,850,785 | G/A | 0.6 | - | - | 0.504 | 1.8*10−16 | +*/+/+ | 0.261 | 9.1*10−12 | +*/+/− | 0.062 | 3.2*10−10 |
|
| |||||||||||||||
| EBF1 | rs11953630 | 5 | 157,777,980 | T/C | 0.37 | - | - | −0.412 | 3.0*10−11 | +/+/+ | −0.281 | 3.8*10−13 | +/+/+ | −0.052 | 1.7*10−7 |
|
| |||||||||||||||
| HFE | rs1799945 | 6 | 26,199,158 | G/C | 0.14 | Y | - | 0.627 | 7.7*10−12 | +/+/− | 0.457 | 1.5*10−15 | +/+/− | 0.095 | 1.8*10−10 |
|
| |||||||||||||||
| BAT2-BAT5 | rs805303 | 6 | 31,724,345 | G/A | 0.61 | Y(p) | Y(+) | 0.376 | 1.5*10−11 | −/−/? | 0.228 | 3.0*10−11 | −/−/+ | 0.054 | 1.1*10−10 |
|
| |||||||||||||||
| CACNB2(5′) | rs4373814 | 10 | 18,459,978 | G/C | 0.55 | - | - | −0.373 | 4.8*10−11 | +/+/− | −0.218 | 4.4*10−10 | −/+/− | −0.046 | 8.5*10−8 |
|
| |||||||||||||||
| PLCE1 | rs932764 | 10 | 95,885,930 | G/A | 0.44 | - | - | 0.484 | 7.1*10−16 | +/+/− | 0.185 | 8.1*10−7 | +/+/− | 0.055 | 9.4*10−9 |
|
| |||||||||||||||
| ADM | rs7129220 | 11 | 10,307,114 | G/A | 0.89 | - | - | −0.619 | 3.0*10−12 | ?/−/+ | −0.299 | 6.4*10−8 | ?/−/+ | −0.044 | 1.1*10−3 |
|
| |||||||||||||||
| FLJ32810-TMEM133 | rs633185 | 11 | 100,098,748 | G/C | 0.28 | - | - | −0.565 | 1.2*10−17 | +*/+/+ | −0.328 | 2.0*10−15 | +*/+/− | −0.07 | 5.4*10−11 |
|
| |||||||||||||||
| FURIN-FES | rs2521501 | 15 | 89,238,392 | T/A | 0.31 | - | Y(−) | 0.65 | 5.2*10−19 | +*/+/+ | 0.359 | 1.9*10−15 | +*/+/+ | 0.059 | 7.0*10−7 |
|
| |||||||||||||||
| GOSR2 | rs17608766 | 17 | 42,368,270 | T/C | 0.86 | - | Y(+) | −0.556 | 1.1*10−10 | +/−/+ | −0.129 | 0.017 | +/−/+ | −0.025 | 0.08 |
|
| |||||||||||||||
| JAG1 | rs1327235 | 20 | 10,917,030 | G/A | 0.46 | - | - | 0.34 | 1.9*10−8 | +*/+/+ | 0.302 | 1.4*10−15 | +*/+*/+ | 0.034 | 4.6*10−4 |
|
| |||||||||||||||
| GNAS-EDN3 | rs6015450 | 20 | 57,184,512 | G/A | 0.12 | Y(p) | - | 0.896 | 3.9*10−23 | ?/+/+ | 0.557 | 5.6*10−23 | ?/+*/+ | 0.11 | 4.2*10−14 |
|
| |||||||||||||||
| MTHFR-NPPB | rs17367504 | 1 | 11,785,365 | G/A | 0.15 | - | Y(−/r) | −0.903 | 8.7*10−22 | +/+/+ | −0.547 | 3.5*10−19 | +/+/+ | −0.103 | 2.3*10−10 |
|
| |||||||||||||||
| ULK4 | rs3774372 | 3 | 41,852,418 | T/C | 0.83 | Y | Y(r/p) | −0.067 | 0.39 | −/−/+ | −0.367 | 9.0*10−14 | +/+/+ | −0.017 | 0.18 |
|
| |||||||||||||||
| FGF5 | rs1458038 | 4 | 81,383,747 | T/C | 0.29 | - | - | 0.706 | 1.5*10−23 | +*/+/+ | 0.457 | 8.5*10−25 | +*/+*/+ | 0.072 | 1.9*10−7 |
|
| |||||||||||||||
| CACNB2(3′) | rs1813353 | 10 | 18,747,454 | T/C | 0.68 | - | - | 0.569 | 2.6*10−12 | +/+/+ | 0.415 | 2.3*10−15 | +/+/+ | 0.078 | 6.2*10−10 |
|
| |||||||||||||||
| C10orf107 | rs4590817 | 10 | 63,137,559 | G/C | 0.84 | - | Y(r) | 0.646 | 4.0*10−12 | −/+/− | 0.419 | 1.3*10−12 | −/−/− | 0.096 | 9.8*10−9 |
|
| |||||||||||||||
| CYP17A1-NT5C2 | rs11191548 | 10 | 104,836,168 | T/C | 0.91 | - | Y(−) | 1.095 | 6.9*10−26 | +*/+*/+ | 0.464 | 9.4*10−13 | +*/+*/+ | 0.097 | 1.4*10−5 |
|
| |||||||||||||||
| PLEKHA7 | rs381815 | 11 | 16,858,844 | T/C | 0.26 | - | - | 0.575 | 5.3*10−11 | +*/+/+ | 0.348 | 5.3*10−10 | +*/−/+ | 0.062 | 3.4*10−6 |
|
| |||||||||||||||
| ATP2B1 | rs17249754 | 12 | 88,584,717 | G/A | 0.84 | - | - | 0.928 | 1.8*10−18 | +*/+*/− | 0.522 | 1.2*10−14 | +*/+*/− | 0.126 | 1.1*10−14 |
|
| |||||||||||||||
| SH2B3 | rs3184504 | 12 | 110,368,991 | T/C | 0.47 | Y | Y(+) | 0.598 | 3.8*10−18 | −/−/+ | 0.448 | 3.6*10−25 | −/−/+ | 0.056 | 2.6*10−6 |
|
| |||||||||||||||
| TBX5-TBX3 | rs10850411 | 12 | 113,872,179 | T/C | 0.7 | - | - | 0.354 | 5.4*10−8 | −/+/− | 0.253 | 5.4*10−10 | −/−/− | 0.045 | 5.2*10−6 |
|
| |||||||||||||||
| CYP1A1-ULK3 | rs1378942 | 15 | 72,864,420 | C/A | 0.35 | - | Y(+) | 0.613 | 5.7*10−23 | +*/+/+ | 0.416 | 2.7*10−26 | +*/+/− | 0.073 | 1.0*10−8 |
|
| |||||||||||||||
| ZNF652 | rs12940887 | 17 | 44,757,806 | T/C | 0.38 | - | Y(−) | 0.362 | 1.8*10−10 | +/−/+ | 0.27 | 2.3*10−14 | +/−/+ | 0.046 | 1.2*10−7 |
Y indicates the BP index SNP is a nsSNP, Y(p) indicates a proxy SNP is a nsSNP. Y(+): indicatesBP index SNP is the strongest known eSNP for a transcript; Y(−): indicates BP index SNP is an eSNP but not strongest known eSNP for any transcript. Y(r): indicates BP index SNP is strongest known eSNP in a regional SNP-RTPCR experiment. Y(p): indicates a proxy SNP (r2 > 0.8) to BP SNP is an eSNP but not the strongest known eSNP. Observed effect directions in East Asian (EA), South Asian (SA), and African (A) ancestry individuals are coded + or − if concordant or discordant with directions in European ancestry results;
denotes significance controlling the FDR at 5% over 58 tests per ancestry (Supplementary Tables 5 and 12). Effect size estimates (beta) correspond to mmHg per coded allele for SBP and DBP and ln(odds) per coded allele for HTN.
