Abstract
In fission yeast, the RNAi pathway is required for centromeric heterochromatin assembly. siRNAs derived from centromeric transcripts are incorporated into the RNA-induced transcriptional silencing (RITS) complex and direct it to nascent homologous transcripts. The RNA-induced transcriptional silencing-bound nascent transcripts further recruit the RNA-directed RNA polymerase complex (RDRC) to promote dsRNA synthesis and siRNA production. Heterochromatin coated with Swi6/Heterochromain Protein 1 is then formed following recruitment of chromatin modification machinery. Swi6 is also required for the upstream production of siRNA, although the mechanism for this has remained obscure. Here, we demonstrate that Swi6 recruits RDRC to heterochromatin through Ers1, an RNAi factor intermediate. An ers1+ mutant allele (ers1-C62) was identified in a genetic screen for mutants that alleviate centromeric silencing, and this phenotype was suppressed by overexpression of either the Hrr1 RDRC subunit or Clr4 histone H3-K9 methyltransferase. Ers1 physically interacts with Hrr1, and loss of Ers1 impairs RDRC centromeric localization. Although Ers1 failed to bind Clr4, a direct interaction with Swi6 was detected, and centromeric localization of Swi6 was enhanced by Clr4 overexpression in ers1-C62 cells. Consistent with this, deletion of swi6+ reduced centromeric localization of Ers1 and RDRC. Moreover, tethering of Ers1 or Hrr1 to centromeric heterochromatin partially bypassed Swi6 function. These findings demonstrate an alternative mechanism for RDRC recruitment and explain the essential role of Swi6/Heterochromain Protein 1 in RNAi-directed heterochromatin assembly.
Keywords: RNA interference, histone H3-lysine 9 methylation
In a eukaryotic cell, the formation of higher order chromatin structure, heterochromatin, is critical for genomic stability, chromosome segregation, and epigenetic gene silencing. Heterochromatic structure is also essential for functional organization of chromosomal domains, such as the centromeres and telomeres, and is defined by specific posttranslational modifications of nucleosome histone tails. The methylation of lysine 9 of histone H3 (H3K9me) is a key marker of heterochromatin and is provided by the methyltransferase Clr4/Suv3-9 (1–3). This H3K9me marker serves as a binding site for Heterochromatin Protein 1 (HP1) family proteins, which provide a platform for recruitment of transacting factors to maintain repressive chromatin structure (4).
In the fission yeast Schizosaccharomyces pombe, the assembly of heterochromatin at centromeres also depends on transcription of the centromeric dg and dh repeats by RNA polymerase II during S phase (5–7) and the subsequent recruitment of RNAi machineries (8–10). In this RNAi system, siRNAs derived from the centromeric repeats and the RNA-induced transcriptional silencing (RITS) complex play a central role to establish H3K9me at centromeric regions (11). RITS complex, containing Argonaute (Ago1), the GW-repeat Tas3 protein, and the chromodomain (CD) protein Chp1, targets nascent centromeric transcripts through base-pairing interactions with Ago1-bound siRNAs and by association of the Chp1 subunit with H3K9me (12, 13). This leads to a recruitment of the RNA-dependent RNA polymerase complex (RDRC), which consists of RNA-dependent RNA polymerase Rdp1, RNA helicase Hrr1, and the polyA polymerase family protein Cid12 (12). RDRC then synthesizes dsRNA, which is processed by Dicer (Dcr1) into siRNAs (12, 14) and loaded initially into the Argonaute siRNA chaperone complex (ARC) (15). Centromeric transcript-derived siRNAs are then transported to RITS complex via the ARC to complete the siRNA amplification cycle. During this process, the chromatin- and RNA-associated RITS complex recruits the Clr4-containing complex (CLRC), which generates the H3K9me heterochromatin marker.
The assembly of these different effector complexes is thought to be mediated by their physical interactions and/or through additional factors (16). RITS complex interacts with RDRC in a Dcr1- and Clr4-dependent manner (12), and this likely facilitates synthesis of dsRNA targets. On the other hand, a direct interaction of Dcr1 with RDRC components and Tas3 (17) suggests that the subsequent dicing process is also coupled with dsRNA synthesis. CLRC also physically associates with RDRC and RITS (18, 19), and the LIM-domain containing protein Stc1 was recently demonstrated to be required for the interaction between Ago1 and CLRC (20).
