Abstract
Summary
The marine intertidal zone is characterized by large variation in temperature, pH, dissolved oxygen and the supply of nutrients and food on seasonal and daily time scales. These oceanic fluctuations drive of ecological processes such as recruitment, competition and consumer-prey interactions largely via physiological mehcanisms. Thus, to understand coastal ecosystem dynamics and responses to climate change, it is crucial to understand these mechanisms.
Here we utilize transcriptome analysis of the physiological response of the mussel Mytilus californianus at different spatial scales to gain insight into these mechanisms. We used mussels inhabiting different vertical locations within Strawberry Hill on Cape Perpetua, OR and Boiler Bay on Cape Foulweather, OR to study inter- and intra-site variation of gene expression.
The results highlight two distinct gene expression signatures related to the cycling of metabolic activity and perturbations to cellular homeostasis. Intermediate spatial scales show a strong influence of oceanographic differences in food and stress environments between sites separated by ~65 km.
Together, these new insights into environmental control of gene expression may allow understanding of important physiological drivers within and across populations.
Keywords: ecological genomics, Mytilus californianus, DNA microarray, spatial variation, gene expression, environmental stress, biogeographic distribution, microarray, mussels, stress
Introduction
In mussel communities along the Oregon coast, basal gene expression patterns as well as physiological responses are likely set by the interplay of multiple, complex abiotic factors. Temperature, and more specifically body temperature, along with food availability have been shown to be two of the most important determinants of survival, growth, and reproduction (e.g., Menge et al. 2008, Helmuth et al. 2006, Halpin et al. 2004). Understanding the underlying mechanisms by which body temperature and food availability drive organismal responses and physiological performance is becoming increasingly imperative as climate change alters habitat temperature (Somero 2010). Until recently we lacked the ability to examine physiological responses at a level of complexity commensurate with that of the changing environment of an organism. Previous studies examining the mechanistic links between the ecology of an organism and its cellular responses have been largely observational, and limited in scope to a small number of abiotic or biotic factors, and the resulting changes of single bioindicators, such as heat shock proteins (Roberts, Hofmann & Somero 1997, Osovitz & Hofmann 2005, Sagarin & Somero 2006), or relatively basic proxies for physiological state, such as growth rate (Levinton & Monahan 1983, Yamahira & Conover 2002), body size (Roy & Martien 2001), reproductive output (Lonsdale & Levinton 1985, Leslie et al. 2005), and mortality (Ebert et al. 1999, Zippay & Hofmann 2010). Now, genome-wide analyses performed within the framework of an ecological context allow us to examine the interplay of multiple drivers and organismal responses (for reviews see Hofmann & Place 2007; Aubin-Horth, Letcher & Hofmann 2009).
Previous analyses of variation in transcript levels have highlighted the potential application of ecological genomics to understand physiological responses of mussel (Mytilus californianus) populations in situ (Place, O’Donnell & Hofmann 2008; Gracey et al. 2008). Given the differences in scale at which these studies were performed, ~2400 km and ~10 meters respectively, it is necessary to understand the role of spatial scales in setting gene expression patterns. In addition, Crawford and colleagues have previously shown substantial variation in gene expression both within and between natural populations of marine fish (Oleksiak, Churchill & Crawford 2002). Furthermore, for intertidal species, microclimates created by differences in wave exposure, slope substrate, and tidepools can lead to ecophysiological differences in organisms occupying similar tide heights (see Dahlhoff & Menge 1996; Helmuth & Hofmann 2000). Such cross-scale variability raises the question of whether or not inter-individual variation within local-scale M. californianus populations could overwhelm large-scale differences in gene level responses to environmental variation. If so, use of these approaches to examine the linkages between environmental variation and species responses could be limited. In this study, we examined the variation in gene expression of M. californianus across three spatial scales to answer two questions:
Micro-scale (among-individual) to small-scale (within-zone) variation: Given that variation in gene expression likely exists between individuals within a single, continuous mussel bed, are patterns of gene expression largely characteristic of vertical location within the intertidal zone or does inter-individual variation obscure potential between-tide height differences in mussels?
Intermediate to regional scales (10’s of km): Are patterns of gene expression similar between sites 65 km apart, suggesting maintenance by large-scale oceanographic processes (e.g., waves, or sea water temperature), or do patterns differ between sites, suggesting acclimation or adaptation to intermediate-scale environmental variation (e.g., differences in food, or air temperature)?
To answer these questions, we profiled the gene expression patterns of mussels inhabiting locations on two separate coastal capes that display a host of ecological and physiological differences, Boiler Bay (BB) located on Cape Foulweather and Strawberry Hill (SH) located on Cape Perpetua. For instance, BB intertidal regions generally have lower mussel recruitment, slower mussel growth, and weaker competition for space than SH (Menge et al. 1997, Menge 2004, Menge, Chan & Lubchenco 2008, Menge et al. 2009). Physiologically, BB mussels reproduce less, display a reduced cellular response to stress, and have lower growth potential than SH mussels (Dahlhoff & Menge 1996, Halpin et al. 2002, Petes et al. 2008a, b). If patterns of gene expression are largely characteristic of vertical location within the intertidal zone, we would expect them to be concordant with basic physiological responses that have been established in previous studies. Thus, we can take advantage of these sites to gain insight into the applicability of these approaches in characterizing the response of mussel populations in situ.
Materials and Methods
Tidal height measurements
Vertical elevation of each plot in the low, mid, and high intertidal zones was determined using survey methods that used the local tidal datum to ensure that mussels collected experienced similar annual submersion times. The low zone was sampled just above the mean lower low water (MLLW), which corresponds to an elevation of 0 m along the eastern North Pacific coast and where organisms are submerged approximately 90% of the time.
Mussel body temperature estimates
To monitor relative mussel body temperatures immediately prior to sample collection, mussel-mimicking temperature data loggers were constructed by embedding ibutton data loggers (Maxim Integrated Products, Sunnyvale, CA USA) into silicone-filled mussel shells. These mussel mimics were securely inserted into each mussel plot (see Helmuth & Hofmann 2000 for details). Approximate mussel body temperature was recorded at 10 minute intervals for 4 days just prior to sample collection.
Chlorophyll-a measurements
Methods for quantification of abundance of phytoplankton (as proxied by chlorophyll-a = Chl-a), a primary food of mussels, followed those summarized in Menge et al. (2008). We collected bottle samples from the shore at each site by filling replicate (n = 3 to 5) acid-washed opaque plastic 250-mL bottles (HDP) at a depth of 30–50 cm below the surface during low tide. Fifty mL of water were filtered through 25-mm pre-combusted Whatman GF/F glass-fiber filters with a pore size of 0.7 mm. Chl-a concentration was determined using a Turner Designs TD-700 fluorometer after extraction in 90% HPLC acetone for 12 h in the dark at –20°C. Prior calibrations of the fluorometer were done using pure Chl-a standards. Samples were taken daily to monthly, both before and during the study period (April through September).
Sample collection
Mussels were sampled on successive mid-day low tides (July 14–15, 2008) by cutting the attaching byssal threads with a scalpel. To investigate the effects of intra-site variation at the same and different elevations at SH, we initially performed horizontal transects at three vertical heights (referred to as SH-low, SH-mid, and SH-high) within a single wave exposed mussel bed. We randomly collected 10 mussels from five replicate plots along the low, mid, and high intertidal transects (see Fig. S1 in Supporting Information). Plots were horizontally spaced at 3 m intervals. Individuals not in immediate contact with each other and with a shell length between 50 – 60 mm were otherwise randomly chosen from each plot (~ 0.5 m × 0.5 m). Plots with mussels inhabiting tide pools or vertical walls were excluded as these microhabitats may impact feeding habits and create a refuge from heat as the angle of incidence of incoming solar radiation is known to directly affect the organism’s thermal properties (Dahlhoff & Menge 1996, Helmuth & Hofmann 2000, Denny, Miller & Harley 2006).
