Abstract
Influenza viruses are the cause of yearly epidemics and occasional pandemics that represent a significant challenge to public health. Current control strategies are imperfect and there is an unmet need for new antiviral therapies. Here, we report the identification of small molecule compounds able to effectively and specifically inhibit growth of influenza A and B viruses in cultured cells through targeting an assembly interface of the viral RNA-dependent RNA polymerase. Using an existing crystal structure of the primary protein–protein interface between the PB1 and PA subunits of the influenza A virus polymerase, we conducted an in silico screen to identify potential small molecule inhibitors. Selected compounds were then screened for their ability to inhibit the interaction between PB1 and PA in vitro using an ELISA-based assay and in cells, to inhibit nuclear import of a binary PB1–PA complex as well as transcription by the full viral ribonucleoprotein complex. Two compounds emerged as effective inhibitors with IC50 values in the low micromolar range and negligible cytotoxicity. Of these, one compound also acted as a potent replication inhibitor of a variety of influenza A virus strains in Madin-Darby canine kidney (MDCK) cells, including H3N2 and H1N1 seasonal and 2009 pandemic strains. Importantly, this included an oseltamivir-resistant isolate. Furthermore, potent inhibition of influenza B viruses but not other RNA or DNA viruses was seen. Overall, these compounds provide a foundation for the development of a new generation of therapeutic agents exhibiting high specificity to influenza A and B viruses.
Influenza A (FluA) and B (FluB) viruses cause highly infectious respiratory diseases, characterized by high morbidity and significant mortality. Both viruses are responsible for seasonal epidemics, which affect up to 20% of the population and result in hundreds of thousands of deaths each year (1). At irregular intervals, antigenically novel strains of FluA provoke pandemic outbreaks with higher attack rates and potentially more severe disease. The 1918 “Spanish” pandemic remains the worst example, causing upwards of 50 million deaths. Thus, both types of virus pose a large threat to public health.
Influenza infections can be controlled by vaccination and antiviral drugs. However, vaccines need regular updating because the virus is antigenically labile and are not always protective. Only two classes of drugs are currently approved for the treatment of influenza: M2 ion channel blockers (adamantanes) and neuraminidase (NA) inhibitors (2). Adamantanes inhibit FluA replication by blocking virus entry. However, they have no activity against FluB viruses, are often associated with serious side effects, and suffer from rapid emergence of drug-resistant viruses (3). NA inhibitors block the release of virions after budding from the host cell (4). They exhibit activity against both FluA and FluB viruses but can also cause side effects and be nullified by resistance (5). Thus, there is a clear need to develop novel influenza virus inhibitors, preferably directed against other viral targets.
The influenza virus RNA polymerase is a heterotrimeric complex of three virus-encoded proteins (PB1, PB2, and PA), all essential for viral RNA synthesis (1). PB1 is the nucleic acid polymerase and forms the backbone of the complex (6, 7). PB2 and PA play accessory roles, best defined for viral transcription (8–10). The three subunits bind each other noncovalently in a set of interactions that are essential for polymerase function. Although the polymerase forms a globular structure (11), the primary protein–protein interactions are via the N terminus of PB1 with the C terminus of PA (12–14) and the C terminus of PB1 with the N terminus of PB2 (14, 15). In contrast to the viral glycoproteins, the polymerase is highly conserved between different viral strains (1). Thus, inhibition of these interactions represents an attractive strategy for the development of drugs with broad efficacy against all influenza virus strains. Recently, two crystallographic structures of a truncated form of PA bound to a PB1-derived peptide have been published (16, 17). These structures revealed that the PA–PB1 binding interface consists of an N-terminal 310 helix from PB1 that binds into a hydrophobic groove in the C terminus of PA. Importantly, the structures showed that relatively few residues drive binding of PB1 to PA, suggesting the potential for small molecule-mediated inhibition.
Using the crystallographic information, we conducted an in silico screening of 3 million small molecule structures to search for inhibitors of the PA–PB1 interaction. From this screening, 32 compounds emerged as candidates. Here, we evaluated the ability of the compounds to disrupt PA–PB1 interactions both in vitro and in cells and thus inhibit viral replication. One compound (compound 1) was identified as a potent and selective inhibitor of both FluA and FluB viruses.
Results
Identification of Hits in an in Silico Screen.
Three million compounds from the ZINC database were screened using FLAP (fingerprints for ligands and proteins) software (18) and the crystal structure of a C-terminal fragment of PA (amino acids 257–716) bound to a PB1-derived peptide (Protein Data Bank code 3CM8) (17) as a template (SI Text and Fig. S1). From the virtual screening, 32 molecules were selected.
Development of an Assay to Identify Inhibitors of the PA–PB1 Interaction.