The HP1 family protein Swi6 specifically binds to H3K9me and forms repressive higher order chromatin by recruiting heterochromatin modification machinery. Although its functional role in the heterochromatin assembly appears to lie downstream of Clr4 and the RNAi pathway, deletion of swi6+ causes unexpected defects in efficient siRNA generation (12, 13, 21, 22). Although Swi6 is hypothesized to support efficient association of RITS and RDRC with centromeric transcripts (12), a direct interaction between Swi6 and RNAi complexes is not detectable in coimmunoprecipitation experiments (12, 23, 24) and the mechanism for this effect has remained unclear.
In a genetic screen for mutants showing heterochromatin defects, we have isolated an allele of ers1+ (ers1-C62) that displayed loss of centromeric silencing. Here, we have identified the Hrr1 subunit of RDRC and Clr4 methyltransferase as a suppressor, specifically, of ers1-C62 and not of the complete ers1Δ null mutant phenotype. Ers1 was shown to interact physically with Hrr1 and Swi6, and these interactions were diminished by ers1-C62 mutation. In the absence of Swi6 or Ers1, RDRC components failed to localize to centromeric DNA repeats. Moreover, artificial tethering of Ers1 or Hrr1 to H3K9me-enriched chromatin partially bypassed Swi6 function. Our findings demonstrate an alternative mechanism for the recruitment of RDRC and an essential role for Swi6/HP1 in RNAi-directed heterochromatin assembly.
Results
To dissect the mechanism of RNAi-directed heterochromatin assembly, a genetic screen was carried out for mutants that displayed a defect in centromere-specific silencing. Complementation analysis showed that 27 of the 31 mutants isolated contained mutations disrupting most known RNAi components (Fig. S1), confirming the integrity of this screen. One of the mutants, denoted as C62, was found to contain an allele of ers1 (also known as rsh1). Ers1 was previously identified in a candidate KO approach and epistasis mapping as being required for RNAi-directed heterochromatic silencing (25, 26), but its function in this process was poorly understood.
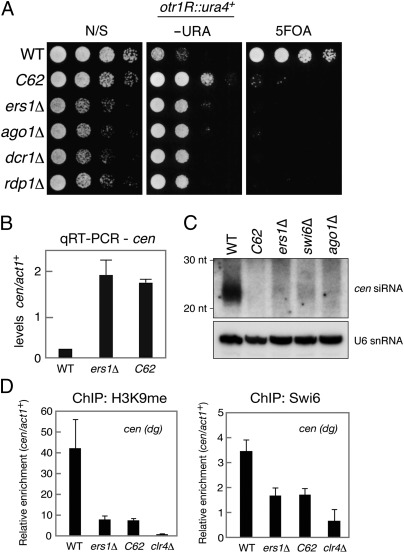
Genome sequencing of the C62 mutant revealed that it possessed a point mutation that converted the serine 234 residue of Ers1 to proline. Because no conserved protein motifs were found in Ers1 (26), it was difficult to infer the likely functional involvement of the Ser234 residue. The phenotype of C62 is similar to that of the ers1Δ null mutant; the C62 mutation resulted in derepression of a ura4+ marker gene integrated into the outermost (otr) pericentromeric repeat of chromosome 1 (otr1R::ura4+) (Fig. 1A), increased levels of noncoding centromeric transcripts (Fig. 1B) and defective siRNA production (Fig. 1C), and reduced levels of H3K9me and Swi6 at centromeric repeats (cen-dg) (Fig. 1D). The C62 mutant cells, however, grew slightly faster on a nonselective medium than those of ers1Δ and other RNAi mutants (Fig. 1A), suggesting that the mutant Ers1C62 protein retained partial function. In addition, C-terminal FLAG-tagged Ers1C62 protein was detected at a similar level as that of WT Ers1 protein (Fig. S2A), confirming that the phenotype was not simply attributable to a difference in protein turnover or accumulation.
Fig. 1.
Characterization of the ers1-C62 mutant. (A) Assay for silencing at otr1R::ura4+. Fivefold serially diluted cultures of WT or mutant strains were spotted onto nonselective (N/S) media, minimal medium lacking uracil (−URA), or counterselection media containing 5FOA. (B) Real-time quantitative RT (qRT)-PCR analysis of centromeric dh transcript levels relative to act1+. (C) Northern blot analysis of centromeric siRNAs prepared from the indicated strains. U6 snRNA was used as a loading control. (D) ChIP analysis of H3K9me2 and Swi6 levels associated with cen (dg), relative to act1+. Immunoprecipitated DNA was subjected to quantitative PCR analysis. Results are the mean ± SD of at least three independent experiments.