To evaluate inter-site variation, horizontal transects were also performed within a single wave-exposed mussel bed at BB during the next low tide series. Five replicate plots separated by 3 m were sampled within the middle and along the upper edge of a continous mussel bed at BB. Initially, we only intended to perform a comparison of mussels from the mid-intertidal zone from each site; however, upon surveying exact tidal height we determined the SH-mid plots were more similar in vertical displacement to the upper edge of the mussel bed within BB (see Table S1 in Supporting Information). Therefore, we sampled both the mid-intertidal zone of the mussel bed, referred to henceforth as BB-mid, and at a tidal height near the upper edge of the mussel bed more closely corresponding to tidal height of SH-mid plots, referred to as BB-high.
To test the effects of emersion stress, ten individuals from each plot were removed from the mussel bed by carefully cutting the attaching byssal threads from the substrate with a scalpel just prior to being inundated by the afternoon high tide. For 5 individuals, gill tissue was immediately excised and flash frozen on dry ice. The remaining 5 individuals were placed in a 20-gallon bucket containing seawater maintained at ambient temperature (~14° C) and O2 saturation by repeated water changes. Mussels were allowed to recover from the emersion stress for 1h. Following the 1h recovery period, gill tissue was excised and flash frozen on dry ice. All samples were maintained on dry ice and transported to UCSB where they were stored at −80 °C until used for analysis.
Comparison of Gene Expression Profiles
To determine if variation in gene expression within a single tide height was smaller than variation across tide heights, we characterized the gene expression profile of fifteen individual samples at each tide height (3 mussels per plot, 5 plots per tide height) from Strawberry Hill using a custom microarray (described below). Three of the 5 mussels sampled from each plot were randomly selected for transcriptome analysis to reduce the total number of arrays needed for this portion of the analysis.
For the comparison between Boiler Bay and Strawberry Hill, RNA was pooled from the 5 biological replicates sampled from within each plot for BB-mid and BB-high, and reverse transcribed for competitive hybridization and compared to the mean log ratio of SH-mid samples (n=5 arrays per tidal height per treatment).
All gene expression profiles were quantified with a 4,992-feature cDNA custom microarray. Microarrays were constructed from a cDNA library prepared from different tissues (gill, adductor muscle, and mantle) of adult M. californianus mussels exposed to a variety of stressors (heat, cold, emersion, hypoxia, hypo-osmolality, cadmium, and low pH) to induce a mRNA pool enriched for as many stress responsive genes as possible. For all competitive hybridizations, total RNA was extracted from the gill tissue of 30 individuals collected from a population of M. californianus mussels in Jalama Beach near Santa Barbara, California. These mussels were lab-acclimated at 14°C for 3 weeks but otherwise untreated, and pooled for use as a source of reference RNA. The use of non-experimentally treated RNA reference samples is commonplace in gene expression analysis and reduces the overall number of arrays needed to perform the analyses while allowing direct comparison of expression values across samples (see Podrabsky & Somero 2004, Place et al. 2008). Putative gene clusters were annotated for identity using NCBI BLAST against the UniRef 90 protein database with an evalue ≤ 10−4.
RNA was extracted from approximately 100 mg of frozen tissue using TRIzol® (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s recommendations. Ten µg of total RNA was reverse transcribed (RT) to cDNA using anchored oligo(dT15) primers and amino-allyl dUTP, fluorescently labeled and hybridized to a cDNA microarray as previously described in Place et al. (2008). Microarrays were printed and a 9-mer Cy3 labeled spot QC and red reflection spot QC were performed at the Oregon State University Center for Genome Research and Biocomputing. All microarrays were stored in a light-protected desiccation cabinet until use. The microarrays were scanned on an AXON GenePix 4000B microarray scanner (Axon Instruments, Molecular Devices, Sunnyvale, CA, USA). Spot intensity data were extracted using GenePix Pro 4.0 software and the ratio of Cy5 to Cy3 fluorescence was quantified for each spot on the arrays (Axon Instruments).
The data discussed in this publication along with microarray printing and QC procedures have been deposited in NCBIs Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), and are accessible through GEO Series accession number GSE21088.
Normalization and statistical analysis of microarray data
For each array, spatial and intensity-based trends in the data were removed by Lowess normalization of fluorescence data using Genespring GX 10.0 software (Agilent). This removes data artifacts introduced by slightly different biochemical properties associated with the dyes (see Draghici 2003). Features that were not represented by a minimum spot intensity in at least 75 % of the samples were excluded from further consideration. In addition, only those features that displayed a signal to noise ratio (SNR) of ≥ 3 were included in further analysis.
Principal component analysis (PCA) was performed on the combined mean log2 ratio (635/532) for biological replicates sampled within a single plot using Genespring GX 10.0. This allowed us to determine the cumulative separation between sites, vertical position on the shore, and recovery, as a function of numerous gene level responses (n= 5 arrays/ tidal height/ treatment). The results of the PCA were projected as a loading plot to illustrate the degree of similarity between transcriptomes.
To assess the impacts of tidal height on gene expression during and after an emersion stress within a site, we used non-parametric significance tests (Mann–Whitney–Wilcoxon) to compare the median log2 ratio (635/532) of paired features from individual mussels taken from SH-low, SH-mid, and SH-high plots, and identified features whose expression displayed significant changes in expression in one or more of the tide heights. All p-values were adjusted with the Benjamini and Hochberg false discovery rate (FDR) correction for multiple testing (FDR = 0.25) and were considered significant if p < 0.05.
As we were interested in examining the interaction of geographic location and vertical position on the shore, we next performed a two-way ANOVA with collection site and tide height as fixed variables. As before, p-values were adjusted with the Benjamini and Hochberg method (FDR = 0.25). We then performed k-means clustering analysis on the log2 ratio data for all genes displaying a significant change in gene expression for tide height or 1h recovery (FDR-corrected p < 0.05). k-means clustering was performed using the Pearson uncentered distance metric within the GeneSpring GX 10.0 gene expression analysis software package.
Biological function of the gene sets described by the clusters identified though the k-means analyses were further characterized by over-representation analysis (ORA) in ErmineJ software package (Lee et al. 2005). Parameters for the ORA were as follows: minimum gene set size =5, maximum gene set size =200, FDR correction Benjamini-Hochberg method. FDR-corrected p-values < 0.1 were considered significant in accordance with recommended practices (Lee et al. 2005).
Results
Tidal height surveys
The mean tidal height at which mussels were sampled from within Boiler Bay (BB) and Strawberry Hill (SH) mussel beds as determined by surveys of elevation were as follows: BB-mid (1.44 ±0.59 m), BB-high (1.68 ±0.69 m), SH-low (1.00 ±0.42 m), SH-mid (1.75 ±0.72 m), and SH-high (2.07 ±0.85 m) above MLLW. Elevations for individual plots are reported in the supplementary materials (Table S1).
