Abstract
Topoisomerases are essential for DNA replication in dividing cells, but their genomic targets and function in postmitotic cells remain poorly understood. Here we show that a switch in the expression from Topoisomerases IIα (Top2α) to IIβ (Top2β) occurs during neuronal differentiation in vitro and in vivo. Genome-scale location analysis in stem cell–derived postmitotic neurons reveals Top2β binding to chromosomal sites that are methylated at lysine 4 of histone H3, a feature of regulatory regions. Indeed Top2β-bound sites are preferentially promoters and become targets during the transition from neuronal progenitors to neurons, at a time when cells exit the cell cycle. Absence of Top2β protein or its activity leads to changes in transcription and chromatin accessibility at many target genes. Top2β deficiency does not impair stem cell properties and early steps of neuronal differentiation but causes premature death of postmitotic neurons. This neuronal degeneration is caused by up-regulation of Ngfr p75, a gene bound and repressed by Top2β. These findings suggest a chromatin-based targeting of Top2β to regulatory regions in the genome to govern the transcriptional program associated with neuronal differentiation and longevity.
Keywords: epigenetic regulation, neurogenesis, gene expression, genomewide assays
Topoisomerases are essential for solving topological problems arising from DNA-templated processes such as replication, transcription, recombination, chromatin remodeling, chromosome condensation, and segregation (1–5). The type I subfamily of topoisomerases achieves this task by passing one strand of the DNA through a break in the opposing strand; proteins in the type II subfamily pass a region of duplex strands from the same or a different molecule through a double-stranded gap generated in DNA (1–5). Mammalian cells encode two isozymes of type II enzymes that have highly homologous N-terminal ATPase and central core domains but differ at their C-termini (6). These two isozymes, Topoisomerases IIα (Top2α) and IIβ (Top2β), have almost identical enzymatic properties in vitro (7, 8); however, their expression patterns are dissimilar. Top2α is the main isoform expressed in proliferating cells, shows high expression in S/G2/M phases of the cell cycle, and plays important roles in DNA replication and chromosome condensation/segregation during the cell cycle (9–12).
The cellular functions of Top2β are much less well understood. It is expressed in all mammalian cells throughout the cell cycle but is up-regulated robustly when cells reach a postmitotic state of terminal differentiation (13–15). For example, the postmitotic granule cells in the external germinal layer of the developing rat cerebellum show a transition from Top2α to Top2β (14), and blocking Top2β catalytic activity affects the expression of about one third of genes induced during differentiation of rat cerebellar granule neurons (16). Genetic deletion of Top2b in mice causes neural defects including aberrant axonal elongation and branching and perinatal death explained by lack of innervation of the skeletal musculature (17). Further support for a functional role for Top2β in brain development comes from the analysis of a brain-specific Top2b deletion in mice; this deletion results in a defect in corticogenesis (13). Based on single-gene analysis, it was hypothesized that Top2β may regulate transcription of neuronal genes by direct binding to their regulatory regions (18, 19). Attempts to purify sites of enzymatic action suggested that Top2β binding is enriched in AT-rich intergenic regions as well as at gene boundaries (20). However, this work was limited to a minor part of the rat genome, leaving open the questions as to where Top2β binds genome-wide and how such binding might explain the defects in brain development observed in Top2β-deficient animals. Also undetermined is how the Top2β chromosomal location relates to the growing repertoire of chromatin modifications that define functional states of genes and regulatory regions (21–23) and contribute to the ability of stem cells to differentiate into specific lineages (24–27).
To gain insight into the molecular function of Topoisomerase II in postmitotic neurons, we used an established neuronal differentiation system that progresses through defined stages with high synchrony and homogeneity (28, 29), and we subsequently validated the observations in vivo. We show that the transition from pluripotent stem cells to postmitotic neurons accompanies up-regulation of Top2β and down-regulation of Top2α. Chromosome-wide analysis shows that Top2β binds preferentially to regions containing H3K4 methylation, a feature of active chromatin. Top2β-target sites are enriched at promoters and become occupied by Top2β during the transition from neuronal progenitors to neurons, at a time when cells exit the cell cycle. Many of these targets show transcriptional changes in absence of Top2β protein or its catalytic activity. As a result of these changes, postmitotic neurons degenerate prematurely. We show that this premature degeneration is accompanied by and caused by an up-regulation of neurotrophin receptor p75, a Top2β target.
Results
Neuronal Differentiation Accompanies a Switch in Topoisomerase II Isoforms.
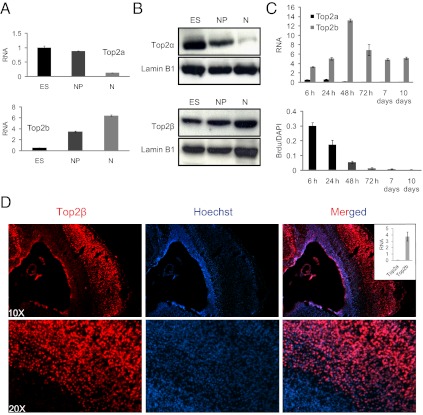
To define Topoisomerase II function in postmitotic cells, we used mouse embryonic stem cells (ESCs) that differentiate under defined conditions into Pax6-positive neural progenitors, which in turn differentiate into postmitotic glutamatergic neurons with high synchrony and purity (28–30). Top2α mRNA expression and protein levels decreased from the stem cell to the neuronal progenitor state and further were down-regulated strongly in postmitotic neurons (Fig. 1 A and B). These expression levels are in line with previous observations in the developing nervous system (31) and are in agreement with the reported high levels of Top2α in proliferative cells such as the ESCs studied here (9, 32, 33). Top2β shows a different pattern: It is up-regulated robustly upon differentiation and reaches its highest levels in neurons, as measured by mRNA and protein analysis (Fig. 1 A and B). At higher temporal resolution, complete disappearance of Top2a (encoding Top2α) was observed 2 d after progenitor plating (Fig. 1C). During the same time period, Top2b (encoding Top2β) levels were highly induced (Fig. 1C). This switch in Topoisomerase II isoforms accompanies cell-cycle exit and rapid cessation of proliferation, as measured by BrdU incorporation (Fig. 1C). These observations with cultured cells were confirmed in vivo, and, in line with a previous report (13), at embryonic day 18.5 (E18.5) we found exclusive and high expression of Top2β in the cortical plate of mouse cortex, which consists of mature neurons (Fig. 1D). We conclude that a switch in Topoisomerase II isoforms from Top2α to Top2β occurs during neuronal differentiation in vitro and in vivo, temporally in parallel with the transition from a pluripotent, rapidly proliferating state to a differentiated, postmitotic state.