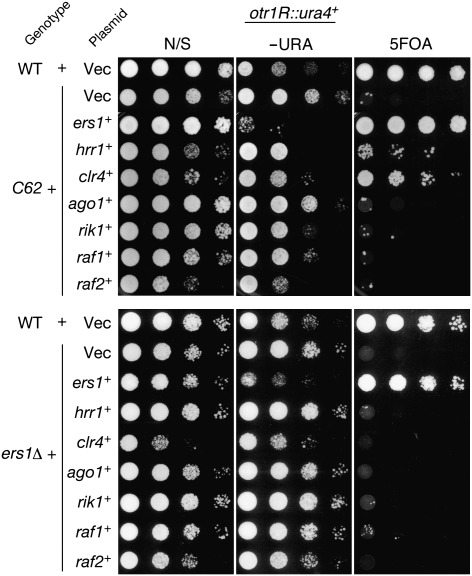
Intriguingly, the silencing defect of ers1-C62 was found to be suppressed by overexpression of hrr1+ or clr4+ in a complementation assay (Fig. 2). A subset of C62 mutant cells expressing hrr1+ or clr4+ grew well on medium containing 5-fluoroorotic acid (5FOA) (Fig. 2). Comparing the complementation by WT ers1+, this suppression was weak; centromeric transcripts were partially silenced (Fig. S2B), and centromeric siRNAs were not efficiently produced in these cells (Fig. S2C). Notably, the above suppression was not observed in the ers1Δ null mutant and was also not observed with other genes involved in the RNAi pathway (Fig. 2). These results suggested that Ers1 is functionally linked with Hrr1 and Clr4, and that the observed suppression was specific to the ers1-C62 allele.
Fig. 2.
Suppression of the C62 mutant phenotype by hrr1+ or clr4+. Multicopy plasmids carrying the indicated genes were introduced into C62 (Upper) and ers1Δ (Lower) mutant cells, and the suppressive function of introduced genes was evaluated by a spotting assay as in Fig. 1A. All indicated genes were expressed under the control of their own promoters. N/S, nonselective; −URA, minimal medium lacking uracil.
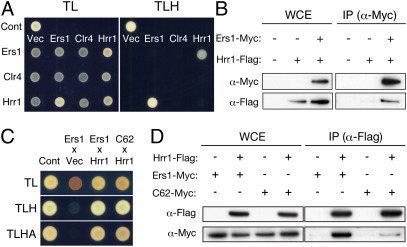
Multicopy suppressors are well known to act through a dosage effect, such that increased levels of the expressed protein complement an impaired protein-protein interaction. To establish whether the functional link between Ers1 and Hrr1 or Clr4 involved a physical association, the potential interaction between these proteins was next examined by a yeast two-hybrid (YTH) assay (Fig. 3A and Fig. S3). Ers1 was found to associate with Hrr1, whereas no interaction between Ers1 and Clr4 or between Hrr1 and Clr4 was detected (Fig. 3A). The interaction between Ers1 and Hrr1 detected in the YTH was further confirmed by coimmunoprecipitation analysis of S. pombe cell lysate (Fig. 3B). Although mutant Ers1C62 associated with Hrr1 with a similar affinity as that of WT Ers1 in the YTH assay (Fig. 3C), the in vivo association between Ers1 and Hrr1 was found to be diminished by the C62 mutation (Fig. 3D). This result suggested that overexpression of Hrr1 complemented a weakened interaction between Hrr1 and mutant Ers1C62.
Fig. 3.
Ers1 physically interacts with Hrr1. (A) YTH analysis of Ers1 and its suppressors. Ers1, Clr4, and Hrr1 were expressed as fusion proteins with either the DNA binding domain or activation domain, and their physical interaction was assayed in diploid strains. Expression of GAD- or GBD-fused proteins was confirmed by Western blot analyses (Fig. S3). p53 and T antigen were used as positive controls, and empty vector was used as a negative control. TL, SD/−Trp/−Leu medium; TLH, SD/−Trp/−Leu/−His medium. (B) Coimmunoprecipitation analyses of Ers1 and Hrr1. IP, immunoprecipitated fraction; WCE, whole-cell extract. (C) YTH analysis showing interaction of WT or mutant Ers1 with Hrr1. TLA, SD/−Trp/−Leu/−Ade medium. (D) Coimmunoprecipitation analysis of mutant Ers1 and Hrr1. Ers1-Myc and Ers1C62-Myc (C62-Myc) were immunoprecipitated with Hrr1-Flag and detected as in B.