Body temperature estimates
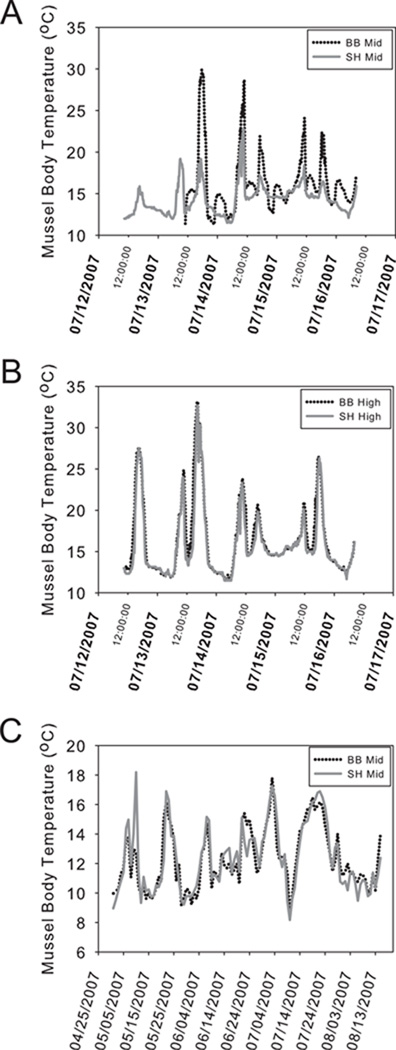
Due to loss of mussel data loggers after the initial deployment at BB and SH, we are unable to provide comparisons for the entire week prior to collection. However, we were able to collect detailed overlapping temperature estimates for mussel body temperature for five days during the experiment. For the five days in which we can directly compare body temperature estimates, BB-mid mussels displayed elevated daily mean body temperature estimates, 15.52–17.09 °C, compared to SH-mid, 13.33–15.10 °C (Fig. 1A). BB-mid mussels also show greater temperature variance during this period (Fig. 1A). Daily minimum temperatures (set by sea surface temperatures) were similar at both sites, ranging between 11–14 °C. But daily maxima were higher at BB-mid, with daily maxima routinely recorded above 20 °C, while daily maximum temperatures recorded for SH-mid rarely reached above 20 °C (Fig. 1A). Alternatively, mussels inhabiting the high intertidal zone displayed more similar daily maximal body temperatures despite differences in vertical location on the shore (Fig. 1B). Seasonally, mussel body temperatures in the mid-intertidal zone of BB and SH are similar with respect to maximal daily temperatures, suggesting differences in physiological responses to temperature changes would be unlikely to result from simple differences in thermal acclimatization for mussels located along these two capes (Fig. 1C). Daily mean, maximum and minimum temperatures for the mid- and high intertidal zone at each site are reported in the supplementary material (see Table S2 in Supporting Information).
Figure 1.
Temperature traces of the estimated body temperature experienced by mussels located in the mid intertidal zone (A) or high intertidal zone (B) at Boiler Bay (Black dotted line) and Strawberry Hill (grey solid line) for the week during the low tide series during which mussels were sampled. (C) Estimated daily maximal body temperature experienced by mussels located in the mid intertidal both before and during the study period (April through August).
Chlorophyll-a measurements
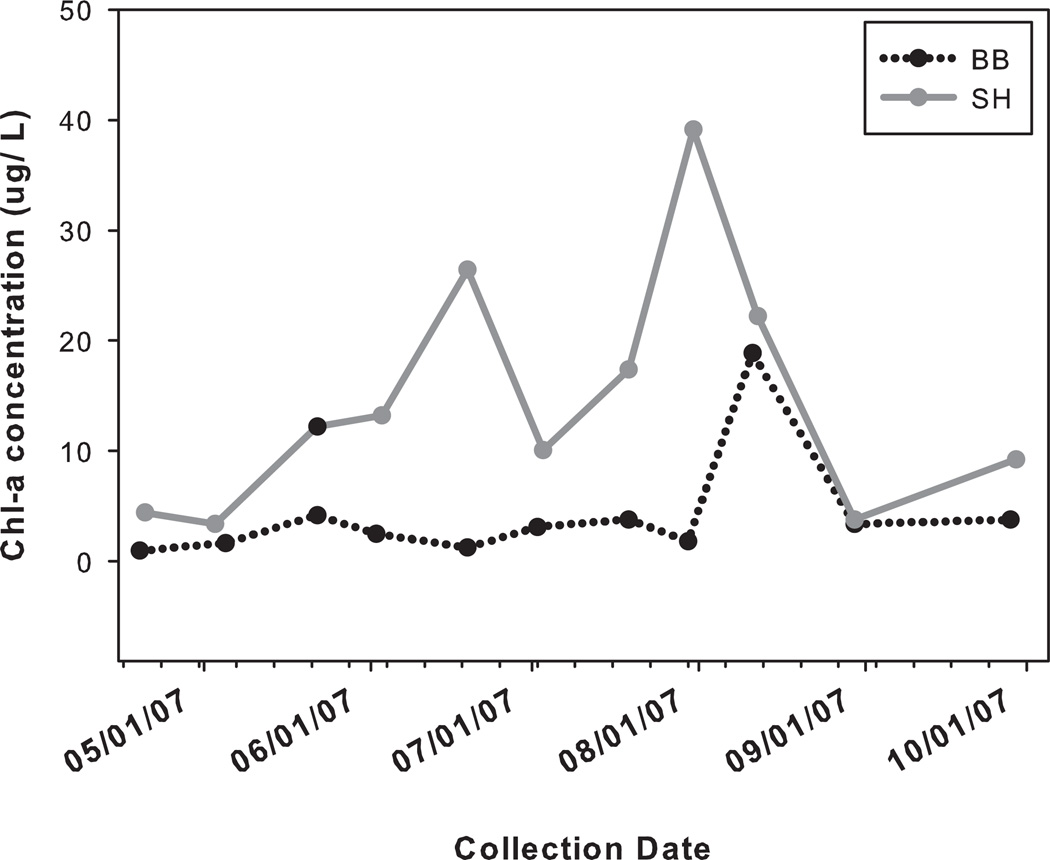
Bottle samples taken from the shore at each site show markedly higher levels of Chl-a in the upper water column at SH when compared to BB (Fig. 2). From May through August, Chl-a concentrations at SH are at least 2-fold higher than at BB, with an increase to nearly 20-fold in mid June (Fig. 2). Chl-a levels were at least 10-fold higher at SH for the month preceding our collection (Fig. 2)
Figure 2.
Chlorophyll-a concentrations taken via bottle sampling performed at Boiler Bay, OR (Black dotted line) and Strawberry Hill, OR (grey solid line). Samples were taken daily to monthly, both before and during the study period (April through September).
Inter-site comparisons of variation in gene expression
To gain a measure of the global variation in gene expression among mussels that are 1) spatially separated within a single mussel bed and 2) experiencing and recovering from mid-day aerial exposure, we performed a principal component analysis (PCA) of the gene level responses in mussels from a single mussel bed with or without a 1h recovery period in ambient seawater (Fig. 3). As expected, mussels inhabiting different vertical heights along the shore show strong variation in their gene level responses during both aerial exposure and after a 1h recovery, specifically with respect to the first principal component (PC1). Annotation of the top 100 features whose expression patterns most contributed to PC1 identified several genes involved in metabolic processes (Fig. 3, see Table S3 in Supporting Information). In contrast, no significant variation in gene expression patterns was detected among mussels inhabiting the same level on the shore during an emersion event even if separated horizontally by as much as 15 meters (Fig. 3). Only SH-high plots given a 1h recovery displayed any variation in their gene level responses with respect to horizontal separation along the shore (Fig. 3, open squares). No discrete shifts in gene expression were detected between SH-low mussels given a 1h recovery in ambient seawater and those sampled immediately after an emersion event (Fig. 3, circles). During a mid-day emersion event, SH-mid and SH-high mussels displayed similar gene expression patterns that deviated from SH-low mussels with respect to PC1 (Fig. 3). After a 1h recovery period, gene expression patterns of SH-mid mussels and 2 of the 5 plots for SH-high mussels were no longer distinguishable from those displayed by SH-low mussels (Fig. 3). Furthermore, for 3 of the 5 SH-high plots sampled, a shift in gene expression patterns with respect to both PC1 and PC2 had occurred after the 1h recovery, resulting in a distinct clustering of these mussels (Fig. 3, open squares).
Figure 3.
Principal component analysis of the cumulative separation in gene expression between mussels within a single mussel bed located at Strawberry Hill, OR. Mussels were sampled from three separate vertical positions on the shore (low – circles, mid – triangles, high – squares). Mussels were sampled during daily emersion (closed symbols) or after a 1h recovery in ambient seawater (open symbols). Plot ordination was performed for the first 2 component loadings - Component 1: x-axis (80.01% of the variation); Component 2: y-axis (12.23% of the variation).