Fig. 1.
Induction of neuronal differentiation accompanies a switch from Top2α to Top2β isoform. (A) Expression analysis of three stages of murine stem-cell differentiation reveals up-regulation of Top2b and down-regulation of Top2a upon neuronal differentiation. mRNA levels as measured by real-time RT-PCR relative to Gapdh are plotted on the y axis. ES, embryonic stem cells; NP, neuronal progenitors; N, neurons. (B) Western blot analysis for Top2α and Top2β protein levels in extracts isolated from the same stages as in A. Lamin B1 serves as a loading control. (C) (Upper) Analysis at a higher temporal resolution shows that Top2a is diminished around neuronal day 2 after plating of progenitor cells, whereas Top2b is up-regulated robustly. mRNA levels are plotted as in A. (Lower) Cell proliferation measured by BrdU incorporation at 6 h, 24 h, 48 h, 3 d, 7 d, and 10 d (stage N in A). (D) Immunofluorescence analysis in neurons using Top2β-specific antibody (red, Left) and Hoechst (blue, Center) for nuclear staining in a section from mouse cortex at embryonic day 18.5 (E18.5). The overlay of both stains (Right) shows that Top2β is expressed abundantly in the differentiated postmitotic neurons that lie in the peripheral zone. Inset shows results of RT-PCR in cortical neurons derived from the peripheral zone. The results are plotted as in A.
Top2β−/− ESCs Exhibit Defects in Neuronal Differentiation.
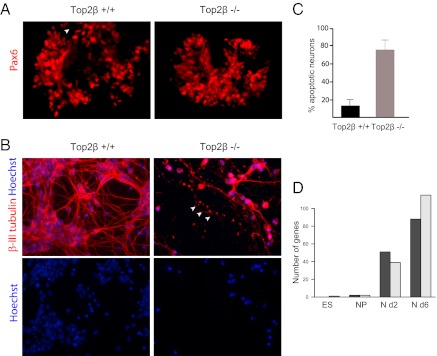
To address the role of Top2β directly, we derived Top2β−/− ESCs from the progeny of Top2β+/− mice (Fig. S1A) (13). Both the growth characteristics and the morphology of Top2β−/− ESCs were indistinguishable from those of wild-type ESCs (Fig. S1B), as confirmed by a global expression analysis that revealed no significant differences in the transcription profile between Top2β−/− and wild-type ESCs (Fig. 2D and Fig. S1C). We conclude that Top2β is dispensable for the stem cell transcription program, including self-renewal.
Fig. 2.
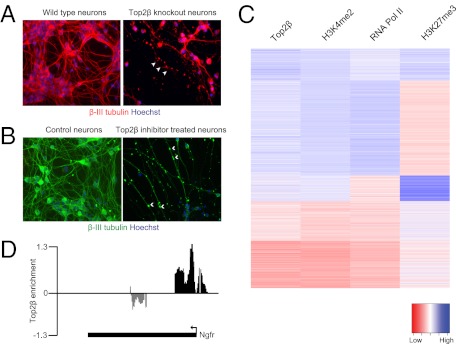
ESC lacking Top2β show impaired neuronal differentiation. (A) Immunofluorescence analysis using an anti-Pax6 antibody (red) in wild-type and Top2β-knockout neuronal progenitor cells reveals no differences in progenitor specification with regard to the expression of this instructive transcription factor. (B) Immunofluorescence analysis in differentiated neurons after 6 d of culture using β-III tubulin–specific antibody (red) shows significant neuronal degeneration in Top2β-knockout cells. Hoechst is used for staining DNA. The arrows mark axonal degeneration. (C) Quantification of apoptotic nuclei after 6 d of neuronal culture showing a significantly increased percentage of neuronal death in Top2β-knockout as compared with wild-type neurons. (D) Graphs depict the number of genes that are expressed differentially in wild-type and Top2β-knockout cells at the embryonic stem cell (ES), neuronal progenitor cell (NP), day 2 neuron (N d2), and day 6 neuron (N d6) stages. Neurons lacking Top2β exhibit the greatest number of deregulated genes. Dark gray bars represent up-regulated genes; light gray bars represent down-regulated genes.
We next examined whether Top2β−/− ESCs retain the potential for lineage commitment and their ability to differentiate into postmitotic neurons. Top2β−/− ESCs generated neuronal progenitors with efficiency similar to that of wild-type ESCs and also expressed characteristic markers such as Pax6 (Fig. 2A). This notion was confirmed and extended at the transcriptome level as measured by Affymetrix analysis (Fig. 2D and Fig. S1C). Although we failed to find a large number of genes that are affected significantly by knockout neurons soon after exit from the cell cycle (Fig. 2D and Fig. S1C), we observed that at later stages the Top2β−/− neurons started to show signs of degeneration and died by apoptosis (Fig. 2 B and C). Transcriptome profiling revealed a number of genes that were statistically significantly deregulated in neurons lacking Top2β as compared with wild-type cells (Fig. 2D and Fig. S1C). The genes down-regulated in Top2β−/− neurons show a strong enrichment for gene ontology (GO) terms associated with “neurogenesis,” whereas genes up-regulated in Top2β−/− neurons were associated with the GO term “cell division” (Fig. S1D). A number of single-gene controls validated these transcriptional changes (Fig. S1E). This observation is not limited to in vitro-differentiated neurons; similar transcriptional changes were observed for cortical neurons isolated from E16.5 Top2β-knockout embryos (Fig. S1C; also see Fig. S4F). Interestingly, Top2a (encoding Top2α) was among the genes that were up-regulated in Top2β−/− neurons both in vitro and in vivo (Fig. S1C). Because Top2α is repressed in nonproliferative cells (34), and its expression is linked exclusively with increased proliferation cycle (1, 10, 35, 36), we speculated that its reactivation in postmitotic neurons may cause neuronal degeneration, e.g., by forcing the neurons to re-enter the cell cycle. To test this possibility, we treated Top2β−/− neurons with meso-2,3-bis(2,6-dioxopiperazin-4-yl)butane (ICRF-193), an established inhibitor of catalytic activity of both Top2 isozymes (37). We found this inhibition did not prevent the degeneration of the neurons lacking Top2β, suggesting that Top2α activity is not causally involved in the observed phenotype.