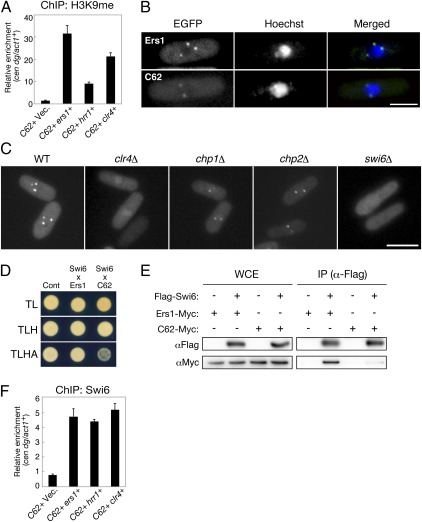
In the YTH assay, no detectable interaction between Ers1 and Clr4 was observed (Fig. 3A). This result argues against in vivo suppression of the C62 phenotype by Clr4 overexpression being attributable to an increased affinity between Clr4 and mutant Ers1C62. It was plausible that Clr4 overexpression might increase the H3K9me levels and affect centromeric association of other chromatin proteins. To test this idea, the levels of H3K9me at the centromeric region were examined in a ChIP assay. ChIP analysis of the clr4+ overexpression cells confirmed that the level of H3K9me was increased at the centromeric region (Fig. 4A). The level of Clr4 protein appeared to be linked to that of the H3K9me levels at heterochromatic regions even in WT cells, because Clr4 overexpression led to an increased H3K9me level at the centromeric repeat locus (Fig. S2D).
Fig. 4.
Heterochromatic localization of Ers1 is dependent on Swi6/HP1. (A) ChIP analysis of H3K9me2 levels associated with cen dg, relative to control act1+ locus. C62 cells expressing ers1+, hrr1+, or clr4+ were picked up from colonies on 5FOA plates and subjected to ChIP analysis by real-time PCR. C62 cells harboring an empty vector (Vec.) were used as a control. Results are the mean ± SD of at least three independent experiments. (B) EGFP-Ers1WT and EGFP-Ers1C62 localization in WT cells. (Scale bar, 2 μm.) Hoechst 33342 was used for staining nuclei of living cells. (C) EGFP-Ers1WT localization in clr4Δ, chp1Δ, chp2Δ, and swi6Δ mutants. (Scale bar, 5 μm.) (D) YTH analysis showing the interaction of WT or mutant Ers1 with Swi6. (E) Coimmunoprecipitation analysis of Ers1 and Swi6 proteins. Myc-tagged WT or mutant (C62) Ers1 was coimmunoprecipitated with Flag-Swi6. IP, immunoprecipitated fraction; WCE, whole-cell extract. (F) ChIP analysis of Swi6 levels associated with cen dg, relative to control act1+ locus. Cells were prepared as in A.
Previous reports showed that Ers1 associates with heterochromatin (25) and that YFP-fused Ers1 displays a characteristic nuclear dot fluorescent pattern (27). To clarify the mechanism for Clr4 suppression of the ers1-C62 phenotype, the subcellular localization of Ers1 was examined. Tagging of Ers1 protein with EGFP did not disrupt the protein function, which was confirmed by complementation of the silencing defect in ers1Δ cells (Fig. S4A). Similar to that previously observed, EGFP-Ers1WT showed a distinct nuclear dot pattern in WT cells consistent with a localization to heterochromatin (Fig. 4B and Fig. S4B). In contrast, EGFP-Ers1C62 showed a more diffuse signal with weak nuclear dots (Fig. 4B and Fig. S4 C and D), suggesting that the correct localization of Ers1 was impaired by the C62 mutation. Moreover, the nuclear dot localization of EGFP-Ers1WT was completely abolished in clr4Δ cells (Fig. 4C), indicating that the activity of Clr4 was also required for Ers1 localization in the nucleus.