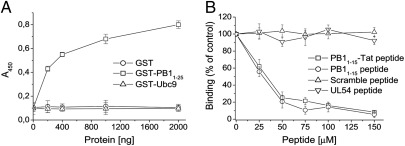
To investigate whether the 32 small molecules selected by virtual screening could indeed inhibit binding between PA and PB1, we developed an ELISA to measure PA–PB1 interactions. Wells coated with 6His–PA239–716, a 6His-tagged form of the PA C-terminal domain were incubated with GST–PB11–25, a fusion protein consisting of the N-terminal 25 residues of PB1 (which are sufficient to bind the PA C-terminal domain) (17, 19), fused to GST. As expected, addition of increasing amounts of GST–PB11–25 resulted in increasing absorbance (Fig. 1A). In contrast, no binding was observed when either GST alone or GST–Ubc9, an irrelevant GST fusion protein, was added. The assay therefore specifically measured interactions between PA239–716 and the PB1-derived peptide.
Fig. 1.
PA–PB1 interaction assay. (A) Increasing amounts of GST–PB11–25 (□), GST–Ubc9 (△), or GST (○) were added to wells coated with 400 ng of 6His–PA239–716. Binding of GST-based proteins was detected with an HRP-conjugated anti-GST antibody, followed by measurement of the absorbance at 450 nm. (B) Increasing concentrations of PB11–15–Tat peptide (□), PB11–15 peptide (○), UL54 peptide (▽), or a scramble peptide (△) were added together with 200 ng of GST–PB11–25 to wells coated with 400 ng of 6His–PA239–716. Binding of GST–PB11–25 was detected as described above. The percentage of the absorbance at 450 nm measured in the presence of inhibitors with respect to that measured in the absence of inhibitors is plotted. Data shown represent the mean ± SD of three independent experiments.
To further validate the assay, we tested a known inhibitor of the PA–PB1 interaction (20), by using increasing concentrations of a synthetic PB1-derived peptide (amino acids 1–15) fused to the translocating domain of HIV Tat protein (PB11–15–Tat peptide). This peptide inhibited the PA–PB1 interaction with an apparent 50% inhibitory concentration (IC50) of 35.5 ± 3.1 μM; a similar IC50 value (28.4 μM) was obtained for an unfused PB11–15 peptide (Fig. 1B). In contrast, a scrambled peptide that contained the same amino acids as PB11–15 but in a random order did not block PA–PB1 binding (Fig. 1B). Similarly, a peptide corresponding to the last 22 residues of human cytomegalovirus (HCMV) UL54 protein (UL54 peptide), which has been shown to inhibit the interaction between the two subunits of HCMV DNA polymerase (21), had no inhibitory effect (Fig. 1B). Thus, this assay could detect specific inhibition of the interaction between PA and the PB1-derived peptide.
Ability of the Compounds to Inhibit the PA–PB1 Interaction.
We then used the ELISA to test the ability of the 32 hits to inhibit binding between PA and PB1. Because their solubility ranged between 200 and 1,000 μM in the experimental conditions, each compound was first screened at a fixed concentration of 50 μM. Fourteen compounds consistently caused a decrease in absorbance (Table S1). The compounds were similarly assayed in an ELISA we previously used to characterize inhibitors of interactions between the HCMV DNA polymerase subunits UL54 and UL44 (21, 22). None of the compounds that inhibited the PA–PB1 interaction affected UL54–UL44 binding (Table S1).
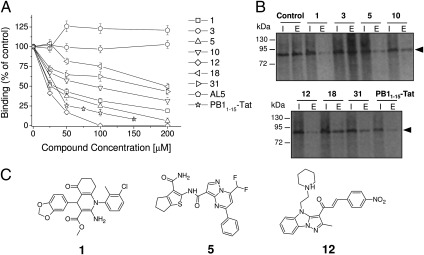
Next, we performed dose–response analyses of the inhibition of the PA–PB1 interaction for the 14 active compounds, and one inactive compound (compound 3) as a negative control. Of these, three compounds—1, 5, and 12—caused a dose-dependent reduction in absorbance with IC50 values similar to or lower than that of the PB11–15–Tat peptide (∼20–30 μM; Fig. 2A and Table 1). Three other compounds—10, 18, and 31—inhibited the PA–PB1 interaction in a titratable manner, but with higher IC50 values (∼91, 200, and 171 μM, respectively; Fig. 2A and Table 1). The remaining compounds did not exhibit reproducible dose-dependent activity (Table S1). As further specificity controls, the compounds were titrated by ELISA for inhibition of HCMV DNA polymerase subunit interactions. None of the compounds exhibited a dose-dependent reduction in absorbance up to a concentration of 200 μM. Additionally, an unrelated small molecule, AL5, previously shown to inhibit the interactions between the HCMV DNA polymerase subunits (22) did not interfere with PA–PB1 binding (Fig. 2A).