Top2β Binds Preferentially to Promoters.
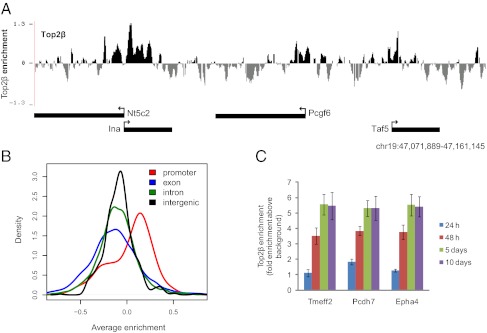
We next explored whether sites of chromosomal binding of Top2β in postmitotic cells relate to the aberrant changes in gene expression and the phenotype observed in Top2β−/− neurons. Toward this goal, we performed ChIP with Top2β-specific antibodies followed by detection using custom tiling arrays. These arrays cover 10% of the mouse genome, including all well-annotated promoters, several large multigene loci, and the complete chromosome 19 (SI Material and Methods). Visual inspection of the chromosomal profiles for Top2β indicated specific enrichment at promoters (Fig. 3A), with biological replicates showing high reproducibility (R = 0.90). Intrigued by this observation, we performed a comprehensive and unbiased analysis by comparing Top2β enrichment along the fully tiled chromosome 19. This analysis revealed a clear enrichment for Top2β binding to promoters as compared with exons, introns, and intergenic regions (Fig. 3B). Furthermore, a time-course analysis for Top2β binding during neuronal differentiation revealed that Top2β targeting to promoters occurs at the time of transition from the progenitor to the neuronal state, a time when Top2β levels are highly induced and cells become postmitotic (Figs. 1C and 3C).
Fig. 3.
Top2β binds preferentially to promoters. (A) Genome browser screenshot from a segment of chromosome 19 showing promoter-specific localization of Top2β in neuronal cells. The y axis represents log2 enrichments (immunoprecipitation/input). (B) Distribution of Top2β enrichments in different genomic regions. The fully tiled chromosome 19 was divided hierarchically into promoter, exon, intron, and intergenic regions, and Top2β enrichments for each region were calculated by averaging over all probes that mapped to the particular region. The distribution of enrichments shows that Top2β is enriched most strongly at promoters. (C) ChIP-qPCR for Top2β enrichment at various identified target promoters using Top2β ChIP material derived 24 h, 48 h, 5 d, and 10 d after plating of progenitors for neuron formation. This analysis shows that Top2β targets gene promoters at neuron day 2 when cells exit the cell cycle (Fig. 1C). Average enrichments from three separate assays are plotted on the y axis as the ratio of precipitated DNA (bound) to the total input DNA and further normalized to a control region. Error bars indicate SEM.
Distinct Histone Modification Patterns Mark Top2β-Bound Promoters.
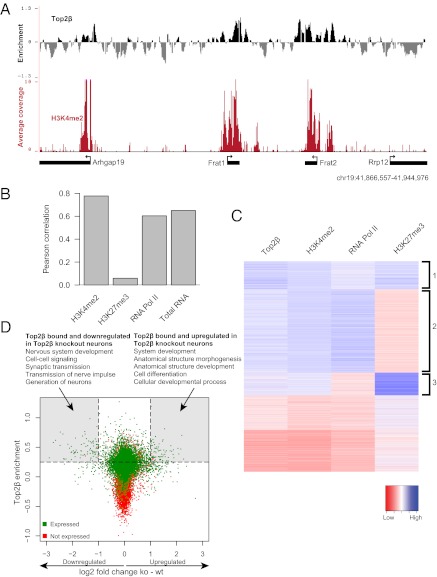
Several recent studies established that promoter sequences in eukaryotic genomes show characteristic patterns of histone modifications that associate with the active or inactive state of the linked gene (21–23). Actively transcribed promoters as well as DNA unmethylated CpG island promoters show uniformly high levels of H3K4 dimethylation (H3K4me2) in mammalian cells (38, 39). H3K4me2 is considered to be a mark of active chromatin, which is involved in the recruitment of many distinct factors involved in chromatin remodeling and gene regulation (21, 40–42). In many cases CpG islands that are inactive harbor both the activating H3K4 mark and the repressive chromatin modification trimethylation of H3K27 (H3K27me3) set by the Polycomb system (39). This bivalency of activating and repressing marks has been implicated in developmental gene regulation (43). To determine if Top2β binding relates to these important marks of chromatin, we compared the Top2β ChIP data with genomewide datasets that were generated by ChIP followed by next-generation sequencing with neurons as starting material (44). This analysis revealed that the active chromatin modification H3K4me2 is present at most Top2β-enriched regions, and vice versa (Fig. S2A). Visual inspection of the chromosomal profiles for Top2β and H3K4me2 suggested their co-occurrence at promoters (Fig. 4A). A genomewide comparison further revealed a high positive correlation between the two datasets at the level of promoters (R = 0.78) that also was evident in a cluster analysis (Fig. 4 B and C and Fig. S2B). Top2β and the Polycomb-associated H3K27me3 mark showed no correlation, because many Top2β targets are not marked by this histone modification (R = 0.06) (Fig. 4 B and C and Fig. S2B). Intriguingly however, most bivalent promoters that have both H3K4me2 and H3K27me3 also were marked by Top2β (Fig. 4C, cluster 3). To relate these patterns to transcriptional activity, we included RNA Polymerase II binding data in this analysis and found that the majority of Top2β-bound promoters also recruit Polymerase (R = 0.63) (Fig. 4 B and C and Fig. S2B). In most cases, this recruitment coincided with active transcription as measured by transcriptome analysis (R = 0.65) (Fig. 4 B and cluster 2 in C and Fig. S2B). Top2β-target bivalent promoters that have both H3K4me2 and H3K27me3 are not occupied by polymerase (Fig. 4C, cluster 3) and are not transcribed (Fig. S2B), in line with the model proposing that this state marks genes that are activated subsequently (43). Extensive single-gene controls with wild-type and Top2β-knockout cells, as well as with brain tissue using two additional antibodies specific for Top2β, validated these genomic binding patterns (Fig. S3 A–C).