Clr4 activity promotes the binding of CD proteins to heterochromatin via H3K9me (1, 3). To test the hypothesis that the nuclear dot localization of Ers1 was attributable to an interaction with H3K9me-bound CD proteins, EGFP-Ers1WT localization was next examined in swi6Δ, chp1Δ, and chp2Δ cells. The localization of EGFP-Ers1WT was clearly abolished in swi6Δ cells similar to that observed in clr4Δ, whereas WT localization patterns were retained in the chp1Δ and chp2Δ cells (Fig. 4C and Fig. S4C). Coimmunoprecipitation, YTH, and direct YTH screening analyses confirmed that Ers1 interacts with Swi6 (Fig. 4 D and E and Table S1), suggesting that a physical association with Swi6, but not the other CD proteins, was required for the heterochromatic localization of Ers1. Moreover, the interaction between Ers1 and Swi6 was weakened by the presence of the C62 mutation (Fig. 4 D and E). Taken together, these results suggested that Ers1 associates with both Hrr1 and Swi6, and that the C62 mutation impairs both associations.
Although overexpression of clr4+ was able to suppress the ers1-C62 silencing defect (Fig. 2), swi6+ overexpression failed to do so (Fig. S2E). Swi6 accumulation at heterochromatin is considered to be limited by the availability of H3K9me binding sites (28), and ChIP analysis of the clr4+-overexpressed C62 cells confirmed that the level of Swi6 was increased at the heterochromatin region (Fig. 4F). The clr4+ suppression phenotype can therefore be explained by the higher H3K9me activity that subsequently leads to enhanced Swi6 binding at heterochromatin regions.
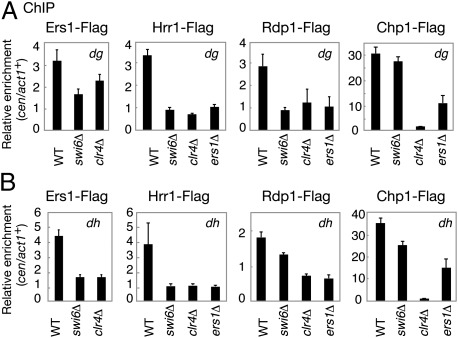
Swi6 was shown previously to be required for efficient siRNA generation (12, 13, 21, 22). However, the mechanism for this effect remained unclear. Based on the observation that Ers1 can associate with both Hrr1 and Swi6, it was speculated that Swi6 is able to recruit RDRC through its interaction with Ers1. This possibility was tested in a ChIP assay of the heterochromatic localization of Ers1 and RDRC components. In agreement with the cytological analysis, the association of Ers1-Flag with the centromeric dg and dh repeats was greatly reduced in clr4Δ and swi6Δ cells (Fig. 5 A and B). The localizations of Hrr1-Flag and Rdp1-Flag at centromeric repeats were also found to be severely compromised in both ers1Δ and swi6Δ mutant cells (Fig. 5 A and B). This is also consistent with a previous report that the deletion of swi6+ decreases the centromeric localization of Rdp1 (14). These results support the idea that the heterochromatic localization of RDRC requires Ers1 and that, in turn, Ers1 localization depends on Swi6.
Fig. 5.
Ers1 and RDRC localization is dependent on Swi6. (A and B) ChIP analysis of the Ers1-Flag, Hrr1-Flag, Rdp1-Flag, and Chp1-Flag levels associated with cen dg and dh loci relative to act1+, normalized to an untagged control strain.
Deleting any of the RNAi components reduces H3K9me and Swi6 recruitment at centromeres (8–10). In contrast to centromeres, redundant mechanisms operate at the mating type locus and telomeres, and H3K9me is retained at these regions in RNAi mutant cells. ChIP analyses showed that Ers1 localization at the mating type locus (cenH) and telomeres was severely decreased in swi6Δ and clr4Δ cells (Fig. S5A). In contrast, a high level of Ers1 was detected at telomeres in hrr1Δ cells (Fig. S5C). These results were consistent with the cytological results that the EGFP-Ers1 nuclear dots were diffused in swi6Δ and clr4Δ cells but partially retained in RNAi mutant cells (Fig. S4 B–D). ChIP analyses also showed that high levels of Swi6 were detected at the mating-type locus and telomeres in ers1Δ and hrr1Δ cells (Fig. S5 B and D), in agreement with the result that EGFP-Swi6 nuclear dots were detected in RNAi mutant cells (Fig. S4 B–D). These results suggested that Swi6 plays an essential role in Ers1 localization at the mating type locus and telomeres.