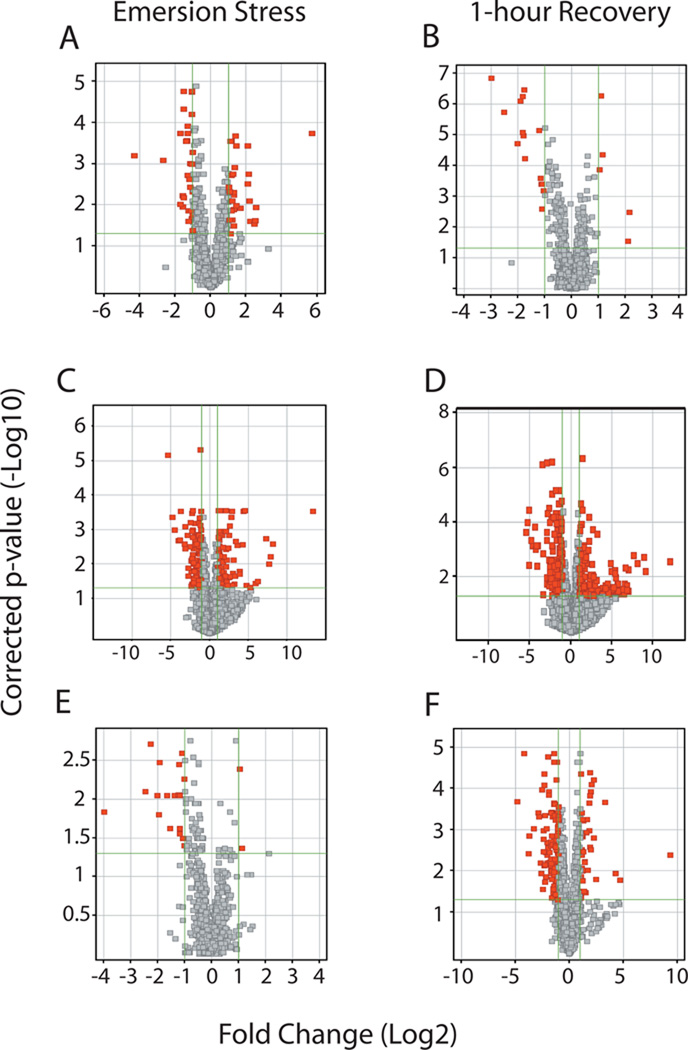
To identify genes that showed significant changes in gene expression, we used non-parametric significance tests to compare the median log2 ratio (635/532) of paired features from individual mussels taken from SH-low, SH-mid, and SH-high plots. Volcano plots were constructed by plotting the negative log10 of the p-value against the log2 of the fold change between the two conditions under consideration (low/mid, low/high, or mid/high). Highlighted data points represent the genes meeting the applied cutoffs (fold change ≥ 2, FDR-corrected p < 0.05) used to identify both biologically (magnitude) and statistically significant changes for each comparison (Fig 4A–F).
Figure 4.
Volcano plots showing the pair-wise comparison of the differential expression of genes between mussels at three separate vertical positions on the shore during an emersion stress or after a 1h recovery in ambient seawater. The red points indicate genes-of-interest that display both large-magnitude fold-changes (FC ≥ 2, x-axis) as well as high statistical significance (p < 0.05, y-axis). Emersion stress: (A) SH-low vs. SH-mid, (C) SH-low vs. SH-high, (E) SH-mid vs. SH-high. 1h recovery: (B) SH-low vs. SH-mid, (D) SH-low vs. SH-high, (F) SH-mid vs. SH-high.
Of the nearly 400 genes that showed differences in gene expression between tide heights (MWW, FDR-corrected p < 0.05), 68 genes from the mid-intertidal population and 167 genes from the high intertidal population displayed at least a 2-fold change in expression relative to the median log2 ratio (635/532) of low intertidal populations within the SH mussel bed (Fig. 4A & 4C, respectively). Classification of these genes by their biological function listed in the Gene Ontology (GO) database verifies the trends in the PCA (Fig. 3), with several genes associated with metabolic processes [Aldehyde dehydrogenase (ALDH), Acetyl-coenzyme A synthetase (ACSA), Low-density lipoprotein receptor-related protein1 (LRP1), Ubiquinol-cytochrome C reductase (Uqcrfs1)] being down-regulated in both SH-mid and SH-high mussels (see Table S4 in Supporting Information). Subsequent to a 1h recovery period in ambient seawater, the number of different genes between the SH-mid and SH-low mussels dropped to only 19 (Fig. 4B). In contrast, the difference between SH-high and SH-low mussels widened, with the number of differentially expressed genes increasing to 336, with a strong up-regulation of multiple stress response genes. Included among these genes were Heat shock cognate 71(Hsc71), Heat shock protein 90-alpha (Hsp90-α), Metallothionein 20-III isoform B (MT-20 IIIb), Ubiquitin (Ub), and stress-induced-phosphoprotein-1 (STI3) (Fig. 4D, Table S4). A similar pattern was seen with respect to SH-mid compared to SH-high intertidal mussels, with the number of genes increasing from 28 during an emersion stress to 103 genes identified after a 1h recovery (Figs. 4E & 4F, respectively).
Intra-site comparisons of variation in gene expression
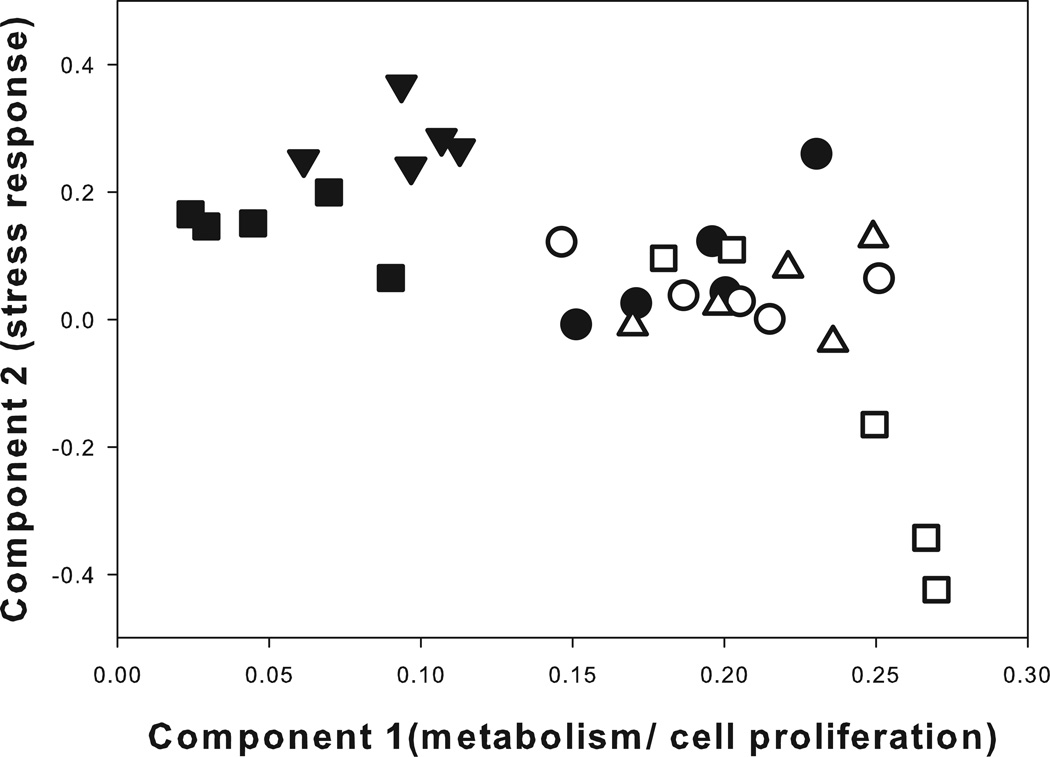
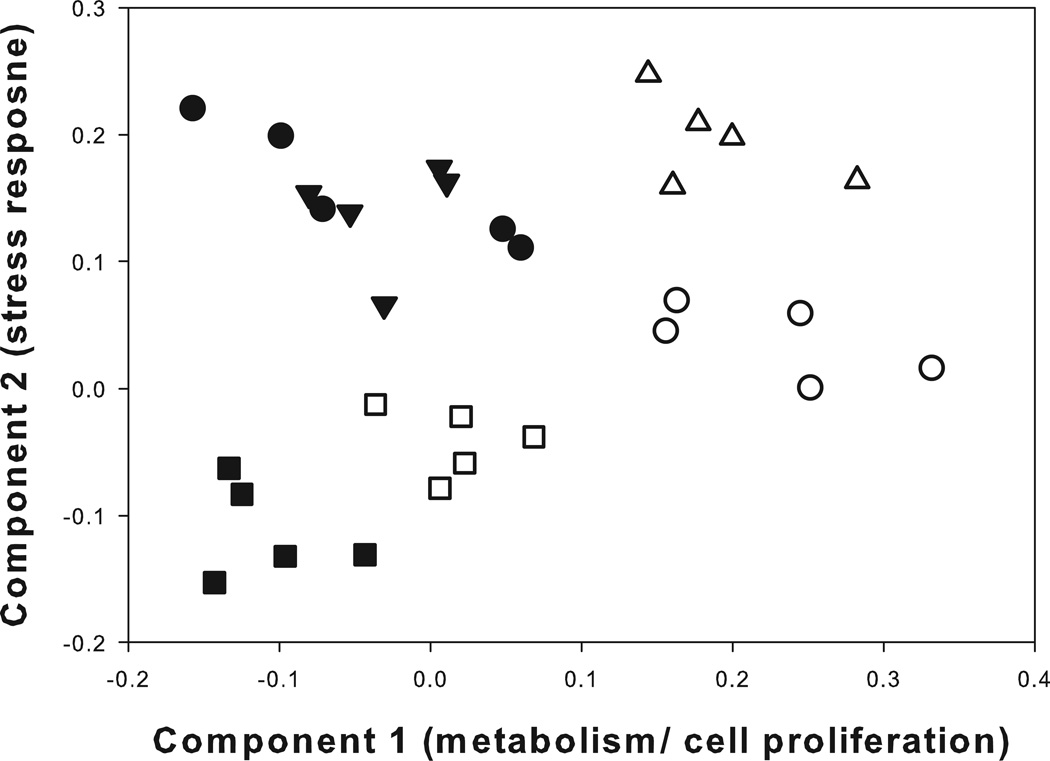
To understand the extent of separation in physiological responses of mussels inhabiting two biogeographically distinct capes, we used PCA to gauge the cumulative variation of the gene level responses. PCA was performed on the gene expression patterns of mussels sampled at 5 different locations along horizontal transects, either immediately prior to inundation by the rising tide or 1h after re-immersion in ambient seawater. The clustering of individuals from within a site is indicative of distinctive gene expression patterns between sites (Fig. 5). Similar to the pattern of gene expression in SH-mid mussels illustrated in the previous PCA plot (Fig. 3), these mussels showed a distinctive shift in gene expression when recovering from emersion with respect to PC1 (Fig. 5). Similarly, mussels sampled from plots located within the mid- and high intertidal zone for BB showed a strong shift with respect to PC1 (Fig. 5). Reinforcing what we observed within the mussel population at SH, mussels sampled from BB-mid and BB-high intertidal plots showed strong similarity in their gene expression signatures during the emersion event (Fig. 5). However, within 1h of recovery at ambient seawater temperatures, the signatures of BB-mid and BB-high mussels were clearly distinguished from one another with respect to PC2 (Fig. 5). Investigation of the top 100 features contributing to PC2 revealed a number of genes with important roles in stress response (see Table S5 in Supporting Information). Although SH-mid intertidal mussels were collected from similar elevations as the BB-high plots, and the same relative location within the mussel bed as the BB-mid plots, the SH-mid mussels showed distinct differences in their gene expression patterns based largely on the magnitude of the gene expression event. In fact, the gene expression patterns of mussels located within BB-high plots were more closely aligned to BB-mid plots despite occupying the same vertical position on the shore as mussels taken from SH-mid plots (Fig. 5).