Fig. 4.
Top2β targets are embedded in H3K4 methylated chromatin, and Top2β occupancy correlates positively with an active transcription state. (A) Genome browser screenshot from a segment of chromosome 19. The upper track shows ChIP-chip data for Top2β. The y axis shows log2 enrichment (immunoprecipitation/input). The lower track shows ChIP-seq data for H3K4me2 in neuronal cells. The y axis represents average coverage per base, averaged over 100 nt. (B) Correlation of Top2β enrichment at TSS in neurons (y axis) with H3K4me2, H3K27me3, RNA Pol II, and mRNA levels. (C) Heat map depicting Top2β enrichment and H3K4me2, H3K27me3, and RNA Pol II levels for each of the promoters. Promoters were clustered using k-means clustering with k = 5. (D) Scatter plot showing Top2β enrichment at promoters (y axis) versus the changes in expression (log2 scale) in wild-type and Top2β-knockout cells at the corresponding genes (x axis). A significant number of genes deregulated in Top2β-knockout neurons are occupied by Top2β at their promoters. The top five hits from GO term analysis for genes from the categories of “Top2β occupied and down-regulated in Top2β KO cells” and “Top2β occupied and up-regulated in Top2β KO cells” are shown. P < 10−2.
These findings indicate that Top2β binds promoters that are characterized by high levels of H3K4 dimethylation and that a majority of these promoters are transcribed actively, whereas many of those that are inactive harbor the additional H3K27me3 mark. This result links Top2β to proximal regulation of transcription and suggests that chromatin modifications are involved in specifying its chromosomal binding and that this binding influences gene expression.
Top2β Binding Modulates Gene Expression.
To determine if Top2β binding to promoter regions influences the transcriptional activity of target genes, we contrasted Top2β occupancy with the transcriptional changes observed in Top2β−/− neurons. We found that a large number of genes that were Top2β bound in wild-type neurons were down-regulated in the knockout cells (Fig. 4D). This dependency of Top2β also was observed for a smaller and less significant set of Top2β targets that showed up-regulation in the mutant cells, whereas a large fraction of bound genes remained unaffected. Importantly, however, a majority (99/171) of the genes showing differential expression in mutant cells were, in fact, bound by Top2β in wild-type cells (P = 9e-19) (Dataset S1). Genes in this group that were down-regulated upon Top2β deletion were associated with gene functions related to neurogenesis (Fig. 4D), whereas genes that were bound and up-regulated were associated with organismal development (Fig. 4D).
Chemical Inhibition of Top2β Catalytic Activity Affects Transcription of Target Genes and Results in a Phenotype Similar to the Absence of Top2β.
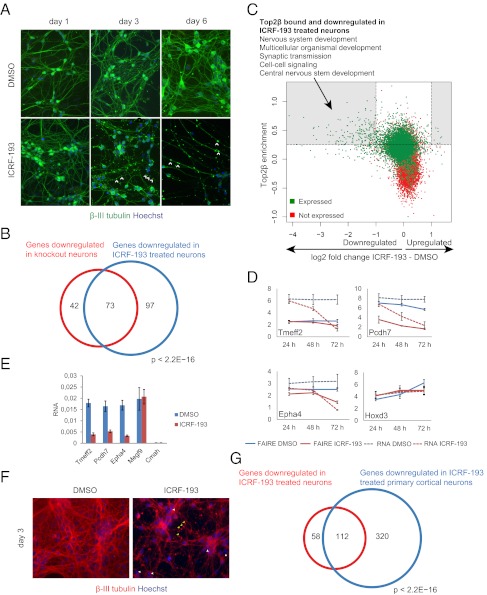
To determine if Top2β catalytic activity is required for its effect on target-gene expression, we again used the inhibitor ICRF-193. ESC-derived neurons were treated for 3 d starting 48 h after plating of neuronal progenitors, when Top2α levels were decreased and Top2β was strongly up-regulated (Fig. 1C). Expression analysis suggested a dose-dependent decrease in activity of Top2β-target genes (Fig. S4A), and ICRF-193–treated cells exhibited neuronal degeneration similar to that observed in Top2β-knockout neurons (Fig. 5A). Transcriptome profiling of cells that were treated with either DMSO (vehicle control) or ICRF-193 revealed significant transcriptional down-regulation and very little transcriptional up-regulation (Fig. S1C). A comparison of the expression changes in ICRF-193–treated and Top2β-knockout ESC-derived neurons revealed a strong overlap between down-regulated genes (Fig. 5B and Fig. S4B). Furthermore, consistent with the findings in Top2β knockout neurons, a significant number of down-regulated genes were direct targets of Top2β in wild-type neurons and were enriched exclusively for neuronal functions (Fig. 5C). To see if ICRF-193–mediated down-regulation of Top2β-target genes accompanies changes in their chromatin accessibility we used the formaldehyde-assisted isolation of regulatory elements (FAIRE) assay (45–47). This approach identifies regions of low nucleosomal occupancy, a chromatin feature of active regulatory regions. Here we observed that ICRF treatment affects the chromatin structure, as detected by the FAIRE assay, only in Top2β-target genes that also are transcriptionally down-regulated (Fig. 5D).
Fig. 5.
Inhibition of Top2β catalytic activity phenocopies the genetic deletion and down-regulates target genes. (A) Immunofluorescence analysis in neurons using β-III tubulin–specific antibody at different time points after treatment with inhibitor or control reveals neuronal degeneration from day 3 onwards as indicated by arrowheads. β-III Tubulin (green) and Hoechst (blue) staining mark neurons and nuclei, respectively. (B) Venn diagram illustrating the overlap between genes down-regulated in knockout and in inhibitor-treated neurons. (C) Scatter plot depicting Top2β enrichment (y axis) versus expression changes (log2 scale) between inhibitor treatment and control (x axis) illustrating that most inhibitor-sensitive genes are Top2β bound. This group of genes is strongly enriched for neuronal gene functions (P < 10−20). (D) Chromatin accessibility assay in neurons treated with the inhibitor for different periods starting at d 2 after plating. Shown are average FAIRE enrichments and RNA levels detected by qPCR for four Top2β-target promoters normalized to a control (y axis). Error bars indicate SEM. (E) Inhibitor treatment of primary cortical neurons leads to similar transcriptional responses in three Top2β-target neuron-specific genes (see Fig. S1C for genome-wide analysis). The graph also shows two non–Top2β-target genes, Megf9 (expressed in neurons) and Cmah (not expressed in neurons) that are not affected by this treatment. Error bars represent SEM. (F) Inhibitor treatment of cortical neurons similarly leads to neuronal degeneration (antibodies are as in A). (G) Venn diagram illustrating the large overlap between genes down-regulated in inhibitor-treated stem cell-derived neurons and inhibitor-treated cortical neurons.