CA = coded allele; NCA = non-coded allele; CAF = coded allele frequency; ? denotes missing data. Genomic positions use NCBI Build 36 coordinates.
Sixteen of these 29 associations were novel (Table 1). Two associations were near the FURIN and GOSR2 genes; prior targeted analyses of variants in these genes suggested they may be BP loci7,8. At the CACNB2 locus we validated association for a previously reported6 SNP rs4373814 and detected a novel independent association for rs1813353 (pairwise r2 =0.015 in HapMap CEU). Of our 13 previously reported associations5,6, only the association at PLCD3 was not supported by the current results (Supplementary Table 4). Some of the associations are in or near genes involved in pathways known to influence BP (NPR3, GUCY1A3-GUCY1B3, ADM, GNAS-EDN3, NPPA-NPPB, and CYP17A1; Supplementary Fig. 4). Twenty-two of the 28 loci did not contain genes that were a priori strong biological candidates.
As expected from prior BP GWAS results, the effects of the novel variants on SBP and DBP were small (Fig. 1 and Table 1). For all variants, the observed directions of effects were concordant for SBP, DBP, and hypertension (Fig. 1, Table 1, Supplementary Fig. 3). Among the genes at the genome-wide significant loci, only CYP17A1, previously implicated in Mendelian congenital adrenal hyperplasia and hypertension, is known to harbour rare variants that have large effects on BP9.
We performed several analyses to identify potential causal alleles and mechanisms. First, we looked up the 29 genome-wide significant index SNPs and their close proxies (r2>0.8) among cis-acting expression SNP (eSNP) results from multiple tissues (Supplementary Materials Section 5). For 13/29 index SNPs, we found association between nearby eSNP variants and expression level of at least one gene transcript (10−4 > p > 10−51, Supplementary Table 6). In 5 cases, the index BP SNP and the best eSNP from a genome-wide survey were identical, highlighting potential mediators of the SNP-BP associations.
Second, because changes in protein sequence are strong a priori candidates to be functional, we sought non-synonymous coding SNPs that were in high LD (r2 >0.8) with the 29 index SNPs. We identified such SNPsat 8 loci (Table 1, Supplementary Materials Section 6, Supplementary Table 7). In addition we performed analyses testing for differences in genetic effect according to body mass index (BMI) or sex, and analyses of copy number variants, pathway enrichment, and metabolomic data, but we did not find any statistically significant results (Supplementary Materials Sections 7–9, Supplementary Tables 8–10).
We evaluated whether the BP variants we identified in Europeans were associated with BP in individuals of East Asian (N=29,719), South Asian (N=23,977), and African (N=19,775) ancestries (Table 1, Supplementary Tables 11–13). We found significant associations in individuals of East Asian ancestry for SNPs at 9 loci and in individuals of South Asian ancestry for SNPs at 6 loci; some have been reported previously (Supplementary Tables 12 and 15). The lack of significant association for individual SNPs may reflect small sample sizes, differences in allele frequencies or LD patterns, imprecise imputation for some ancestries using existing reference samples, or a genuinely different underlying genetic architecture. Because of limited power to detect effects of individual variants in the smaller non-European samples, we created genetic risk scores for SBP and DBP incorporating all 29 BP variants weighted according to effect sizes observed in the European samples. In each non-European ancestry group, risk scores were strongly associated with SBP (P=1.1×10−40 in East Asian, P=2.9×10−13 in South Asian, P=9.8×10−4 in African ancestry individuals) and DBP (P=2.9×10−48, P=9.5×10−15, and P=5.3×10−5, respectively; Supplementary Table 13).
We also created a genetic risk score to assess association of the variants in aggregate with hypertension and with clinical measures of hypertensive complications including left ventricular mass, left ventricular wall thickness, incident heart failure, incident and prevalent stroke, prevalent coronary artery disease (CAD), kidney disease, and measures of kidney function, using results from other GWAS consortia (Table 2, Supplementary Materials Sections 10–11, Supplementary Table 14). The risk score was weighted using the average of SBP and DBP effects for the 29 SNPs. In an independent sample of 23,294 women10, an increase of 1 standard deviation in the genetic risk score was associated with a 21% increase in the odds of hypertension (95% CI 19%–28%; Table 2, Supplementary Table 14). Among individuals in the top decile of the risk score, the prevalence of hypertension was 29% compared with 16% in the bottom decile (odds ratio 2.09, 95% CI 1.86–2.36). Similar results were observed in an independent hypertension case-control sample (Table 2). In our study, individuals in the top compared to bottom quintiles of genetic risk score differed by 4.6 mm Hg SBP and 3.0 mm Hg DBP, differences that approach population-averaged BP treatment effects for a single antihypertensive agent11. Epidemiologic data have shown that differences in SBP and DBP of this magnitude, across the population range of BP, are associated with an increase in cardiovascular disease risk3. Consistent with this and in line with findings from randomized trials of BP-lowering medication in hypertensive patients12,13, the genetic risk score was positively associated with left ventricular wall thickness (P=6.0×10−6), occurrence of stroke (P=3.3×10−5) and CAD (P=8.1×10−29). The same genetic risk score was not, however, significantly associated with chronic kidney disease or measures of kidney function, even though these renal outcomes were available in a similar sample size as for the other outcomes (Table 2). The absence of association with kidney phenotypes could be explained by a weaker causal relation of BP with kidney phenotypes than with CAD and stroke. This finding is consistent with the mismatch between observational data that show a positive association of BP with kidney disease, and clinical trial data that show inconsistent evidence of benefit of BP lowering on kidney disease prevention in patients with hypertension14. Thus, several lines of evidence converge to suggest that BP elevation may in part be a consequence rather than a cause of sub-clinical kidney disease.
Table 2. Genetic risk score and cardiovascular outcome association results.
Association of genetic risk score (using all 29 SNPs at 28 loci, parameterised using the average of SBP and DBP effects [=(SBP effect + DBP effect)/2] from the discovery analysis), tested in results from other GWAS consortia.