Figure 5.
Principal component analysis of the cumulative separation in gene expression between mussel populations located at Strawberry Hill and Boiler Bay OR. Mussels were sampled from the mid intertidal zone at SH (squares) and two separate vertical positions on the shore at BB (mid – triangles, high – circles). Mussels were sampled during daily emersion (closed symbols) or after a 1h recovery in ambient seawater (open symbols). Plot ordination was performed for the first 2 component loadings - Component 1: x-axis (35.13% of the variation); Component 2: y-axis (30.87% of the variation).
Pairwise comparison of BB-mid to SH-mid plots identified 102 differentially expressed features during the emersion event and 188 differentially expressed features after a 1h recovery in ambient seawater (see Fig. S2 & Table S6 in Supporting Information). Consistent with the lower body temperatures recorded in SH-mid mussels (Fig. 1A), examination of the biological processes associated with the genes differentially expressed after a 1h recovery revealed a general down-regulation of stress response genes, such as Hsc71, 78 kDa glucose-regulated protein (Grp78), and Ub, in SH-mid mussels (Table S6). In addition, SH-mid mussels displayed an up-regulation of metabolic genes, such as ALDH and LRP1, compared to BB-mid mussels (Table S6), consistent with the increased food availability at SH (Fig. 2).
Gene interactions and functional classification
In all, 648 features displayed significant changes in gene expression in response to at least one of the two parameters tested (Collection site & Tide-height). The relationships and interaction terms of genes identified within each condition are illustrated in Figure S3 (see Fig. S3 in Supporting Information). Of 648 features identified, most features (451) varied with geographic location, 167 features varied with vertical location within the two mussel beds, and variation of only 30 features was context-dependent (interaction terms were significant; Fig. S3). Of the 30 features displaying an interaction, two-thirds were found to overlap with the gene list for collection site (7), tidal height (5), or both (8) (Fig. S3).
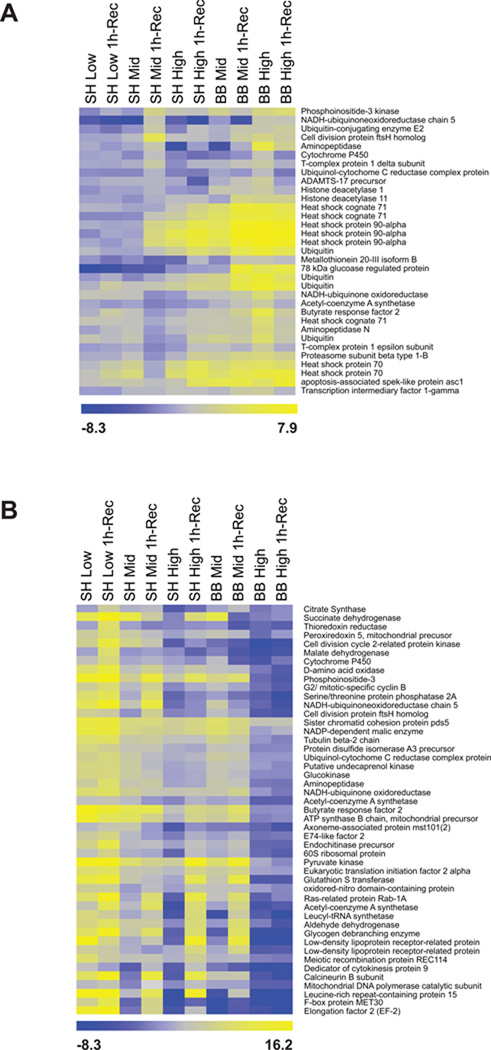
K-means clustering of the 648 features identified in the variance analysis described above produced 8 gene set clusters that displayed similar characteristics of gene expression (data not shown). Functional characterization of these 8 gene sets by over-representation analysis (ORA) revealed key functional categories that were enriched in only 2 of the 8 clusters. Six of the clusters were functionally unclassifiable, potentially due to the large number of unknown features. Cluster A was enriched for protein folding (GO:0006457, FDR-corrected p = 0.0463) while cluster C was enriched for metabolic processes (GO:0008152, FDR-corrected p = 0.0814). Annotated heat maps of the genes for which function could be assigned highlight the clustering and normalized expression of gene sets with enrichment for GO biological process classification (Fig. 6). The genes that show the largest difference in response to emersion stress are members of the molecular chaperone family Hsc71, Hsp70, and Hsp90-α (Fig. 6A). Even though mussel body temperatures from the high intertidal plots were similar between BB and SH during the mid-day exposure (Fig. 1B, Table S1), we saw an exaggerated response in genes associated with cellular stress in cluster A (Fig. 6A). Consistent with previous trends, expressions of genes associated with metabolism appear suppressed during emersion, especially in the high intertidal plots where the down-regulation of these genes appeared to continue into the first hour of re-immersion in ambient seawater (Fig. 6B). Lastly, the level of expression for genes associated with cell cycle control, G2/ mitotic-specific cyclin B, Cell division cycle 2-related protein kinase, and cell division protein ftsH homolog remain low even in mid-intertidal mussels in which up-regulation of metabolic genes has resumed (Fig. 6B).