Fig. 2.
Inhibition of the PA–PB1 interaction in vitro. (A) Dose-dependent activities of the compounds in ELISA PA–PB1 interaction assays. Increasing concentrations of the indicated inhibitors were added together with 200 ng of GST–PB11–25 to wells coated with 400 ng of 6His–PA239–716. Binding of GST–PB11–25 was quantified as before (Fig. 1). Data shown represent the mean ± SD of three independent experiments. (B) Activities of the compounds in His-pulldown assays. Purified 6His–PA239–716 was incubated with in vitro translated radiolabeled PB1 and the indicated inhibitors and allowed to bind to Ni-NTA columns. The columns were washed and proteins eluted with imidazole. Radiolabeled PB1 protein (arrowed) was visualized by autoradiography after SDS/PAGE. Lane I, input; lane E, eluted. (C) Chemical structures of the three compounds that most efficiently inhibited the PA–PB1 interaction.
Table 1.
Summary of activities of hit compounds against FluA and FluB viruses
| Compound | ELISA interaction assay (IC50, μM) | FluA minireplicon assay (IC50, μM) | FluA plaque reduction assay (ED50, μM) | FluA viral yield reduction assay (ED50, μM) |
FluB plaque reduction assay (ED50, μM) | Cytotoxicity (MTT) assay (CC50, μM)* | |
| 12 h p.i. | 48 h p.i. | ||||||
| 1 | 30.4 ± 4.5 | 18.5 ± 3.8 | 12.2 ± 2.6/22.5 ± 3.7† | 1.5 ± 0.9 | 19.4 ± 3.6 | 12.5 ± 2.2/21.0 ± 2.8† | >1,000 |
| 3 | >200 | >100 | >100 | >100 | >100 | >100 | >250 |
| 5 | 25.4 ± 3.9 | 31.4 ± 4.2 | 75.5 ± 8.8/>100† | 30.7 ± 4.1 | >100 | >100 | >1,000 |
| 10 | 90.7 ± 2.4 | >100 | >100 | ND | ND | ND | >250 |
| 18 | 199.5 ± 5.3 | >100 | >100 | ND | ND | ND | >250 |
| 31 | 170.6 ± 4.7 | >100 | >100 | ND | ND | ND | >250 |
| PB11–15-Tat peptide | 35.5 ± 3.1 | 15.5 ± 2.6 | 48.4 ± 3.7 | ND | ND | ND | >250 |
| RBV | ND | ND | 8.4 ± 2.3/18.6 ± 2.9† | 9.9 ± 0.8 | 9.3 ± 1.2 | 14.3 ± 5.1/20.2 ± 3.4† | >250 |
ND, not determined; RBV, ribavirin.
*Reported CC50 values are those determined in MDCK cells.
†ED50 reported for compounds 1, 5, and RBV represents the range of values determined in plaque reduction assays with different FluA and FluB virus strains.
We then tested whether the compounds that inhibited the interaction between PA and the PB1-derived peptide inhibited binding of PA to full-length PB1. To this end, we developed a pulldown assay using the 6His–PA239–716 fusion protein and in vitro translated, full-length PB1. Consistent with the ELISA studies, the PB11–15–Tat peptide and compounds 1, 5, 10, 12, 18, and 31 interfered with the interaction between PA and PB1, whereas compound 3 did not (Fig. 2B). A good correlation between the amount of PB1 pulled down in these assays and the degree of inhibition in the ELISAs was seen, suggesting that the inhibitory compounds affected PA binding to full-length PB1 with similar efficiency to that seen for the PB1-derived peptide.
Thus, we identified six small molecules that inhibit the PA–PB1 interaction specifically and in a dose-dependent manner. Fig. 2C reports the structures of the most active compounds.
Cytotoxicity of the Compounds.
Next, we tested the cytotoxicity of the test compounds, as well as known antiinfluenza drugs as a reference, in a panel of cell lines. Ribavirin (RBV) and oseltamivir showed CC50 (concentration that causes a decrease of cell viability of 50%) values >250 μM in all tested cell lines (Table S2), as previously reported (23, 24). Compound 12 exhibited elevated cytotoxicity; thus it was excluded from further analysis. In contrast, compounds 1, 3, 5, 10, 18, and 31 showed no significant cytotoxicity up to concentrations of 250–1,000 μM (Table S2).
Functional Inhibition of PA–PB1 Interactions in Cells.