RDRC can physically interact with RITS complex, and it was suggested to be recruited to activated RITS on nascent transcripts through this interaction (12). In swi6Δ cells, Chp1 association with the centromeric dg or dh repeat locus decreased partially (70–80%) but was still much greater than that observed in clr4Δ cells (Fig. 5 A and B). This was in contrast to Hrr1 or Rdp1, whose association was severely reduced in swi6Δ cells (10–20%) to levels comparable to those of clr4Δ cells. These observations suggest that in addition to direct association with the RITS complex, there are alternative pathways to recruit RDRC to heterochromatic regions; moreover, they suggest that a Swi6- and Ers1-mediated interaction plays a critical role in RDRC recruitment.
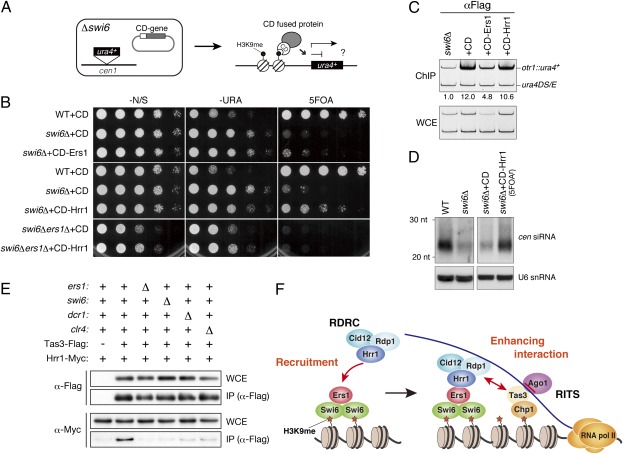
To test directly whether the siRNA production defect in swi6Δ cells (Fig. 1C) was caused by the delocalization of Ers1 and RDRC, Ers1 and Hrr1 were fused to a CD to tether the proteins directly to H3K9me (Fig. 6A) and expressed in swi6Δ cells containing the otr1R::ura4+ heterochromatin marker gene (Fig. 6B). The swi6 mutation does not change in H3K9me at the otr1R::ura4+ marker locus (3, 29). ChIP analysis confirmed that CD-Ers1 and CD-Hrr1 indeed targeted the H3K9me-enriched heterochromatic region (Fig. 6C). As a result of repression of the ura4+ gene, WT cells grow poorly on media without uracil but showed normal growth on media containing the counterselective agent 5FOA [Figs. 1A (WT) and 6B (WT + CD)]. Deletion of swi6+ caused derepression of otr1R::ura4+; thus, the swi6Δ cells grew well on the media without uracil, whereas growth on 5FOA media was inhibited (Fig. 6B and Fig. S6; swi6Δ + CD). Notably, the expression of CD-Ers1 or CD-Hrr1 gave rise to 5FOA-resistant colonies, indicating partial repression of otr1R::ura4+ (Fig. 6B and Fig. S6). Although CD-Ers1 expression did not have a noticeable effect on siRNA levels, CD-Hrr1 expression resulted in a marked increase in the level of centromeric siRNAs (Fig. 6D). These results suggested that the tethering of Hrr1 at the pericentromeric region was sufficient to activate the RNAi pathway and that the siRNA reduction in swi6Δ cells is caused, at least in part, by delocalization of RDRC. The difference between CD-Ers1 and CD-Hrr1 is likely to reflect a hierarchical relationship between Hrr1 and Ers1 and to be attributable to a difference in efficiency of RDRC recruitment to H3K9me-enriched heterochromatic regions.
Fig. 6.
Tethering of Ers1 and Hrr1 to heterochromatin promotes centromeric silencing and siRNA generation. (A) Schematic diagram of the constructs used. The vector expressed CD-fused proteins and a ura4+ marker gene inserted at the cen1 locus. (B) Assay for the silencing of otr1R::ura4+. CD alone or CD-fused Ers1 or Hrr1 was ectopically expressed in WT or swi6Δ cells. Other independently isolated transformants were also analyzed (Fig. S6). (C) ChIP analysis of the levels of CD-fused proteins associated with the otr1R::ura4+ locus in swi6Δ cells. The otr1R::ura4+ signal enrichment relative to the control minigene (ura4DS/E) signal is shown beneath each lane. WCE, whole-cell extract. (D) Northern blot analysis of centromeric siRNAs. The level of cellular siRNAs isolated from FOA-resistant, CD-Hrr1–expressing swi6Δ cells was compared with that of control swi6Δ cells expressing CD alone. U6 snRNA was used as a loading control. (E) Coimmunoprecipitation assay of Tas3-Flag and Hrr1-Myc in WT or indicated mutant background. (F) Model for Swi6-mediated siRNA generation at pericentromeric heterochromatin. Details are provided in the main text.