| Phenotype | Source | Effect | SE | P-value | # SNPs | Contrast top vs. bottom | N case/control or total | |||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| (per SD of genetic risk score) | quintiles | deciles | ||||||||
| Blood pressure phenotypes | ||||||||||
| SBP [mmHg] | WGHS | 1.645 | 0.098 | (a) | 6.5*10−63 | 29 | 4.61 | 5.77 | (a) | 23,294 |
|
| ||||||||||
| DBP [mmHg] | WGHS | 1.057 | 0.067 | (a) | 8.4*10−57 | 29 | 2.96 | 3.71 | (a) | 23,294 |
|
| ||||||||||
| Prevalent hypertension | WGHS | 0.211 | 0.018 | (b) | 3.1*10−33 | 29 | 1.80 | 2.09 | (b) | 5,018/18,276 |
|
| ||||||||||
| Prevalent hypertension | BRIGHT | 0.287 | 0.031 | (b) | 7.7*10−21 | 29 | 2.23 | 2.74 | (b) | 2,406/1,990 |
|
| ||||||||||
|
Dichotomous endpoints
| ||||||||||
| Incident heart failure | CHARGE-HF | 0.035 | 0.021 | (c) | 0.10 | 29 | 1.10 | 1.13 | (c) | 2,526/18,400 |
|
| ||||||||||
| Incident stroke | NEURO-CHARGE | 0.103 | 0.028 | (c) | 0.0002 | 28 | 1.34 | 1.44 | (c) | 1,544/18,058 |
|
| ||||||||||
| Prevalent stroke | UK-US Stroke Collaborative Group(SCG) | 0.075 | 0.037 | (b) | 0.05 | 29 | 1.23 | 1.30 | (b) | 1,473/1,482 |
|
| ||||||||||
| Stroke (combined, incident and prevalent) | CHARGE & SCG | NA | NA | NA | 3.3*10−5 | NA | NA | NA | NA | 3,017/19,540 |
|
| ||||||||||
| Prevalent CAD | CARDIoGRAM | 0.092 | 0.010 | (b) | 1.6*10−19 | 28 | 1.29 | 1.38 | (b) | 22,233/64,726 |
|
| ||||||||||
| Prevalent CAD | C4D ProCARDIS | 0.132 | 0.022 | (b) | 2.2*10−9 | 29 | 1.45 | 1.59 | (b) | 5,720/4,381 |
|
| ||||||||||
| Prevalent CAD | C4D HPS | 0.083 | 0.027 | (b) | 0.002 | 29 | 1.26 | 1.34 | (b) | 2,704/2,804 |
|
| ||||||||||
| Prevalent CAD (combined) | CARDIoGRAM & C4D | 0.100 | 0.009 | (b) | 8.1*10−29 | 29 | 1.32 | 1.42 | (b) | 30,657/71,911 |
|
| ||||||||||
| Prevalent chronic kidney disease | CKDGen | 0.014 | 0.015 | (b) | 0.35 | 29 | 1.04 | 1.05 | (b) | 5,807/61,286 |
|
| ||||||||||
| Prevalent microalbuminuria | CKDGen | 0.008 | 0.019 | (b) | 0.68 | 29 | 1.02 | 1.03 | (b) | 3,698/27,882 |
|
| ||||||||||
|
Continuous measures oftarget organ damage
| ||||||||||
| Left ventricular mass [g] | EchoGen | 0.822 | 0.317 | (a) | 0.01 | 29 | 2.30 | 2.89 | (a) | 12,612 |
|
| ||||||||||
| Left ventricular wall thickness[cm] | EchoGen | 0.009 | 0.002 | (a) | 6.0*10−6 | 29 | 0.03 | 0.03 | (a) | 12,612 |
|
| ||||||||||
| Serum creatinine | KidneyGen | −0.001 | 0.001 | (d) | 0.24 | 29 | 1.00 | 1.00 | (d) | 23,812 |
|
| ||||||||||
| eGFR (4 parameter MDRD equation) | CKDGen | −0.0001 | 0.0009 | (d) | 0.93 | 29 | 1.00 | 1.00 | (d) | 67,093 |
|
| ||||||||||
| Urinary albumin/creatinine ratio | CKDGen | 0.005 | 0.007 | (d) | 0.43 | 29 | 1.01 | 1.02 | (d) | 31,580 |
Units are the unit of phenotypic measurement, either per SD of genetic risk score, or as a difference between top/bottom quintiles or deciles.
Units are ln(odds) per SD of genetic risk score, or odds ratio between top/bottom quintiles or deciles.
Units are ln(hazard) per SD of genetic risk score, or hazard ratio between top/bottom quintiles or deciles.
Units are ln(phenotype) per SD of genetic risk score, or phenotypic ratio between top/bottom quintiles or deciles.
Our discovery meta-analysis (Supplementary Fig. 2) suggests an excess of modestly significant (10−5 <P <10−2) associations likely arising from common BP variants of small effect. By dividing our principal GWAS dataset into non-overlapping discovery (N≈56,000) and validation (N≈14,000) subsets, we found robust evidence for the existence of such undetected common variants (Supplementary Fig. 5, Supplementary Materials Section 12). We estimate15 that there are 116 (95% CI 57–174) independent BP variants with effect sizes similar to those reported here, which collectively explain ≈2.2% of the phenotypic variance for SBP and DBP, compared with 0.9% explained by the 29 associations discovered thus far (Supplementary Fig. 6, Supplementary Materials Section 13).
Most of the 28 BP loci harbour multiple genes (Supplementary Table 15, Supplementary Fig. 4), and although substantial research is required to identify the specific genes and variants responsible for these associations, several loci contain highly plausible biological candidates. The NPPA and NPPB genes at the MTHFR-NPPB locus encode precursors for atrial- and B-type natriuretic peptides (ANP, BNP), and previous work has identified SNPs, modestly correlated with our index SNP at this locus, that are associated with plasma ANP, BNP, and BP16. We found the index SNP at this locus was associated with opposite effects on BP and on ANP/BNP levels, consistent with a model in which the variants act through increased ANP/BNP production to lower BP16 (Supplementary Materials Section 14).
Two other loci identified in the current study harbour genes involved in natriuretic peptide and related nitric oxide signalling pathways,17,18 both of which act to regulate cyclic guanosine monophosphate (cGMP). The first locus contains NPR3, which encodes the natriuretic peptide clearance receptor (NPR-C). NPR3 knockout mice exhibit reduced clearance of circulating natriuretic peptides and lower BP19. The second locus includes GUCY1A3 and GUCY1B3, encoding the alpha and beta subunits of soluble guanylatecyclase (sGC); knockout of either gene in murine models results in hypertension20.
Another locus contains ADM, encoding adrenomedullin, which has natriuretic, vasodilatory, and BP-lowering properties21. At the GNAS-EDN3 locus, ZNF831 is closest to the index SNP, but GNAS and EDN3 are two nearby compelling biological candidates (Supplementary Fig. 4, Supplementary Table 15).
We identified two loci with plausible connections to BP via genes implicated in renal physiology or kidney disease. At the first locus, SLC4A7 is an electro-neutral sodium bicarbonate co-transporter expressed in the nephron and in vascular smooth muscle22. At the second locus, PLCE1 (phospholipase-C-epsilon-1 isoform) is important for normal podocyte development in the glomerulus; sequence variation in PLCE1 has been implicated in familial nephrotic syndromes and end-stage kidney disease23.
Missense variants in two genes involved in metal ion transport were associated with BP in our study. The first encodes a His/Asp change at amino acid 63 (H63D) in HFE and is a low penetrance allele for hereditary hemochromatosis24. The second is an Ala/Thr polymorphism located in exon 7 of SLC39A8, which encodes a zinc transporter that also transports cadmium and manganese25. The same allele of SLC39A8 associated with BP in our study has recently been associated with high-density lipoprotein (HDL) cholesterol levels26 and BMI27 (Supplementary Table 15).
In conclusion, we have shown that 29 independent genetic variants influence BP in people of European ancestry. The variants reside in 28 loci, 16 of which were novel, and we confirmed association of several of them in individuals of non-European ancestry. A risk score derived from the 29 variants was significantly associated with BP-related organ damage and clinical cardiovascular disease, but not kidney disease. These loci improve our understanding of the genetic architecture of BP, provide new biological insights into BP control and may identify novel targets for the treatment of hypertension and the prevention of cardiovascular disease.