Figure 6.
Annotated heat maps showing the relative expression (down-regulated [blue]; up-regulated [yellow]) of gene sets identified in the k-means clustering analysis with overrepresentation of genes linked to a specific Gene Ontology (GO) biological classification. Over-representation analysis of the k-means clustering results identified to key functional categories that were significantly overrepresented: (A) protein folding (GO:0006457, FDR corrected p = 0.0463) and (B) metabolic processes (GO:0008152, FDR corrected p = 0.0814).
Discussion
In our analysis of gene level responses of M. californianus, we identified two trends in biological function that offer insight into the mechanisms linking the environment to organismal response at the population level. First, significant variation of the transcriptome can be attributed to changes in genes involved in primary metabolic pathways. For mussels inhabiting different elevations within the rocky intertidal, both within a single continuous mussel bed as well as populations that span biogeographically distinct capes, metabolic processes were associated with the principal component that accounts for the greatest fraction of variation in gene expression. Temporally, mussels inhabiting two different capes separated by ~65 km display remarkably similar metabolic responses despite occupying different tide heights on shore. Differences between them were primarily in the magnitude of responses, rather than in the pattern of responses. These data suggest that oceanographic level processes can link physiological responses across intermediate scales and may highlight the important role that metabolic state may play in the response of these populations. Second, expression of genes involved in the highly conserved cellular stress response (CSR), a coordinated set of genomic responses that protect the cell against environmental stress (Kültz 2003; 2005), consistently differentiated mussels across both small and intermediate spatial scales, and may have secondary impacts on the oscillation of the expression of genes associated with metabolic pathways observed in these populations.
Our results suggest that within a mussel bed, vertical position on the shore may be the dominant determinant of gene expression patterns, and potentially physiological responses. This supports our hypothesis that gene expression patterns are more characteristic of absolute rather than effective tide-height. Although microhabitat impacts individual physiological traits and can result in “effective tide height”, or the stimulation of responses that are associated with absolute elevation, (Okamura 1992, Williams & Somero 1996, Helmuth & Hofmann 2000, Halpin et al. 2002), our data suggest vertical height on the shore provides a strong metric for predicting local organismal responses and limits to changing climates.
When the latitudinal separation is increased from meters to tens of kilometers, our data suggest the dominant drivers of variation at the level of the transcript shifts from vertical tide-height to oceanographic processes varying over intermediate spatial scales. Our expression profiles identified over 600 features that differed in the relative level of expression when collection site or tide height was considered. Collection site alone accounted for most of the features showing at least a 2-fold change in expression. In addition, analysis of the expression profiles of mussels sampled from plots at both SH and BB show stronger similarities in expression signatures for mussels taken within the same site, even when sampled from different tide heights. Hence, strong physical linkages formed between mussel populations and intermediate-scale oceanographic processes may overwhelm local-scale differences in tidal elevation. These findings only partially support our second hypothesis that gene expression patterns are set by large-scale oceanographic processes, as it appears that the mechanism by which these processes act are themselves dependent on scale.
Food availability and metabolism
Historically, phytoplankton abundance has been lower at BB compared to SH (Menge et al 1997, Menge et al. 2009), a trend that held during our study. In addition to the underlying oceanographic differences resulting in lower food availability at BB, local variation across smaller spatial scales can also impact availability and utilization of resources. Among other factors, vertical location within the intertidal zone significantly impacts the window of time sessile organisms have to feed and thus may be a strong environmental driver of the physiological response of mussels within a population. From an ecological perspective, food availability at a given geographic location, in combination with the feeding window has long been implicated in differences in growth rates and reproductive output (Wingfield & Sapolsky 2003, Petes et al. 2008a). Feeding state also influences circadian rhythms (Sonoda et al. 2007) and thus has been proposed as a mechanism that may act to gate the metabolic response of an intertidal organism subjected to daily tidal variation (Gracey et al. 2008). Therefore, if the magnitude and frequency of transcriptional variance is indeed linked to environmental parameters, we would expect the variation in metabolic responses be to be reflected both in food availability, and the feeding window of the mussels as a function of elevation within the intertidal zone. Indeed, when we investigate the metabolic response of mussels inhabiting different elevations as a proxy for feeding window, we find qualitative responses of metabolic genes that are largely reflective of this scenario.
At the level of the transcript, low intertidal mussels altered their physiology very little with respect to the tide cycle. Thus, although these mussels were emersed during daylight spring tides, the frequency and duration of exposure evidently was not sufficient to result in meaningful variation in gene expression patterns. Although we only tracked gene expression changes that occurred with the first hour of re-submergence and cannot rule out the presence of longer-term variation, these data are consistent with the lack of transcriptional changes reported for M. californianus inhabiting the low intertidal zone of Monterey Bay over the course of a 3-day tide series (Gracey et al. 2008).
For mussels inhabiting the mid- and high intertidal zone, a reduction in the expression of a cluster of genes involved in metabolic processes such as pyruvate kinase, ATP synthase β chain, LDL receptor-related protein 1 and acetyl-coA synthetase suggests that their metabolic capacity was suppressed during emersion. For SH-mid mussels, the relative expression of this gene cluster reached levels comparable to those seen in low intertidal mussels within an hour of re-immersion. Although the links between gene expression changes and functional responses need to be further verified in this system, these gene expression patterns suggest that SH-mid mussels quickly resume normal respiration despite reducing metabolic capacity during an emersion event.
As seen with the initial intra-site analysis, inspection of the top 100 features from PC1 of the inter-site PCA revealed several genes whose biological function was associated with metabolic activities. Unlike the gene expression signatures seen in SH, mussels sampled from BB-mid and BB-high plots yielded similar metabolic responses both prior to, and 1h after re-immersion. Furthermore, when the gene level responses of mussels from BB are compared to mussels sampled from the same tidal height in SH, the expression patterns display similar qualitative responses but the magnitude of the response was exaggerated in mussels sampled from BB. This variation may be evidence of direct links between metabolic state and physiological perception of environmental differences in food availability at intermediate scales.
Dahlhoff and Menge (1996) suggested that the effects of stress might be amplified in organisms if the occurrence of abiotic stress coincides with times of low productivity. Two more recent studies provide further empirical evidence of this interaction. In bivalves, hypoxia alone was found to have a weak effect on RNA, scope for growth, and morphometric indices (Norkko et al. 2005). However, when food availability was reduced, the impact of hypoxia was amplified (Norkko et al. 2005). Similar interactions were reported in a study assessing the effects of food availability and elevated body temperature on the survival of two Mytilus congeners (Schneider, Van Thiel & Helmuth 2010). In this study, Schneider et al. (2010) reported a 24% difference in survival at 30 °C between mussels given a low or high food treatment. These findings are similar to ours for the potential role of food availability in setting gene expression patterns at different scales. Our data suggest the variation in gene expression of metabolic genes is likely driven in large part by phytoplankton abundance at intermediate scales, in addition to feeding windows at different tidal heights within a site. Taken together, these data highlight the different mechanisms by which environmental parameters can act, and continue to underscore the need to understand physiological responses across multiple scales.