To establish the in vivo relevance of these observations, we isolated cortical neurons from E16.5 mouse embryos and plated them on polyornithine- and laminin- coated dishes. Top2a is not expressed in these cells, whereas Top2b is transcribed robustly (Fig. 1D). Expression analysis indicated a decrease in the expression of Top2β-target genes upon ICRF-193 treatment in cortical neurons (Fig. 5E), similar to the results described for ESC-derived neurons (Fig. S4A). Furthermore, ICRF-193 treatment led to neuronal degeneration (Fig. 5F) similar to the degeneration we observed in Top2β−/− neurons (Fig. 2B) and in ICRF-193–treated ESC-derived neurons (Fig. 5A). We next profiled the transcriptome of cortical neuronal cells treated with either DMSO (vehicle control) or ICRF-193 and again observed mostly transcriptional down-regulation of tested genes (Fig. S1C). A comparison of the expression changes with ICRF-193 treatment in cortical neurons and in stem cell-derived neurons revealed a strong overlap among down-regulated genes (Fig. 5G and Fig. S4C). These data suggest that Top2β inhibition results in transcriptional as well as phenotypic changes, similar to the changes observed with neurons generated from Top2β-knockout ESCs. Furthermore, a substantial overlap was observed among genes that were down-regulated upon ICRF-193 treatment of cortical neurons and Top2β-knockout cortical neurons (Fig. S4D). It is important to note that only a few of the genes that are up-regulated in Top2β-knockout neurons show a similar response upon enzymatic inhibition (Fig. S4 B and D). This result might indicate either that in most cases transcriptional up-regulation is an indirect effect or that it depends on the presence of Top2β but not on its catalytic activity. Among the genes that were Top2β bound and up-regulated in mutant neurons and those treated with ICRF-193, we concentrated on the nerve growth factor receptor (Ngfr) (Fig. S1C), commonly referred to as the “neurotrophin receptor p75,” a member of the TNF receptor family and previously implicated in neuronal death (48–50).
Up-Regulation of the Neurotrophin Receptor p75 Is Involved in the Death of Neurons Lacking Top2β.
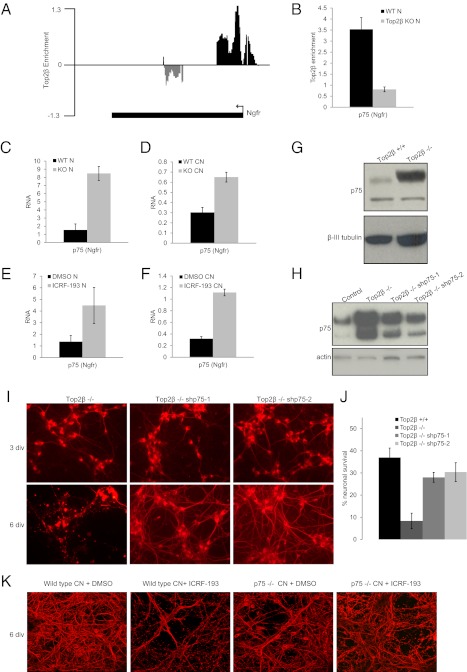
The neurotrophin receptor p75 is a direct target of Top2β (Fig. 6 A and B) and is up-regulated in both Top2β-deficient ESC-derived neurons and cortical neurons (Fig. 6 C and D and Figs. S1C and S4 B–F). It also is up-regulated in ICRF-193–treated wild-type ESC-derived neurons and cortical neurons (Fig. 6 E and F and Fig. S1C). This transcriptional up-regulation led to higher protein levels as measured by Western blot analysis (Fig. 6G and Fig. S5).
Fig. 6.
Loss of Top2β-mediated repression of p75 Ngfr mediates premature death of Top2β-knockout neurons. (A) Genome browser screenshot showing enrichment of Top2β at p75 Ngfr promoter in neuronal cells. The y axis represents log2 enrichment (immunoprecipitation/input). (B) PCR validation of Top2β enrichment at the Ngfr promoter in neurons. Average enrichments from three separate assays are plotted on the y axis as the ratio of precipitated (bound) DNA to the total input DNA and further normalized to a control region. Error bars indicate SEM. (C) Expression analysis of Ngfr showing its up-regulation in Top2β-knockout versus wild-type neurons (N). mRNA levels as measured by real-time RT-PCR relative to Gapdh are plotted on the y axis. Error bars indicate SEM. (D) Ngfr is up-regulated similarly in cortical neurons (CN) isolated from E16.5 Top2β-knockout mouse embryos. The graph is plotted as in C. (E) Up-regulation of Ngfr transcript levels through ICRF-193–mediated inhibition of Top2β catalytic activity in neuronal cells (N). The graph is plotted as in C. (F) Up-regulation of Ngfr upon ICRF-193 inhibition in cortical neurons (CN). The graph is plotted as in C. (G) p75 protein levels as detected by Western blotting of extracts isolated from wild-type and Top2β-knockout neurons confirm its increase at the protein level in cells lacking Top2β. β-III Tubulin serves as loading control. (H) Western blot analysis for p75 levels in protein extracts isolated from control and Top2β-knockout neurons as well as neurons with shRNA-mediated depletion of p75 confirm a significant knockdown of p75 in the two p75 shRNA clones. Actin serves as a loading control. (I) Immunofluorescence analysis using β-III tubulin–specific antibody (red) after 3 and 6 d in vitro (div) neuronal culture shows survival of Top2β-knockout neurons depleted of p75 as compared with Top2β-knockout neurons that degenerate. (J) The percentage of neurons surviving (y axis) is calculated as the ratio between the neurons surviving after 6 d of culture and the number of progenitors used for neuron formation. The data indicate a marked increase in cell survival in Top2β-knockout neurons depleted of p75 as compared with Top2β-knockout neurons. (K) Immunofluorescence analysis using β-III tubulin–specific antibody (red) in wild-type and p75−/− primary cortical neurons 3 d after treatment with control (DMSO) or inhibitor (ICRF-193) reveals markedly reduced neuronal death in inhibitor-treated p75−/− neurons as compared with similarly treated wild-type cells.