Methods summary
Supplementary Materials provide complete methods and include the following sections: study recruitment and phenotyping, adjustment for antihypertensive medications, genotyping, data quality control, genotype imputation, within-cohort association analyses, meta-analyses of discovery and validation stages, stratified analyses by sex and BMI, identification of eSNPs and nsSNPs, metabolomic and lipidomic analyses, CNV analyses, pathway analyses, analyses for non-European ancestries, association of a risk score with hypertension and cardiovascular disease, estimation of numbers of undiscovered variants, measurement of natriuretic peptides, and brief literature reviews and GWAS database lookups of all validated BP loci.
Supplementary Material
Acknowledgments
A number of the participating studies and authors are members of the CHARGE and Global BPgen consortia. Many funding mechanisms by NIH/NHLBI, European, and private funding agencies contributed to this work and a full list is provided in Section 21 of the Supplementary Materials.
Authors
Georg B. Ehret1,2,3,*, Patricia B. Munroe4,*,#, Kenneth M. Rice5,*, Murielle Bochud2,*, Andrew D. Johnson6,7,*, Daniel I. Chasman8,9,*, Albert V. Smith10,11,*, Martin D. Tobin12, Germaine C. Verwoert13,14,15, Shih-Jen Hwang6,16,7, Vasyl Pihur1, Peter Vollenweider17, Paul F. O'Reilly18, Najaf Amin13, Jennifer L Bragg-Gresham19, Alexander Teumer20, Nicole L. Glazer21, Lenore Launer22, Jing Hua Zhao23, Yurii Aulchenko13, Simon Heath24, Siim Sõber25, Afshin Parsa26, Jian'an Luan23, Pankaj Arora27, Abbas Dehghan13,14,15, Feng Zhang28, Gavin Lucas29, Andrew A. Hicks30, Anne U. Jackson31, John F Peden32, Toshiko Tanaka33, Sarah H. Wild34, Igor Rudan35,36, Wilmar Igl37, Yuri Milaneschi33, Alex N. Parker38, Cristiano Fava39,40, John C. Chambers18,41, Ervin R. Fox42, Meena Kumari43, Min Jin Go44, Pim van der Harst45, Wen Hong Linda Kao46, Marketa Sjögren39, D. G. Vinay47, Myriam Alexander48, Yasuharu Tabara49, Sue Shaw-Hawkins4, Peter H. Whincup50, Yongmei Liu51, Gang Shi52, Johanna Kuusisto53, Bamidele Tayo54, Mark Seielstad55,56, Xueling Sim57, Khanh-Dung Hoang Nguyen1, Terho Lehtimäki58, Giuseppe Matullo59,60, Ying Wu61, Tom R. Gaunt62, N. Charlotte Onland-Moret63,64, Matthew N. Cooper65, Carl G.P. Platou66, Elin Org25, Rebecca Hardy67, Santosh Dahgam68, Jutta Palmen69, Veronique Vitart70, Peter S. Braund71,72, Tatiana Kuznetsova73, Cuno S.P.M. Uiterwaal63, Adebowale Adeyemo74, Walter Palmas75, Harry Campbell35, Barbara Ludwig76, Maciej Tomaszewski71,72, Ioanna Tzoulaki77,78, Nicholette D. Palmer79, CARDIoGRAM consortium80, CKDGen Consortium80, KidneyGen Consortium80, EchoGen consortium80, CHARGE-HF consortium80, Thor Aspelund10,11, Melissa Garcia22, Yen-Pei C. Chang26, Jeffrey R. O'Connell26, Nanette I. Steinle26, Diederick E. Grobbee63, Dan E. Arking1, Sharon L. Kardia81, Alanna C. Morrison82, Dena Hernandez83, Samer Najjar84,85, Wendy L. McArdle86, David Hadley50,87, Morris J. Brown88, John M. Connell89, Aroon D. Hingorani90, Ian N.M. Day62, Debbie A. Lawlor62, John P. Beilby91,92, Robert W. Lawrence65, Robert Clarke93, Rory Collins93, Jemma C Hopewell93, Halit Ongen32, Albert W. Dreisbach42, Yali Li94, J H. Young95, Joshua C. Bis21, Mika Kähönen96, Jorma Viikari97, Linda S. Adair98, Nanette R. Lee99, Ming-Huei Chen100, Matthias Olden101,102, Cristian Pattaro30, Judith A. Hoffman Bolton103, Anna Köttgen104,103, Sven Bergmann105,106, Vincent Mooser107, Nish Chaturvedi108, Timothy M. Frayling109, Muhammad Islam110, Tazeen H. Jafar110, Jeanette Erdmann111, Smita R. Kulkarni112, Stefan R. Bornstein76, Jürgen Grässler76, Leif Groop113,114, Benjamin F. Voight115, Johannes Kettunen116,126, Philip Howard117, Andrew Taylor43, Simonetta Guarrera60, Fulvio Ricceri59,60, Valur Emilsson118, Andrew Plump118, Inês Barroso119,120, Kay-Tee Khaw48, Alan B. Weder121, Steven C. Hunt122, Yan V. Sun81, Richard N. Bergman123, Francis S. Collins124, Lori L. Bonnycastle124, Laura J. Scott31, Heather M. Stringham31, Leena Peltonen119,125,126,127, Markus Perola125, Erkki Vartiainen125, Stefan-Martin Brand128,129, Jan A. Staessen73, Thomas J. Wang6,130, Paul R. Burton12,72, Maria Soler Artigas12, Yanbin Dong131, Harold Snieder132,131, Xiaoling Wang131, Haidong Zhu131, Kurt K. Lohman133, Megan E. Rudock51, Susan R Heckbert134,135, Nicholas L Smith134,136,135, Kerri L Wiggins137, Ayo Doumatey74, Daniel Shriner74, Gudrun Veldre25,138, Margus Viigimaa139,140, Sanjay Kinra141, Dorairajan Prabhakaran142, Vikal Tripathy142, Carl D. Langefeld79, Annika Rosengren143, Dag S. Thelle144, Anna Maria Corsi145, Andrew Singleton83, Terrence Forrester146, Gina Hilton1, Colin A. McKenzie146, Tunde Salako147, Naoharu Iwai148, Yoshikuni Kita149, Toshio Ogihara150, Takayoshi Ohkubo149,151, Tomonori Okamura148, Hirotsugu Ueshima152, Satoshi Umemura153, Susana Eyheramendy154, Thomas Meitinger155,156, H.-Erich Wichmann157,158,159, Yoon Shin Cho44, Hyung-Lae Kim44, Jong-Young Lee44, James Scott160, Joban S. Sehmi160,41, Weihua Zhang18, Bo Hedblad39, Peter Nilsson39, George Davey Smith62, Andrew Wong67, Narisu Narisu124, Alena Stančáková53, Leslie J. Raffel161, Jie Yao161, Sekar Kathiresan162,27, Chris O'Donnell163,27,9, Stephen M. Schwartz134, M. Arfan Ikram13,15, W. T. Longstreth Jr.164, Thomas H. Mosley165, Sudha Seshadri166, Nick R.G. Shrine12, Louise V. Wain12, Mario A. Morken124, Amy J. Swift124, Jaana Laitinen167, Inga Prokopenko51,168, Paavo Zitting169, Jackie A. Cooper69, Steve E. Humphries69, John Danesh48, Asif Rasheed170, Anuj Goel32, Anders Hamsten171, Hugh Watkins32, Stephan J.L. Bakker172, Wiek H. van Gilst45, Charles S. Janipalli47, K. Radha Mani47, Chittaranjan S. Yajnik112, Albert Hofman13, Francesco U.S. Mattace-Raso13,14, Ben A. Oostra173, Ayse Demirkan13, Aaron Isaacs13, Fernando Rivadeneira13,14, Edward G Lakatta174, Marco Orru175,176, Angelo Scuteri174, Mika Ala-Korpela177,178,179, Antti J Kangas177, Leo-Pekka Lyytikäinen58, Pasi Soininen177,178, Taru Tukiainen180,181,177, Peter Würtz177,18,180, Rick Twee-Hee Ong56,57,182, Marcus Dörr183, Heyo K. Kroemer184, Uwe Völker20, Henry Völzke185, Pilar Galan186, Serge Hercberg186, Mark Lathrop24, Diana Zelenika24, Panos Deloukas119, Massimo Mangino28, Tim D. Spector28, Guangju Zhai28, James F. Meschia187, Michael A. Nalls83, Pankaj Sharma188, Janos Terzic189, M. J. Kranthi Kumar47, Matthew Denniff71, Ewa Zukowska-Szczechowska190, Lynne E. Wagenknecht79, F. Gerald R. Fowkes191, Fadi J. Charchar192, Peter E.H. Schwarz193, Caroline Hayward70, Xiuqing Guo161, Charles Rotimi74, Michiel L. Bots63, Eva Brand194, Nilesh J. Samani71,72, Ozren Polasek195, Philippa J. Talmud69, Fredrik Nyberg68,196, Diana Kuh67, Maris Laan25, Kristian Hveem66, Lyle J. Palmer197,198, Yvonne T. van der Schouw63, Juan P. Casas199, Karen L. Mohlke61, Paolo Vineis200,60, Olli Raitakari201, Santhi K. Ganesh202, Tien Y. Wong203,204, E Shyong Tai205,57,206, Richard S. Cooper54, Markku Laakso53, Dabeeru C. Rao207, Tamara B. Harris22, Richard W. Morris208, Anna F. Dominiczak209, Mika Kivimaki210, Michael G. Marmot210, Tetsuro Miki49, Danish