Temperature and thermal windows of performance
In their consideration of thermal tolerance windows, Friedrich and Pörtner (2000) discuss three distinct physiological transitions that may define the progression of temperature impacts in ectothermic organisms. The first transition that occurs has been termed the ‘pejus’ temperature (Tp) and is marked by the onset of oxygen limitations in aerobic scope, which is followed by passive tolerance tactics induced by a critical temperature (Tc). The last transition, brought on by exposure to denaturing temperatures (Td), is characterized by loss of structural integrity of macromolecules and the induction of protective mechanisms such as the CSR (Frederich & Pörtner 2000). The relative small number of gene expression changes observed in SH-low mussels during an extreme low tide would suggest that body temperatures rarely exceed the thermal windows for optimal growth and performance in these mussels. In contrast, during low tide emersion, gene expression patterns observed in mussels sampled from the mid- and high intertidal plots may be an early indicator that the Tp threshold has been reached. Lastly, during the first hour of re-immersion, we see induction of pathways associated with protein rescue, cellular repair, and protein degradation, in addition to genes involved in oxidative stress pathways in a majority of SH-high mussels. This may be an indication that a large number of SH-high mussels may cross the Td threshold during low tide emersion and incur significant energetic debts due to macromolecular damage.
Gene expression patterns also appear to be strongly influenced by temperature across intermediate scales, but mussels sampled from BB did not show the strong influence of zonation on thermal windows as was seen at SH. Unlike SH-mid mussels that display gene expression patterns that would suggest they remain below the Td threshold, both BB-mid and BB-high mussels display gene expression patterns that may indicate that they have surpassed the Td threshold. This, despite the fact that they were existing at vertical elevations equivalent to, and in some cases lower than, mussels within the mid-intertidal zone of SH. The increased solar heating seen in BB-mid mussels is likely the primary factor underlying the clear distinction between the gene expression signatures of mussels sampled from SH-mid and BB-mid plots with respect to stress response genes. However, the expression patterns seen in BB-high mussels are not as easily explained. Despite experiencing similar body temperatures during the mid-day exposure, BB-high mussels exhibit a more pronounced stress response compared to SH-high mussels. These data suggest the capacity for maintaining cellular homeostasis may be reduced in BB populations and that growth and fitness could be compromised for extended periods even at moderate elevations.
In addition to thermal insults, reactive oxygen species (ROS) also appear to be important modulators of cellular responses during the first hour of recovery from aerial exposure. A host of organisms show metabolic depression during cellular stress events and it has been postulated by some that this metabolic depression is related to an attempt to reduce the accumulation of ROS (see Lesser 2006 for review). Production of ROS have been implicated in the transcriptional regulation of CYP isoforms and may serve as negative feedback regulators of cytochrome P450 (Barouki & Morel 2001). During aerial emersion, we observed down-regulation of genes involved in aerobic respiration that was accompanied by a nearly 15-fold lower expression level of cytochrome P450 mRNA in SH-high mussels relative to SH-low mussels. Furthermore, within an hour of re-immersion, SH-high mussels showed up-regulation of the antioxidant genes, Perodoxin 5 and glutathione S-transferase. These data are consistent with reported high levels of carotenoids, believed to play an important role in mitigating oxidative stress in high intertidal mussels (Petes et al. 2008a). More recently, transcriptomic analysis of the thermal stress response of lab-acclimated Mytilus congeners has described similar patterns of simultaneous down-regulation of metabolic genes and up-regulation of oxidative stress genes (Lockwood, Sanders & Somero 2010). Parallel proteomic analysis of these mussels verifies similar functional changes at the level of the protein, supporting the link between changes at the level of transcript and physiological changes at the level of the organism (Tomanek & Zuzow 2010). Taken together, these data provide further support that SH-high mussels demonstrate cellular responses consistent with an organism that has crossed the Td threshold.
Conclusions
The complex linkages between physiological responses and environmental variation present a major challenge to understanding the consequences of global climate change for species. In this study we have provided an initial glimpse into some of these complex interactions, raising the possibility that the mechanisms by which food availability and temperature work to set gene expression patterns can differ at small and intermediate scales. In addition, our results highlight potentially compounding effects of temperature and food availability, pointing to a mechanism by which biotic and abiotic variables may work together to lower physiological limits and set geographic boundaries. For instance, the induction of stress response pathways may come at a substantial energetic cost to other biological functions, and that gene expression may result in a trade-off between increased fitness and growth rate (e.g., Lang, Murray & Botstein 2009). If this energetic cost is accrued within a low or poor nutrient environment, an organism may lack sufficient energetic reserves to mount a proper cellular defense, resulting in a magnification of the insult. As a result of these compounding effects, mussel populations inhabiting waters characterized by low productivity may be living closer to their thermal physiological limits than previously estimated. These results could have important implications for future efforts aimed at predicting the impact of climate change on the connectivity of populations across large biogeographic scales where distinct oceanographic features work to constrain range limits through physiological tolerance.
Supplementary Material
Acknowledgements
We thank all the members of the Hofmann and Menge Lab groups for assistance with sample collection, especially Dr. Michael O’Donnell for his contributions to field collections. Dr. Francis Chan oversaw collection of the phytoplankton samples and provided the data for Fig. 2. The authors acknowledge the U.S. National Science Foundation for financial support during the course of this writing project (NSF grants OCE-0425107 to G.E.H. and OCE-0726983 to B.A.M.). S.P.P. was supported by a U.S. National Institutes of Health NIH-NRSA Postdoctoral Fellowship during a portion of this project. B.A.M. was also supported by grants from the D. and L. Packard Foundation, the G. and B. Moore Foundation, the A. W. Mellon Foundation, and the Valley Foundation. This is contribution number XXX from PISCO, the Partnership for Interdisciplinary Studies of Coastal Oceans funded primarily by the Gordon and Betty Moore Foundation and David and Lucile Packard Foundation.