To investigate whether p75 up-regulation is responsible for the degeneration in Top2β−/− neurons, we reduced its endogenous levels by shRNA-mediated knockdown in stably transfected clones of Top2β−/− cells. Western blot analysis confirmed efficient knockdown in neurons in two clones expressing three different shRNAs against p75 (Fig. 6H), and this reduction markedly delayed neuronal death (Fig. 6 I and J). To support our observations further, we derived cortical neurons from E16.5 p75 wild-type and p75-knockout mouse embryos (51) and treated them with ICRF-193. Under these conditions, p75-knockout neurons showed significantly reduced death compared with the wild-type neurons (Fig. 6K). Together these findings suggest that up-regulation of the Top2β target p75 is causal for the premature death of neurons lacking Top2β protein or its enzymatic activity.
Discussion
Mammals have two isoforms of Topoisomerase II with similar enzymatic properties in vitro (7, 8). Both isoforms can complement the single form of the enzyme in yeast (52), but in mammals only Top2α can provide the type II topoisomerase functions required for DNA propagation, such as chromosome condensation and segregation (32, 53). Thus Top2α is necessary for the survival of cycling cells (54). By comparison, Top2β was recognized mostly for what it is unable to do: It does not support mitotic chromosome segregation (32, 53, 55) and appears dispensable for cycling cells (56). However, mice lacking Top2β exhibit a perinatal death because of defects in neuronal development (17). This phenotype has been linked to Top2β function in the transcriptional regulation of neuron-specific genes (13, 18, 31). Further hints came from the observations that type II topoisomerase activity is involved in steroid-stimulated gene activation of the pS2 promoter (19) and that it relaxes nucleosomal chromatin in vitro (5). These observations led to the hypothesis that Top2β has a function in chromatin condensation/decondensation and in local changes of chromatin architecture. Our results provide strong support for this hypothesis, because we show at the level of the genome that Top2β binds selectively to chromosomal regions marked by H3K4 dimethylation, a histone modification characteristic of active regulatory regions. Notably we did not observe consistent changes in chromatin organization of Top2β targets upon ICRF-193 treatment, as measured by the FAIRE assay. Instead we observed reduced accessibility only at targets that were transcriptionally down-regulated. It is important to note, however, that the currently available assays to measure chromatin organization are limited in scope and sensitivity (57). Thus, how Top2β activity contributes to the local chromatin organization of target genes remains an open question.
Enzymes that catalyze H3K4 methylation were first identified genetically in the fruit fly as activators of Hox loci, and subsequent studies have found that H3K4 methylation has an activating role in gene regulation (21–23, 58). Although there is only a single H3K4 methyltransferase in yeast, several enzymes methylate and demethylate this residue in higher eukaryotes (21). Further complexity is generated by a growing list of proteins that selectively bind methylated H3K4 (58), leading to the concept that H3K4me functions in tethering a multitude of proteins to chromatin and repelling others. For example, a component of the NURF chromatin-remodeling complex, BPTF, recognizes H3K4 methyl marks via a PHD domain, which in turn recruits the SNF2L ATPase to activate gene expression (59), whereas a DNA methyltransferase is inhibited by this histone modification (60). Similarly a number of other H3K4me-binding proteins with enzymatic activities have been discovered; among these proteins are JMJD2A, CHD1, and WDR5 (61–64). Each of these activities is shown to modulate chromatin structure during the process of gene regulation. It is conceivable that the involved DNA binding of proteins and/or remodeling of nucleosomes create structural constraints that require topoisomerase activity to ensure proper gene regulation.
A large group of H3K4me2-decorated promoters are transcriptionally active. Many inactive ones harbor the repressive mark H3K27me3 as well. This coexistence of active and repressive marks at gene promoters has been defined as a “bivalent chromatin state” and is suggested to be an important signal for activation at later developmental stages (43). The specific genomic location of Top2β is compatible with a chromatin-based recruitment mechanism functioning at sites of chromatin remodeling and gene regulation. Compatible with this hypothesis is the finding that human TopIIβ, but not TopIIα, copurifies with the chromatin-remodeling factor ACF (65).
The preferential location to regulatory regions in the genome challenges the notion that type II topoisomerases in mammalian cells are merely structural components of chromosomes (66, 67) that show enriched binding at AT-rich intergenic regions (20) and instead links them with gene regulation in postmitotic cells. Indeed, expression of many bound genes was found to be altered in Top2β−/− neurons both in vitro and in vivo, with most being down-regulated. These genes respond similarly to ICRF-193 treatment of wild-type cells suggesting specific function of the catalytic activity of Top2β in maintenance of expression of bound targets. Importantly, Top2α, even though up-regulated, cannot compensate for absence of Top2β in the knockout cells, suggesting that Top2β has a genuine function in these cells. This model raises the question as to why Top2β appears dispensable in cycling stem cells. At this point we only can speculate that the structural constraints solved by Top2β might arise in both cycling and postmitotic cells but can accumulate only in postmitotic cells, because they are resolved during mitotic division in cycling cells and thus do not accumulate and cause no phenotype. In line with this model, Top2β targets promoters during the transition from progenitor to neuronal state at a time when cells become postmitotic and when Top2β is strongly up-regulated.