Saleheen170,48, Giriraj R. Chandak47, Josef Coresh211, Gerjan Navis212, Veikko Salomaa125, Bok-Ghee Han44, Xiaofeng Zhu94, Jaspal S. Kooner160,41, Olle Melander39, Paul M Ridker8,213,9, Stefania Bandinelli214, Ulf B. Gyllensten37, Alan F. Wright70, James F. Wilson34, Luigi Ferrucci33, Martin Farrall32, Jaakko Tuomilehto215,216,217,218, Peter P. Pramstaller30,219, Roberto Elosua29,220, Nicole Soranzo119,28, Eric J.G. Sijbrands13,14, David Altshuler221,115, Ruth J.F. Loos23, Alan R. Shuldiner26,222, Christian Gieger157, Pierre Meneton223, Andre G. Uitterlinden13,14,15, Nicholas J. Wareham23, Vilmundur Gudnason10,11, Jerome I. Rotter161, Rainer Rettig224, Manuela Uda175, David P. Strachan50, Jacqueline C.M. Witteman13,15, Anna-Liisa Hartikainen225, Jacques S. Beckmann105,226, Eric Boerwinkle227, Ramachandran S. Vasan6,228, Michael Boehnke31, Martin G. Larson6,229, Marjo-Riitta Järvelin18,230,231,232,233, Bruce M. Psaty21,135,*, Gonçalo R Abecasis19,*, Aravinda Chakravarti1,*,#, Paul Elliott18,233,*, Cornelia M. van Duijn13,234,*, Christopher Newton-Cheh27,115,*,#, Daniel Levy6,16,7,*,#, Mark J. Caulfield4,*,#, Toby Johnson4,*
Footnotes
contributed equally
to whom correspondence should be addressed: aravinda@jhmi.edu; m.j.caulfield@qmul.ac.uk; levyd@nhlbi.nih.gov; p.b.munroe@qmul.ac.uk; cnewtoncheh@partners.org
Center for Complex Disease Genomics, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
Institute of Social and Preventive Medicine (IUMSP), Centre Hospitalier Universitaire Vaudois and University of Lausanne, Bugnon 17, 1005 Lausanne, Switzerland
Cardiology, Department of Specialties of Internal Medicine, Geneva University Hospital, Rue Gabrielle-Perret-Gentil 4, 1211 Geneva 14, Switzerland
Clinical Pharmacology and The Genome Centre, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK
Department of Biostatistics, University of Washington, Seattle, WA, USA
Framingham Heart Study, Framingham, MA, USA
National Heart Lung, and Blood Institute, Bethesda, MD, USA
Division of Preventive Medicine, Brigham and Women’s Hospital, 900 Commonwealth Avenue East, Boston MA 02215, USA
Harvard Medical School, Boston, MA, USA
Icelandic Heart Association, Kopavogur, Iceland
University of Iceland, Reykajvik, Iceland
Department of Health Sciences, University of Leicester, University Rd, Leicester LE1 7RH, UK
Department of Epidemiology, Erasmus Medical Center, PO Box 2040, 3000 CA, Rotterdam, The Netherlands
Department of Internal Medicine, Erasmus Medical Center, Rotterdam, The Netherlands
Netherlands Consortium for Healthy Aging (NCHA), Netherland Genome Initiative (NGI), The Netherlands
Center for Population Studies, National Heart Lung, and Blood Institute, Bethesda, MD, USA
Department of Internal Medicine, Centre Hospitalier Universitaire Vaudois, 1011 Lausanne, Switzerland
Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, Norfolk Place, London W2 1PG, UK
Center forStatistical Genetics, Department of Biostatistics, University of Michigan School of Public Health, Ann Arbor, MI 48103, USA
Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Cardiovascular Health Research Unit, Departments of Medicine, Epidemiology and Health Services, University of Washington, Seattle, WA, USA
Laboratory of Epidemiology, Demography, Biometry, National Institute on Aging, National Institutes of Health, Bethesda, Maryland 20892, USA
MRC Epidemiology Unit, Institute of Metabolic Science, Cambridge CB2 0QQ, UK
Centre National de Génotypage, Commissariat à L’Energie Atomique, Institut de Génomique, Evry, France
Institute of Molecular and Cell Biology, University of Tartu, Riia 23, Tartu 51010, Estonia
University of Maryland School of Medicine, Baltimore, MD, USA, 21201, USA
Center for Human Genetic Research, Cardiovascular Research Center, Massachusetts General Hospital, Boston, Massachusetts, 02114, USA
Department of Twin Research & Genetic Epidemiology, King’s College London, UK
Cardiovascular Epidemiology and Genetics, Institut Municipal d’Investigacio Medica, Barcelona Biomedical Research Park, 88 Doctor Aiguader, 08003 Barcelona, Spain
Institute of Genetic Medicine, European Academy Bozen/Bolzano (EURAC), Viale Druso 1, 39100 Bolzano, Italy -Affiliated Institute of the University of Lübeck, Germany
Department of Biostatistics, Center for Statistical Genetics, University of Michigan, Ann Arbor, Michigan, 48109, USA
Department of Cardiovascular Medicine, The Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, OX3 7BN, UK
Clinical Research Branch, National Institute on Aging, Baltimore MD 21250, USA
Centre for Population Health Sciences, University of Edinburgh, EH89AG, UK
Centre for Population Health Sciences and Institute of Genetics and Molecular Medicine, College of Medicine and Vet Medicine, University of Edinburgh, EH8 9AG, UK
Croatian Centre for Global Health, University of Split, Croatia
Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, SE-751 85 Uppsala, Sweden
Amgen, 1 Kendall Square, Building 100, Cambridge, MA 02139, USA
Department of Clinical Sciences, Lund University, Malmö, Sweden
Department of Medicine, University of Verona, Italy
Ealing Hospital, London, UB1 3HJ, UK
Department of Medicine, University of Mississippi Medical Center, USA
Genetic Epidemiology Group, Epidemiology and Public Health, UCL, London, WC1E 6BT, UK
Center for Genome Science, National Institute of Health, Seoul, Korea
Department of Cardiology, University Medical Center Groningen, University of Groningen, The Netherlands
Departments of Epidemiology and Medicine, Johns Hopkins University, Baltimore MD, USA
Centre for Cellular and Molecular Biology (CCMB), Council of Scientific and Industrial Research (CSIR), Uppal Road, Hyderabad 500 007, India
Department of Public Health and Primary Care, University of Cambridge, CB1 8RN, UK
Department of Basic Medical Research and Education, and Department of Geriatric Medicine, Ehime University Graduate School of Medicine, Toon, 791-0295, Japan
Division of Community Health Sciences, St George’s University of London, London, SW17 0RE, UK
Epidemiology & Prevention, Division of Public Health Sciences, Wake Forest University School of Medicine, Winston-Salem, NC 27157, USA
Division of Biostatistics and Department of Genetics, School of Medicine, Washington University in St. Louis, Saint Louis, Missouri 63110, USA
Department of Medicine, University of Eastern Finland and Kuopio University Hospital, 70210 Kuopio, Finland
Department of Preventive Medicine and Epidemiology, Loyola University Medical School, Maywood, IL, USA
Department of Laboratory Medicine & Institute of Human Genetics, University of California San Francisco, 513 Parnassus Ave. San Francisco CA 94143, USA
Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore, 138672, Singapore
Centre for Molecular Epidemiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 117597, Singapore
Department of Clinical Chemistry, University of Tampere and Tampere University Hospital, Tampere, 33521, Finland
Department of Genetics, Biology and Biochemistry, University of Torino, Via Santena 19, 10126, Torino, Italy