Given that Ers1 associates with both Swi6 and Hrr1, one hypothesis is that Ers1 acts solely as a bridge between Swi6 and Hrr1. However, the expression of CD-Hrr1 did not induce centromeric silencing in swi6Δers1Δ double-mutant cells (Fig. 6B), indicating that Ers1 has a role in the RNAi pathway in addition to connecting Swi6 and Hrr1. A potential role for Ers1 in the interaction between RITS and RDRC was thus examined next. As previously observed (12), coimmunoprecipitation analyses showed that Hrr1 associates with the Tas3-containing RITS complex in WT cells, which was severely impaired in dcr1Δ or clr4Δ mutant cells (Fig. 6E). Strikingly, ers1Δ or swi6Δ also abolished the association between Hrr1 and Tas3. These findings suggest that Ers1 and Swi6 not only recruit RDRC but that they also further promote its association with RITS.
To gain further insight into the mechanisms underlying the assembly of the effector complexes, the interactions between each effector complex component and Ers1 were next examined in a combinatorial two-hybrid assay. This analysis confirmed the finding that Ers1 physically interacts with Swi6 and Hrr1, and revealed mutual interactions among the RDRC components (Fig. S7). The GW-rich domain of Tas3 was previously shown to associate with Ago1 (30, 31), but this interaction was not detected in the current assay, suggesting that some additional factors and/or posttranslational modifications may be required. Interestingly, our assay uncovered previously uncharacterized interactions between Tas3 and two RDRC components, Rdp1 and Cid12 (Fig. S7), which may be used to enhance stable interactions between RDRC and RITS in the later stages of the assembly process.
Discussion
Ers1 was previously identified in a candidate KO approach and epistasis mapping as being required for RNAi-directed processes (25, 26), although its function remained unclear. In this study, a mutant allele for ers1+ (ers1-C62) was isolated from a genetic screen for heterochromatic silencing mutants and revealed a functional link with both Clr4 and Hrr1. Coimmunoprecipitation analyses showed that the C62 mutation impairs the physical interaction identified between Ers1 and both Swi6 and Hrr1. Because no conserved motif was found in Ers1, it remains unclear exactly how the C62 mutation affects Ers1 function, and further biochemical analyses to map interacting domain(s) will be important to elucidate the molecular mechanism by which Ers1 links Swi6 and Hrr1.
Although the Swi6 levels at centromeric regions were restored by the clr4+ overexpression, siRNAs were not efficiently produced in the cells overexpressing clr4+ (Fig. S2C). It is likely that the elevated levels of chromatin-bound Swi6 also contributed to repress the otr1R::ura4+ expression apart from enhancing its binding to Ers1C62. Considering that the ers1-C62 allele is required for the suppression by clr4+ overexpression (Fig. 2), this suppression appears to be mediated by a combined effect of increased Swi6 dosage and enhanced interaction between Swi6 and Ers1C62.
Although several groups have isolated Swi6-interacting factors, peptides for Ers1 have not been identified in purified fractions to date (23, 24). This suggests that Ers1 may not be a stable binding partner of Swi6 and that although Swi6 plays a primary role for Ers1 localization, additional protein-protein interactions may be required to target Ers1 to heterochromatic regions.
A previous report suggested that RDRC is recruited to nascent transcripts through its physical interaction with the RITS complex (12). The above results demonstrate that direct interaction alone is not sufficient to achieve this and that the recruitment of RDRC by Swi6 and Ers1 is a prerequisite for its targeting to heterochromatin, and presumably to the nascent transcripts. Ers1 may form a platform with Swi6 to recruit RDRC to H3K9me-enriched heterochromatin after H3K9me is provided by Clr4 during S-phase progression (32) (Fig. 6F). It is conceivable that Tas3 forms another platform with Chp1 for siRNA-bound Ago1 and that the components recruited for these two platforms are assembled by physically associating with each other and/or by associating with nascent transcripts. Although H3K9me is the primary marker defining heterochromatic regions, detailed mechanisms linking nascent RNA and chromatin modification remain to be defined in many organisms. The present study has uncovered a previously undescribed link between H3K9me and RDRC demonstrating that conserved H3K9me-binding proteins play a primary role in assembling essential RNA processing factors.