The findings of our comprehensive analysis are compatible with previous single-gene studies (18, 19) but reveal an unexpected universal recruitment of this enzyme to gene sites that are proximal for gene regulation and greatly expand the set of target genes. Notably, a small subset of Top2β-bound genes was found to respond by transcriptional up-regulation because of the absence of Top2β or to ICRF treatment. It is conceivable that these genes are actively repressed and that maintenance of this repression requires structural integrity of the DNA template, as is the case for gene activation. The most prominently affected gene was Ngfr p75, a key regulator of cell death during development of the central and peripheral nervous system (48–50). Although Ngfr p75 is widely expressed during development, it subsequently is down-regulated and is reexpressed in the adult only in response to injury, where it has consistently been shown to be responsible for cell death accompanying neurodegeneration (68, 69). We previously showed that prevention of its down-regulation in ESC-derived neurons causes cell death (70), thus indicating that a higher level of this receptor alone is sufficient to cause the phenotype reported here with the same cellular system. We find that the depletion of p75 in Top2β-knockout neurons led to a marked delay in their death. Furthermore, p75-knockout neurons showed significantly reduced death compared with the wild-type neurons upon chemical inhibition of Top2β activity. These data strongly argue that up-regulation of the Top2β-target gene p75 is causal for the premature death of neurons lacking Top2β function. Furthermore, little is known about the molecular mechanisms controlling the expression of Ngfr p75, and thus the finding that Top2β binds and likely acts on the Ngfr p75 promoter is a significant step toward understanding its regulation.
Taken together, our findings provide insights into the regulatory function and genomic localization of Top2β in postmitotic cells. Our results suggest a model whereby Top2β occupies active regulatory regions in the genome where it modulates transcription of target genes that control neuronal differentiation and survival.
Materials and Methods
Derivation of Wild-Type and Top2β-Knockout ESCs.
Blastocyst embryos were obtained from superovulated Top2β+/− female mice mated to Top2β+/− males. Embryos were harvested in FHM medium (Chemicon) by flushing the uterus (blastocysts) at blastocyst stage and were cultured on mouse embryonic fibroblast feeders as described by Bryja et al. (71). Later, individual clones were genotyped. The primer sequences used for genotyping are available on request. Housing and handling of mice conformed to the Swiss Animal Protection Ordinance, chapter 1.
Cell Culture.
Wild-type and Top2β-knockout ESC (129Sv) were cultured and differentiated as previously described (29). ESC expressing shRNA constructs against p75 were generated as previously described (72).
Western Blot Analysis.
Western blot analysis was performed with purified protein extracts using 1/1,000 dilutions of Top2β, Top2α, p75, lamin B1, and tubulin III antibodies. Blots were developed with ECL reagent (GE Healthcare).
ChIP.
ChIP experiments were performed as described previously (39), starting with 70 μg of chromatin and 5 μg of the following antibodies: anti-Top2b: H286 (Santa Crutz), anti-RNA Pol II: N-20 (Santa Crutz), anti-H3K4me2: Ab32356 (Abcam), and anti-H3K27me3: 07-449 (Millipore). The ChIP libraries processed for Illumina sequencing were prepared with the Illumina ChIP-Seq DNA Sample Prep Kit (catalog no. IP-102-1001) according to Illumina's instructions and were sequenced on the Genome Analyzer 2 following the manufacturer's protocols. ChIP-real time PCR was performed using SYBR Green chemistry (ABI) and ChIP (1:40) or input (1:100) DNA per PCR. Primer sequences are available upon request.
FAIRE assay.
Immunocyto- and Immunohisto-Chemistry.
Cells or embryos were fixed in 4% (wt/vol) PFA prepared in phosphate buffered saline (PBS) for 20 min or 5 h respectively. Fixed embryos were then cryopreserved overnight in 30% (wt/vol) sucrose prepared in PBS and were sectioned into 14-μm sections using a cryostat. Cells or sections were blocked and permeabilized for 1 h in PBS supplemented with 10% (vol/vol) horse serum and 0.2% Triton X-100. Subsequently, they were incubated overnight at 40 °C with the primary antibody, prepared in the same solution. Following several washes in PBS, they were then incubated for 1 h in PBS containing the corresponding secondary antibody and the nuclear dye Hoechst, were then rinsed in PBS and mounted. Images were collected using confocal microscopy. The following primary antibodies were used: anti-Pax6 (DSHB, 1:100), anti-b-III tubulin (Covance, 1:2000), and anti-doublecortin (Chemicon, 1:500).
Bioinformatic Analysis.
See SI Materials and Methods for ChIP-chip array and expression microarray design, hybridization, and analysis and for ChIP-seq and RNA-seq analysis.
Datasets.
Microarray and ChIP-chip raw data have been deposited at the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under accession nos. GSE27245 and GSE27246, respectively. Deep sequencing data were deposited at the GEO database under submission number GSE25533. The reviewer's access links for each of the submissions are provided below:
Supplementary Material
Acknowledgments
We thank Frédéric Zilbermann for technical assistance, Dr. Ken Tsutsui (Okayama University) for providing Top2β (3B6) antibodies, Yi Lisa Lyu (University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School) for providing Top2β+/− mice, and Tim Roloff, Patrick Kopp, and Jean-Francois Spetz from facilities of the Friedrich Miescher Institute. We thank Christian Beisel and Ina Nissen from the Laboratory of Quantitative Genomics at the Department of Biosystems Science and Engineering, Swiss Federal Institute of TechnologyZürich for Illumina sequencing. We also thank members of the D.S. laboratory for critical comments about the manuscript. V.K.T. was supported by a Marie Curie International Incoming Fellowship and a European Molecular Biology Organization long-term postdoctoral fellowship. Research in the laboratory of Y.-A.B. is supported by European Union Grant EuTRACC and a Sinergia project from the Swiss National Foundation. Work in the laboratory of C.M. and F.B. was supported by Deutsche Forschungsgemeinschaft Grant SFB 728-A1. Research in the laboratory of D.S. is supported by the Novartis Research Foundation, European Union Grants LSHG-CT-2004-503433 and LSHG-CT-2006-037415, European Research Council Grant ERC-204264, the Swiss Initiative for Systems Biology (Project Cell Plasticity), and, together with Y.-A.B., by a Sinergia project from the Swiss National Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Microarray and ChIP-chip raw data reported in this paper have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE27245 (microarray) and GSE27246 (ChIP-chip)]. Deep sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE25533).
See Author Summary on page 5923 (volume 109, number 16).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119798109/-/DCSupplemental.