Human Genetics Foundation (HUGEF), Via Nizza 52, 10126, Torino, Italy
Department of Genetics, University of North Carolina, Chapel Hill, NC, 27599, USA
MRC Centre for Causal Analyses in Translational Epidemiology, School of Social & Community Medicine, University of Bristol, Bristol BS8 2BN, UK
Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Heidelberglaan 100, 3508 GA Utrecht, The Netherlands
Complex Genetics Section, Department of Medical Genetics -DBG, University Medical Center Utrecht, 3508 GA Utrecht, The Netherlands
Centre for Genetic Epidemiology and Biostatistics, University of Western Australia, Crawley, WA, Australia
HUNT Research Centre, Department of Public Health and General Practice, Norwegian University of Science and Technology, 7600 Levanger, Norway
MRC Unit for Lifelong Health & Ageing, London, WC1B 5JU, UK
Occupational and Environmental Medicine, Department of Public Health and Community Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, 40530 Gothenburg, Sweden
Centre for Cardiovascular Genetics, University College London, London WC1E 6JF, UK
MRC Human Genetics Unit and Institute of Genetics and Molecular Medicine, Edinburgh, EH2, UK
Department of Cardiovascular Sciences, University of Leicester, Glenfield Hospital, Leicester, LE3 9QP, UK
Leicester NIHR Biomedical Research Unit in Cardiovascular Disease, Glenfield Hospital, Leicester, LE3 9QP, UK
Studies Coordinating Centre, Division of Hypertension and Cardiac Rehabilitation, Department of Cardiovascular Diseases, University of Leuven, Campus Sint Rafaël, Kapucijnenvoer 35, Block D, Box 7001, 3000 Leuven, Belgium
Center for Research on Genomics and Global Health, National Human Genome Research Institute, Bethesda, MD 20892, USA
Columbia University, NY, USA
Department of Medicine III, Medical Faculty Carl Gustav Carus at the Technical University of Dresden, 01307 Dresden, Germany
Epidemiology and Biostatistics, School of Public Health, Imperial College, London, W2 1PG, UK
Clinical and Molecular Epidemiology Unit, Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece
Wake Forest University Health Sciences, Winston-Salem, NC 27157, USA
A list of consortium members is supplied in the Supplementary Materials
Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI 48109, USA
Division of Epidemiology, Human Genetics and Environmental Sciences, School of Public Health, University of Texas at Houston Health Science Center, 12 Herman Pressler, Suite 453E, Houston, TX 77030, USA
Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD 20892, USA
Laboratory of Cardiovascular Science, Intramural Research Program, National Institute on Aging, NIH, Baltimore, Maryland, USA
Washington Hospital Center, Division of Cardiology, Washington DC, USA
ALSPAC Laboratory, University of Bristol, Bristol, BS8 2BN, UK
Pediatric Epidemiology Center, University of South Florida, Tampa, FL, USA
Clinical Pharmacology Unit, University of Cambridge, Addenbrookes Hospital, Hills Road, Cambridge CB2 2QQ, UK
University of Dundee, Ninewells Hospital &Medical School, Dundee, DD1 9SY, UK
Genetic Epidemiology Group, Department of Epidemiology and Public Health, UCL, London WC1E 6BT, UK
Pathology and Laboratory Medicine, University of Western Australia, Crawley, WA, Australia
Molecular Genetics, PathWest Laboratory Medicine, Nedlands, WA, Australia
Clinical Trial Service Unit and Epidemiological Studies Unit, University of Oxford, Oxford, OX3 7LF, UK
Department of Epidemiology and Biostatistics, Case Western Reserve University, 2103 Cornell Road, Cleveland, OH 44106, USA
Department of Medicine, Johns Hopkins University, Baltimore, USA
Department of Clinical Physiology, University of Tampere and Tampere University Hospital, Tampere, 33521, Finland
Department of Medicine, University of Turku and Turku University Hospital, Turku, 20521, Finland
Department of Nutrition, University of North Carolina, Chapel Hill, NC, 27599, USA
Office of Population Studies Foundation, University of San Carlos, Talamban, Cebu City 6000, Philippines
Department of Neurology and Framingham Heart Study, Boston University School of Medicine, Boston, MA, 02118, USA
Department of Internal Medicine II, University Medical Center Regensburg, 93053 Regensburg, Germany
Department of Epidemiology and Preventive Medicine, University Medical Center Regensburg, 93053 Regensburg, Germany
Department of Epidemiology, Johns Hopkins University, Baltimore MD, USA
Renal Division, University Hospital Freiburg, Germany
Département de Génétique Médicale, Université de Lausanne, 1015 Lausanne, Switzerland
Swiss Institute of Bioinformatics, 1015 Lausanne, Switzerland
Division of Genetics, GlaxoSmithKline, Philadelphia, Pennsylvania 19101, USA
International Centre for Circulatory Health, National Heart & Lung Institute, Imperial College, London, UK
Genetics of Complex Traits, Peninsula Medical School, University of Exeter, UK
Department of Community Health Sciences & Department of Medicine, Aga Khan University, Karachi, Pakistan
Medizinische Klinik II, Universität zu Lübeck, Lübeck, Germany
Diabetes Unit, KEM Hospital and Research Centre, Rasta Peth, Pune-411011, Maharashtra, India
Department of Clinical Sciences, Diabetes and Endocrinology Research Unit, University Hospital, Malmö, Sweden
Lund University, Malmö 20502, Sweden
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, Massachusetts, 02139, USA
Department of Chronic Disease Prevention, National Institute for Health and Welfare, FIN-00251 Helsinki, Finland
William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK
Merck Research Laboratory, 126 East Lincoln Avenue, Rahway, NJ 07065, USA
Wellcome Trust Sanger Institute, Hinxton, CB10 1SA, UK
University of Cambridge Metabolic Research Labs, Institute of Metabolic Science Addenbrooke’s Hospital, CB2 OQQ, Cambridge, UK
Division of Cardiovascular Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI, USA
Cardiovascular Genetics, University of Utah School of Medicine, Salt Lake City, UT, USA
Department of Physiology and Biophysics, Keck School of Medicine, University of Southern California, Los Angeles, California 90033, USA
National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland 20892, USA
National Institute for Health and Welfare, 00271 Helsinki, Finland
FIMM, Institute for Molecular Medicine, Finland, Biomedicum, P.O. Box 104, 00251 Helsinki, Finland
Broad Institute, Cambridge, Massachusetts 02142, USA
Leibniz-Institute for Arteriosclerosis Research, Department of Molecular Genetics of Cardiovascular Disease, University of Münster, Münster, Germany
Medical Faculty of the Westfalian Wilhelms University Muenster, Department of Molecular Genetics of Cardiovascular Disease, University of Muenster, Muenster, Germany
Division of Cardiology, Massachusetts General Hospital, Boston, MA, USA
Georgia Prevention Institute, Department of Pediatrics, Medical Collegeof Georgia, Augusta, GA, USA
Unit of Genetic Epidemiology and Bioinformatics, Department of Epidemiology, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands
Department of Biostatical Sciences, Division of Public Health Sciences, Wake Forest University School of Medicine, Winston-Salem, NC 27157, USA
Department of Epidemiology, University of Washington, Seattle, WA, 98195, USA
Group Health Research Institute, Group Health Cooperative, Seattle, WA, USA
Seattle Epidemiologic Research and Information Center, Veterans Health Administration Office of Research & Development, Seattle, WA 98108, USA
Department of Medicine, University of Washington, 98195, USA
Department of Cardiology, University of Tartu, L. Puusepa 8, 51014 Tartu, Estonia
Tallinn University of Technology, Institute of Biomedical Engineering, Ehitajate tee 5, 19086 Tallinn, Estonia
Centre of Cardiology, North Estonia Medical Centre, Sütiste tee 19, 13419 Tallinn, Estonia
Division of Non-communicable disease Epidemiology, The London School of Hygiene and Tropical Medicine London, Keppel Street, London WC1E 7HT, UK
South Asia Network for Chronic Disease, Public Health Foundation of India, C-1/52, SDA, New Delhi 100016, India
Department of Emergency and Cardiovascular Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, 41685 Gothenburg, Sweden
Department of Biostatistics, Institute of Basic Medical Sciences, University of Oslo, 0317 Oslo, Norway
Tuscany Regional Health Agency, Florence, Italy
Tropical Medicine Research Institute, University of the West Indies, Mona, Kingston, Jamaica
University of Ibadan, Ibadan, Nigeria
Department of Genomic Medicine, and Department of Preventive Cardiology, National Cerebral and Cardiovascular Research Center, Suita, 565-8565, Japan
Department of Health Science, Shiga University of Medical Science, Otsu, 520-2192, Japan
Department of Geriatric Medicine, Osaka University Graduate School of Medicine, Suita, 565-0871, Japan
Tohoku University Graduate School of Pharmaceutical Sciences and Medicine, Sendai, 980-8578, Japan
Lifestyle-related Disease Prevention Center, Shiga University of Medical Science, Otsu, 520-2192, Japan
Department of Medical Science and Cardiorenal Medicine, Yokohama City University School of Medicine, Yokohama, 236-0004, Japan
Department of Statistics, Pontificia Universidad Catolicade Chile, Vicuña Mackena 4860, Santiago, Chile
Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Centre for Environmental Health, 85764 Neuherberg, Germany
Institute of Human Genetics, Klinikum rechts der Isar, Technical University of Munich, 81675 Munich, Germany
Institute of Epidemiology, Helmholtz Zentrum Munich, German Research Centre for Environmental Health, 85764 Neuherberg, Germany
Chair of Epidemiology, Institute of Medical Informatics, Biometry and Epidemiology, Ludwig-Maximilians-Universität, 81377 Munich, Germany
Klinikum Grosshadern, 81377 Munich, Germany
National Heart and Lung Institute, Imperial College London, London, UK, W12 0HS, UK
Medical Genetics Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA
Medical Population Genetics, Broad Institute of Harvard and MIT, 5 Cambridge Center, Cambridge MA 02142, USA
National Heart, Lung and Blood Institute and its Framingham Heart Study, 73 Mount Wayte Ave., Suite #2, Framingham, MA 01702, USA
Department of Neurology and Medicine, University of Washington, Seattle, USA
Department of Medicine (Geriatrics), University of Mississippi Medical Center, Jackson, MS, USA
Department of Neurology, Boston University School of Medicine, USA
Finnish Institute of Occupational Health, Finnish Institute of Occupational Health, Aapistie 1, 90220 Oulu, Finland
Wellcome Trust Centre for Human Genetics, University of Oxford, UK
Lapland Central Hospital, Department of Physiatrics, Box 8041, 96101 Rovaniemi, Finland
Center for Non-Communicable Diseases Karachi, Pakistan
Atherosclerosis Research Unit, Department of Medicine, Karolinska Institute, Stockholm, Sweden
Department of Internal Medicine, University Medical Center Groningen, University of Groningen, The Netherlands
Department of Medical Genetics, Erasmus Medical Center, Rotterdam, The Netherlands
Gerontology Research Center, National Institute on Aging, Baltimore, MD 21224, USA
Istituto di Neurogenetica e Neurofarmacologia, Consiglio Nazionale delle Ricerche, Cittadella Universitaria di Monserrato, Monserrato, Cagliari, Italy
Unita’ Operativa Semplice Cardiologia, Divisione di Medicina, Presidio Ospedaliero Santa Barbara, Iglesias, Italy
Computational Medicine Research Group, Institute of Clinical Medicine, University of Oulu and Biocenter Oulu, 90014 University of Oulu, Oulu, Finland
NMR Metabonomics Laboratory, Department of Biosciences, University of Eastern Finland, 70211 Kuopio, Finland
Department of Internal Medicine and Biocenter Oulu, Clinical Research Center, 90014 University of Oulu, Oulu, Finland
Institute for Molecular Medicine Finland FIMM, 00014 University of Helsinki, Helsinki, Finland
Department of Biomedical Engineering and Computational Science, School of Science and Technology, Aalto University, 00076 Aalto, Espoo, Finland
NUS Graduate School for Integrative Sciences & Engineering (NGS) Centre for Life Sciences (CeLS), Singapore, 117456, Singapore
Department of Internal Medicine B, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Institute of Pharmacology, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Institute for Community Medicine, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
U557 Institut National de la Santé et de la Recherche Médicale, U1125 Institut National de la Recherche Agronomique, Université Paris 13, Bobigny, France
Department of Neurology, Mayo Clinic, Jacksonville, FL, USA
Imperial College Cerebrovascular Unit (ICCRU), Imperial College, London, W6 8RF, UK
Faculty of Medicine, University of Split, Croatia
Department of Internal Medicine, Diabetology, and Nephrology, Medical University of Silesia, 41-800, Zabrze, Poland
Public Health Sciences section, Division of Community Health Sciences, University of Edinburgh, Medical School, Teviot Place, Edinburgh, EH8 9AG, UK
School of Science and Engineering, University of Ballarat, 3353 Ballarat, Australia
Prevention and Care of Diabetes, Department of Medicine III, Medical Faculty Carl Gustav Carus at the Technical University of Dresden, 01307 Dresden, Germany
University Hospital Münster, Internal Medicine D, Münster, Germany
Department of Medical Statistics, Epidemiology and Medical Informatics, Andrija Stampar School of Public Health, University of Zagreb, Croatia
AstraZeneca R&D, 431 83 Mölndal, Sweden
Genetic Epidemiology & Biostatistics Platform, Ontario Institute for Cancer Research, Toronto
Samuel Lunenfeld Institute for Medical Research, University of Toronto, Canada
Faculty of Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, UK
Department of Epidemiology and Public Health, Imperial College, Norfolk Place London W2 1PG, UK
Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku and the Department of Clinical Physiology, Turku University Hospital, Turku, 20521, Finland
Department of Internal Medicine, Division of Cardiovascular Medicine, University of Michigan Medical Center, Ann Arbor, Michigan, USA
Singapore Eye Research Institute, Singapore, 168751, Singapore