References Cited
- Aubin-Horth N, Letcher BH, Hofmann HA. Gene-expression signatures of Atlantic salmon’s plastic life cycle. General and Comparative Endocrinology. 2009;163:278–284. doi: 10.1016/j.ygcen.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlhoff EP, Menge BA. Influence of phytoplankton concentration and wave exposure on the ecophysiology of Mytilus californianus. Marine Ecology-Progress Series. 1996;144:97–107. [Google Scholar]
- Denny MW, Miller LP, Harley CDG. Thermal stress on intertidal limpets: long-term hindcasts and lethal limits. Journal of Experimental Biology. 2006;209:2420–2431. doi: 10.1242/jeb.02258. [DOI] [PubMed] [Google Scholar]
- Draghici S. Data analysis tools for DNA microarrays. Boca Raton: Chapman & Hall/ CRC; 2003. Data processing and normalization; pp. 311–341. [Google Scholar]
- Ebert TA, Dixon JD, Schroeter SC, et al. Growth and mortality of red sea urchins Strongylocentrotus franciscanus across a latitudinal gradient*. Marine Ecology Progress Series. 1999;190:189–209. [Google Scholar]
- Frederich M, Pörtner HO. Oxygen limitation of thermal tolerance defined by cardiac and ventilatory performance in spider crab, Maja squinado. American Journal of Physiology-Regulatory Integrative and Comparative Physiology. 2000;279:R1531–R1538. doi: 10.1152/ajpregu.2000.279.5.R1531. [DOI] [PubMed] [Google Scholar]
- Gracey AY, Chaney ML, Boomhower JP, et al. Rhythms of Gene Expression in a Fluctuating Intertidal Environment. Current Biology. 2008;18:1501–1507. doi: 10.1016/j.cub.2008.08.049. [DOI] [PubMed] [Google Scholar]
- Halpin PM, Sorte CJ, Hofmann GE, Menge BA. Patterns of variation in levels of Hsp70 in natural rocky shore populations from microscales to mesoscales. Integrative and Comparative Biology. 2002;42:815–824. doi: 10.1093/icb/42.4.815. [DOI] [PubMed] [Google Scholar]
- Halpin PM, Strub P, Petersen WT, Baumgartner TR. An overview of interactions among oceanography, marine ecosystems, climatic and human disruptions along the eastern margins of the Pacific Ocean. Revista Chilena de Historia Natural. 2004;77:371–409. [Google Scholar]
- Helmuth B, Broitman BR, Blanchette CA, et al. Mosaic patterns of thermal stress in the rocky intertidal zone: Implications for climate change. Ecological Monographs. 2006;76:461–479. [Google Scholar]
- Helmuth B, Hofmann GE. Defining thermal stress in the rocky intertidal: linking ecology and physiology through biophysics. American Zoologist. 2000;40:1051–1051. [Google Scholar]
- Hofmann GE, Place SP. Genomics-enabled research in marine ecology: challenges, risks and pay-offs. Marine Ecology-Progress Series. 2007;332:249–255. [Google Scholar]
- Kültz D. Evolution of the cellular stress proteom: from monophyletic origin to ubiquitous function. J. Exp Biol. 2003;206:3119–3124. doi: 10.1242/jeb.00549. [DOI] [PubMed] [Google Scholar]
- Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness trade-off in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5755–5760. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Braynen W, Keshav K, Pavlidis P. EmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie HM, Breck EN, Chan F, Lubchenco J, Menge BA. Barnacle reproductive hotspots linked to nearshore ocean conditions. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10534–10539. doi: 10.1073/pnas.0503874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser MP. Oxidative stress in marine environments: Biochemistry and physiological ecology. Annual Review of Physiology. 2006;68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- Levinton JS, Monahan RK. The Latitudinal Compensation Hypothesis - Growth Data and a Model of Latitudinal Growth-Differentiation Based Upon Energy Budgets .2. Intraspecific Comparisons between Subspecies of Ophryotrocha-Puerilis (Polychaeta, Dorvilleidae) Biological Bulletin. 1983;165:699–707. doi: 10.2307/1541472. [DOI] [PubMed] [Google Scholar]
- Lockwood BL, Sanders JG, Somero GN. Transcriptomic response to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. Journal of Experimental Biology. 2010;213:3548–3558. doi: 10.1242/jeb.046094. [DOI] [PubMed] [Google Scholar]
- Lonsdale DJ, Levinton JS. Latitudinal Differentiation in Copepod Growth - an Adaptation to Temperature. Ecology. 1985;66:1397–1407. [Google Scholar]
- Menge BA. Bottom-up:top-down determination of rocky intertidal shorescape dynamics. In: GA Polis, ME Power, G Huxel., editors. Food webs at the landscape level. Chicago, Illinois: University of Chicago Press; 2004. pp. 62–81. [Google Scholar]
- Menge BA, Chan F, Lubchenco J. Response of a rocky intertidal ecosystem engineer and community dominant to climate change. Ecology Letters. 2008;11:151–162. doi: 10.1111/j.1461-0248.2007.01135.x. [DOI] [PubMed] [Google Scholar]
- Menge BA, Chan F, Nielsen KJ, Di Lorenzo E, Lubchenco J. Climatic variation alters supply-side ecology: impact of climate patterns on phytoplankton and mussel recruitment. Ecological Monographs. 2009;79:379–395. [Google Scholar]
- Menge BA, Daley BA, Wheeler PA, Strub PT. Rocky intertidal oceanography: An association between community structure and nearshore phytoplankton concentration. Limnology and Oceanography. 1997;42:57–66. [Google Scholar]
- Norkko J, Pilditch CA, Thrush SF, Wells RMG. Effects of food availability and hypoxia on bivalves: the value of using multiple parameters to measure bivalve condition in environmental studies. Marine Ecology Progress Series. 2005;298:205–218. [Google Scholar]
- Okamura B. Microhabitat Variation and Patterns of Colony Growth and Feeding in a Marine Bryozoan. Ecology. 1992;73:1502–1513. [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nature Genetics. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Osovitz CJ, Hofmann GE. Thermal history-dependent expression of the hsp70 gene in purple sea urchins: Biogeographic patterns and the effect of temperature acclimation. Journal of Experimental Marine Biology and Ecology. 2005;327:134–143. [Google Scholar]
- Petes LE, Menge BA, Harris AL. Intertidal mussels exhibit energetic trade-offs between reproduction and stress resistance. Ecological Monographs. 2008a;78:387–402. [Google Scholar]
- Petes LE, Mouchka ME, Milston-Clements RH, Momoda TS, Menge BA. Effects of environmental stress on intertidal mussels and their sea star predators. Oecologia. 2008b;156:671–680. doi: 10.1007/s00442-008-1018-x. [DOI] [PubMed] [Google Scholar]
- Place SP, O’Donnell MJ, Hofmann GE. Gene expression in the intertidal mussel Mytilus californianus: physiological response to environmental factors on a biogeographic scale. Marine Ecology-Progress Series. 2008;356:1–14. [Google Scholar]
- Podrabsky JE, Somero GN. Changes in gene expression associated with acclimation to constant temperatures and fluctuating daily temperatures in an annual killifish Austrofundulus limnaeus. Journal of Experimental Biology. 2004;207:2237–2254. doi: 10.1242/jeb.01016. [DOI] [PubMed] [Google Scholar]
- Roberts DA, Hofmann GE, Somero GN. Heat-shock protein expression in Mytilus californianus: Acclimatization (seasonal and tidal-height comparisons) and acclimation effects. Biological Bulletin. 1997;192:309–320. doi: 10.2307/1542724. [DOI] [PubMed] [Google Scholar]
- Roy K, Martien KK. Latitudinal distribution of body size in north-eastern Pacific marine bivalves. Journal of Biogeography. 2001;28:485–493. [Google Scholar]
- Sagarin RD, Somero GN. Complex patterns of expression of heat-shock protein 70 across the southern biogeographical ranges of the intertidal mussel Mytilus californianus and snail Nucella ostrina. Journal of Biogeography. 2006;33:622–630. [Google Scholar]
- Schneider KR, Van Thiel LE, Helmuth B. Interactive effects of food availability and aerial body temperature on the survival of two intertidal Mytilus species. Journal of Thermal Biology. 2010;35:161–166. [Google Scholar]
- Somero GN. The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. Journal of Experimental Biology. 2010;213:912–920. doi: 10.1242/jeb.037473. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1 beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanek L, Zuzow MJ. The Proteomic response of the mussel congeners Mytilus gallaprovincialis and M. trossulus to acute heat stress: implications for thermal tolerance limits and metabolic costs of thermal stress. Journal of Experimental Biology. 2010;213:3559–3574. doi: 10.1242/jeb.041228. [DOI] [PubMed] [Google Scholar]
- Williams EE, Somero GN. Seasonal-, tidal-cycle- and microhabitat-related variation in membrane order of phospholipid vesicles from gills of the intertidal mussel Mytilus californianus. Journal of Experimental Biology. 1996;199:1587–1596. doi: 10.1242/jeb.199.7.1587. [DOI] [PubMed] [Google Scholar]
- Yamahira K, Conover DO. Intra- vs interspecific latitudinal variation in growth: Adaptation to temperature or seasonality? Ecology. 2002;83:1252–1262. [Google Scholar]
- Wingfield JC, Sapolsky RM. Reproduction and resistance to stress: when and how. Journal of Neuroendocrinology. 2003;15:711–724. doi: 10.1046/j.1365-2826.2003.01033.x. 2003. [DOI] [PubMed] [Google Scholar]
- Zippay M, Hofmann G. Physiological tolerances across latitudes: thermal sensitivity of larval marine snails (Nucella spp.) Marine Biology. 2010;157:707–714. doi: 10.1007/s00227-009-1354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.