References
- 1.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roca J. Topoisomerase II: A fitted mechanism for the chromatin landscape. Nucleic Acids Res. 2009;37:721–730. doi: 10.1093/nar/gkn994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin CA, Sng JH, Patel S, Fisher LM. Novel HeLa topoisomerase II is the II beta isoform: Complete coding sequence and homology with other type II topoisomerases. Biochim Biophys Acta. 1993;1172:283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 7.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase II beta. Bioessays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Drake FH, et al. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J Biol Chem. 1987;262:16739–16747. [PubMed] [Google Scholar]
- 9.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J Cell Biol. 2003;160:645–655. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- 11.Villman K, Ståhl E, Liljegren G, Tidefelt U, Karlsson MG. Topoisomerase II-alpha expression in different cell cycle phases in fresh human breast carcinomas. Mod Pathol. 2002;15:486–491. doi: 10.1038/modpathol.3880552. [DOI] [PubMed] [Google Scholar]
- 12.Goswami PC, Roti Roti JL, Hunt CR. The cell cycle-coupled expression of topoisomerase IIalpha during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol Cell Biol. 1996;16:1500–1508. doi: 10.1128/mcb.16.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proc Natl Acad Sci USA. 2003;100:7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsutsui K, Tsutsui K, Hosoya O, Sano K, Tokunaga A. Immunohistochemical analyses of DNA topoisomerase II isoforms in developing rat cerebellum. J Comp Neurol. 2001;431:228–239. doi: 10.1002/1096-9861(20010305)431:2<228::aid-cne1067>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe M, Tsutsui K, Tsutsui K, Inoue Y. Differential expressions of the topoisomerase II alpha and II beta mRNAs in developing rat brain. Neurosci Res. 1994;19:51–57. doi: 10.1016/0168-0102(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 16.Tsutsui K, Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIbeta in neuronal differentiation. J Biol Chem. 2001;276:5769–5778. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 18.Lyu YL, et al. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 20.Sano K, Miyaji-Yamaguchi M, Tsutsui KM, Tsutsui K. Topoisomerase IIbeta activates a subset of neuronal genes that are repressed in AT-rich genomic environment. PLoS ONE. 2008;3:e4103. doi: 10.1371/journal.pone.0004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 25.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 28.Bibel M, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 29.Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 30.Plachta N, Bibel M, Tucker KL, Barde YA. Developmental potential of defined neural progenitors derived from mouse embryonic stem cells. Development. 2004;131:5449–5456. doi: 10.1242/dev.01420. [DOI] [PubMed] [Google Scholar]
- 31.Tsutsui K, Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIbeta in neuronal differentiation. J Biol Chem. 2001;276:5769–5778. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- 32.Grue P, et al. Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J Biol Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 33.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 34.Isaacs RJ, Harris AL, Hickson ID. Regulation of the human topoisomerase IIalpha gene promoter in confluence-arrested cells. J Biol Chem. 1996;271:16741–16747. doi: 10.1074/jbc.271.28.16741. [DOI] [PubMed] [Google Scholar]
- 35.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 36.Turley H, et al. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br J Cancer. 1997;75:1340–1346. doi: 10.1038/bjc.1997.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 39.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Chan DW, et al. Unbiased proteomic screen for binding proteins to modified lysines on histone H3. Proteomics. 2009;9:2343–2354. doi: 10.1002/pmic.200800600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 42.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 44.Lienert F, et al. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet. 2011;7:e1002090. doi: 10.1371/journal.pgen.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci USA. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dechant G, Barde YA. The neurotrophin receptor p75(NTR): Novel functions and implications for diseases of the nervous system. Nat Neurosci. 2002;5:1131–1136. doi: 10.1038/nn1102-1131. [DOI] [PubMed] [Google Scholar]
- 49.Frade JM. NRAGE and the cycling side of the neurotrophin receptor p75. Trends Neurosci. 2000;23:591–592. doi: 10.1016/s0166-2236(00)01704-5. [DOI] [PubMed] [Google Scholar]
- 50.Underwood CK, Coulson EJ. The p75 neurotrophin receptor. Int J Biochem Cell Biol. 2008;40:1664–1668. doi: 10.1016/j.biocel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 51.von Schack D, et al. Complete ablation of the neurotrophin receptor p75NTR causes defects both in the nervous and the vascular system. Nat Neurosci. 2001;4:977–978. doi: 10.1038/nn730. [DOI] [PubMed] [Google Scholar]
- 52.Jensen AD, Svejstrup JQ. Purification and characterization of human topoisomerase I mutants. Eur J Biochem. 1996;236:389–394. doi: 10.1111/j.1432-1033.1996.00389.x. [DOI] [PubMed] [Google Scholar]
- 53.Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol Biol Cell. 2004;15:5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akimitsu N, et al. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 55.Linka RM, et al. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007;35:3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khélifa T, Casabianca-Pignède MR, René B, Jacquemin-Sablon A. Expression of topoisomerases II alpha and beta in Chinese hamster lung cells resistant to topoisomerase II inhibitors. Mol Pharmacol. 1994;46:323–328. [PubMed] [Google Scholar]
- 57.Bell O, Tiwari VK, Thomä NH, Schübeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 58.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 60.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 62.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 63.Sims RJ, 3rd, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 65.LeRoy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 66.Razin SV. Functional architecture of chromosomal DNA domains. Crit Rev Eukaryot Gene Expr. 1996;6:247–269. doi: 10.1615/critreveukargeneexpr.v6.i2-3.70. [DOI] [PubMed] [Google Scholar]
- 67.Earnshaw WC, Heck MM. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985;100:1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller FD, Kaplan DR. Neurotrophin signalling pathways regulating neuronal apoptosis. Cell Mol Life Sci. 2001;58:1045–1053. doi: 10.1007/PL00000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nykjaer A, Willnow TE, Petersen CM. p75NTR—live or let die. Curr Opin Neurobiol. 2005;15:49–57. doi: 10.1016/j.conb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 70.Plachta N, et al. Identification of a lectin causing the degeneration of neuronal processes using engineered embryonic stem cells. Nat Neurosci. 2007;10:712–719. doi: 10.1038/nn1897. [DOI] [PubMed] [Google Scholar]
- 71.Bryja V, et al. An efficient method for the derivation of mouse embryonic stem cells. Stem Cells. 2006;24:844–849. doi: 10.1634/stemcells.2005-0444. [DOI] [PubMed] [Google Scholar]
- 72.Nikoletopoulou V, et al. Neurotrophin receptor-mediated death of misspecified neurons generated from embryonic stem cells lacking Pax6. Cell Stem Cell. 2007;1:529–540. doi: 10.1016/j.stem.2007.08.011. [DOI] [PubMed] [Google Scholar]