Materials and Methods
Strains and Plasmids.
The strains used in this study are listed in Table S2. Deletion and chromosomal-tagged strains were generated by a PCR-based targeting protocol. Ers1 cDNA was inserted into the EGFP expression vector pYB228 (29) to observe EGFP-Ers1 localization. Multicopy plasmids carrying RNAi genes were obtained from a pTN-L1 fission yeast genomic library (pAL-KS library; National BioResource Project). To express CD-fused ers1+ or hrr1+ from its native promoters, two copies of the coding sequence of Swi6CD (78–136 aa) were tandemly cloned with the sequence for 5× Flag tag. The resultant fragment (Flag-CD2) was then ligated with the ers1+ promoter (Pers1) and ers1+ cDNA, or the hrr1+ promoter (Phrr1) and hrr1+ cDNA, and introduced into a pRE vector harboring the LEU2 marker (28). Plasmids carrying Pers1-Flag-CD2 and Phrr1-Flag-CD2 were used as controls for each spot assay.
Silencing Assay.
Silencing assays were performed using unsaturated cultures grown in yeast extract with adenine (YEA) medium. Serial dilutions (5-fold) were prepared from a culture of 1 × 107 cells/mL, and 5 μL was spotted on plates with YEA medium, minimal nonselective medium, minimal medium lacking uracil, or minimal medium containing 5FOA. The plates were then incubated at 30 °C for 2.5–4 d.
YTH Analysis.
YTH analysis was performed using the MatchMaker Yeast Two-Hybrid system (Clontech). The coding sequences of ers1+, hrr1+, clr4+, swi6+, and other RNAi genes were cloned into YTH vectors (pGADT7 and pGBKT7). These plasmids were then transformed into two-hybrid strains (AH109 and Y187) to detect the proteins’ interactions.
ChIP.
ChIP was performed as described previously (29). Anti-FLAG M2 affinity gel (Sigma), purified polyclonal anti-Swi6, and anti-H3K9me2 monoclonal antibodies (29) were used for immunoprecipitation. The primers used in this study are listed in Table S3. The PCR products were separated and analyzed on 10% (wt/vol) polyacrylamide gels (ATTO), and the LAS3000 system (Fujifilm) was used for the quantification analysis. Alternatively, the immunoprecipitated DNAs were directly analyzed by real-time PCR (7300 Real-Time PCR system; ABI).
Additional information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Derek Goto for kind advice on detecting siRNAs and for critical reading of the manuscript. We also thank Terry Orr-Weaver for critical reading, our laboratory members at the Institute of Physical and Chemical Research Center for Development Biology for helpful discussions, and S. Seno for excellent secretarial work. This research was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116972109/-/DCSupplemental.
References
- 1.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 2.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 4.Hiragami K, Festenstein R. Heterochromatin protein 1: A pervasive controlling influence. Cell Mol Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato H, et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 6.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 7.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20(2):134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloc A, Martienssen R. RNAi, heterochromatin and the cell cycle. Trends Genet. 2008;24:511–517. doi: 10.1016/j.tig.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motamedi MR, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 13.Bühler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buker SM, et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- 16.Goto DB, Nakayama JI. RNA and epigenetic silencing: Insight from fission yeast. Dev Growth Differ. 2011;54(1):129–141. doi: 10.1111/j.1440-169X.2011.01310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 19.Gerace EL, Halic M, Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell. 2010;39:360–372. doi: 10.1016/j.molcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayne EH, et al. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iida T, Nakayama J, Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell. 2008;31:178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motamedi MR, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer T, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roguev A, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rougemaille M, Shankar S, Braun S, Rowley M, Madhani HD. Ers1, a rapidly diverging protein essential for RNA interference-dependent heterochromatic silencing in Schizosaccharomyces pombe. J Biol Chem. 2008;283:25770–25773. doi: 10.1074/jbc.C800140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 28.Sadaie M, et al. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partridge JF, et al. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Till S, et al. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 32.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.