Department of Ophthalmology, National University of Singapore, Singapore, 119074, Singapore
Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 119074, Singapore
Duke-National University of Singapore Graduate Medical School, Singapore, 169857, Singapore
Division of Biostatistics, Washington University School of Medicine, Saint Louis, MO, 63110, USA
Department of Primary Care & Population Health, UCL, London, UK, NW3 2PF, UK
BHF Glasgow Cardiovascular Research Centre, University of Glasgow, 126 University Place, Glasgow, G12 8TA, UK
Epidemiology Public Health, UCL, London, UK, WC1E 6BT, UK
Departments of Epidemiology, Biostatistics, and Medicine, Johns Hopkins University, Baltimore MD, USA
Division of Nephrology, Department of Internal Medicine, University Medical Center Groningen, University of Groningen, The Netherlands
Division of Cardiology, Brigham and Women’s Hospital, 900 Commonwealth Avenue East, Boston MA 02215, USA
Geriatric Rehabilitation Unit, Azienda Sanitaria Firenze (ASF), Florence, Italy
National Institute for Health and Welfare, Diabetes Prevention Unit, 00271 Helsinki, Finland
Hjelt Institute, Department of Public Health, University of Helsinki, 00014 Helsinki, Finland
South Ostrobothnia Central Hospital, 60220 Seinäjoki, Finland
Red RECAVA Grupo RD06/0014/0015, Hospital UniversitarioLa Paz, 28046 Madrid, Spain
Department of Neurology, General Central Hospital, 39100 Bolzano, Italy
CIBER Epidemiología y Salud Pública, 08003 Barcelona
Department of Medicine and Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA
Geriatric Research and Education Clinical Center, Veterans Administration Medical Center, Baltimore, MD, USA
U872 Institut National de la Santé et de la Recherche Médicale, Centre de Recherche des Cordeliers, Paris, France
Institute of Physiology, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Institute of Clinical Medicine/Obstetrics and Gynecology, University of Oulu, Finland
Service of Medical Genetics, Centre Hospitalier Universitaire Vaudois, 1011 Lausanne, Switzerland
Human Genetics Center, 1200 Hermann Pressler, Suite E447 Houston, TX 77030, USA
Division of Epidemiology and Prevention, Boston University School of Medicine, Boston, MA, USA
Department of Mathematics, Boston University, Boston, MA, USA
Institute of Health Sciences, University of Oulu, BOX 5000, 90014 University of Oulu, Finland
Biocenter Oulu, University of Oulu, BOX 5000, 90014 University of Oulu, Finland
National Institute for Health and Welfare, Box 310, 90101 Oulu, Finland
MRC-HPA Centre for Environment and Health, School of Public Health, Imperial College London, Norfolk Place, London W2 1PG, UK
Centre of Medical Systems Biology (CMSB 1-2), NGI Erasmus Medical Center, Rotterdam, The Netherlands
Note added in proof: Since this manuscript was submitted, Kato et al published a BP GWAS in East Asians that identified a SNP highly correlated to the SNP we report at the NPR3-c5orf23 locus28.
Author contributions
Full author contributions and roles are listed in the Supplementary Materials Section 19.
References
- 1.Levy D, et al. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 2.Kearney PM, et al. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 3.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 4.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 5.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer TE, et al. GOSR2 Lys67Arg is associated with hypertension in whites. Am J Hypertens. 2009;22:163–168. doi: 10.1038/ajh.2008.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li N, et al. Associations between genetic variations in the FURIN gene and hypertension. BMC Med Genet. 2010;11:124. doi: 10.1186/1471-2350-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mussig K, et al. 17alpha-hydroxylase/17,20-lyase deficiency caused by a novel homozygous mutation (Y27Stop) in the cytochrome CYP17 gene. J Clin Endocrinol Metab. 2005;90:4362–4365. doi: 10.1210/jc.2005-0136. [DOI] [PubMed] [Google Scholar]
- 10.Ridker PM, et al. Rationale, design, and methodology of the Women’s Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem. 2008;54:249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 11.Burt VL, et al. Trends in the prevalence, awareness, treatment, and control of hypertension in the adult US population. Data from the health examination surveys, 1960 to 1991. Hypertension. 1995;26:60–69. doi: 10.1161/01.hyp.26.1.60. [DOI] [PubMed] [Google Scholar]
- 12.Turnbull F, et al. Effects of different regimens to lower blood pressure on major cardiovascular events in older and younger adults: meta-analysis of randomised trials. BMJ. 2008;336:1121–1123. doi: 10.1136/bmj.39548.738368.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Law MR, Morris JK, Wald NJ. Use of blood pressure lowering drugs in the prevention of cardiovascular disease: meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies. BMJ. 2009;338:b1665. doi: 10.1136/bmj.b1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis JB. Blood pressure control in chronic kidney disease: is less really more? J Am Soc Nephrol. 2010;21:1086–1092. doi: 10.1681/ASN.2010030236. [DOI] [PubMed] [Google Scholar]
- 15.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton-Cheh C, et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41:348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schenk DB, et al. Purification and subunit composition of atrial natriuretic peptide receptor. Proc Natl Acad Sci U S A. 1987;84:1521–1525. doi: 10.1073/pnas.84.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt HH, Walter U. NO at work. Cell. 1994;78:919–925. doi: 10.1016/0092-8674(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 19.Matsukawa N, et al. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci U S A. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friebe A, Mergia E, Dangel O, Lange A, Koesling D. Fatal gastrointestinal obstruction and hypertension in mice lacking nitric oxide-sensitive guanylyl cyclase. Proc Natl Acad Sci U S A. 2007;104:7699–7704. doi: 10.1073/pnas.0609778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimitsu T, Ono H, Minami J, Matsuoka H. Pathophysiologic and therapeutic implications of adrenomedullin in cardiovascular disorders. Pharmacol Ther. 2006;111:909–927. doi: 10.1016/j.pharmthera.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Pushkin A, et al. Cloning, tissue distribution, genomic organization, and functional characterization of NBC3, a new member of the sodium bicarbonate cotransporter family. J Biol Chem. 1999;274:16569–16575. doi: 10.1074/jbc.274.23.16569. [DOI] [PubMed] [Google Scholar]
- 23.Hinkes B, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 24.Feder JN, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 25.He L, Wang B, Hay EB, Nebert DW. Discovery of ZIP transporters that participate in cadmium damage to testis and kidney. Toxicology and applied pharmacology. 2009;238:250–257. doi: 10.1016/j.taap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato N, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.