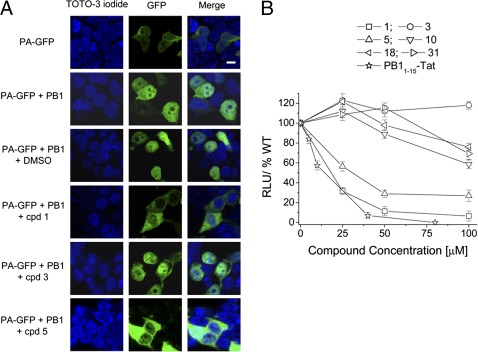
We then tested the ability of the active compounds to interfere with functional interactions between PA and PB1 in a cellular context. First, we investigated whether the inhibitors could affect PA–PB1 binding in the cell cytosol and consequently block the intranuclear translocation of PA, which requires formation of a PA–PB1 complex (25). Thus, we transfected HEK 293T cells with plasmids expressing PB1 and a PA–GFP fusion protein and analyzed the intracellular localization of PA–GFP in the presence or absence of test compounds. As previously shown (25), individually expressed PA–GFP was largely cytoplasmic, whereas coexpression of PA–GFP with PB1 resulted in marked nuclear accumulation of PA (Fig. 3A). Treatment of PA–PB1 coexpressing cells with compounds 1 and 5 reduced PA nuclear localization, whereas DMSO vehicle or compound 3 had no effect (Fig. 3A). Weak inhibition was observed upon treatment with compounds 10, 18, and 31 (Fig. S2).
Fig. 3.
Effects of the compounds on PA–PB1 interactions in a cellular context. (A) Effect of compounds on intranuclear localization of the PA–PB1 complex. HEK 293T cells were transfected with plasmids expressing PB1 and a PA–GFP fusion protein in the presence of test compounds or DMSO as a control. Cells transfected with the PA–GFP-expressing plasmid alone served as a negative control. At 24 h posttransfection, cells were examined by confocal laser scanning microscopy. Individual green (GFP) and blue (TOTO-3 iodide) channels and merged images are shown. (Scale bar, 5 μm.) (B) Activities of the compounds in FluA virus minireplicon assays. HEK 293T cells were transfected with plasmids encoding PB1, PB2, PA, NP and a firefly luciferase reporter gene flanked by the noncoding sequences of FluA segment 8. The transfection mixtures also contained a plasmid constitutively expressing Renilla luciferase, which served to normalize variations in transfection efficiency. Luciferase activity was quantified at 24 h posttransfection. Activity observed with transfection reaction mixtures containing DMSO instead of test compounds was set at 100% and relative light units (RLU) were calculated. Omission of PB2 served as a negative control. Data shown represent the mean ± SD from at least six independent experiments.
We next analyzed the ability of the compounds to inhibit the activity of FluA virus RNA polymerase by a minireplicon assay (26). Cells were cotransfected with plasmids encoding the three polymerase subunits and the viral nucleoprotein (NP) along with a plasmid containing the firefly luciferase reporter gene flanked by the noncoding regions of A/WSN/33 segment 8, and treated with test or control compounds. In the absence of inhibitor, the polymerase and NP proteins transcribed the viral-like RNA expressed by the reporter plasmid into mRNA, resulting in luciferase expression. A strong decrease in reporter gene activity was observed in the presence of the PB11–15–Tat peptide. Treatment of transfected cells with compounds 1 and 5 resulted in dose-dependent inhibition of luciferase activity with IC50 of 18.5 ± 3.8 μM and 31.4 ± 4.2 μM, respectively (Fig. 3B and Table 1). Compounds 10, 18, and 31 exhibited weak inhibitory activity at the highest concentrations, resulting in IC50 values >100 μM, whereas compound 3 had no effect.
Thus, two of the six compounds that inhibited PA–PB1 binding in vitro also interfered effectively in cells with PA intranuclear translocation and with the catalytic activity of the viral polymerase.
Activity of the Compounds Against FluA Virus Replication.
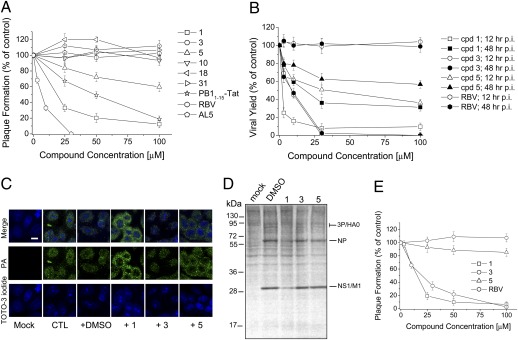
We then investigated the antiviral effects of the compounds in FluA virus-infected MDCK cells. First, we tested the compounds in plaque reduction assays with the A/PR/8/34 (PR8) strain. RBV, a known inhibitor of RNA viruses, exhibited a 50% effective dose (ED50) of 8.4 ± 2.3 μM, consistent with a previous report (27). Compound 1 inhibited plaque formation with an ED50 of 18.6 ± 4.1 μM (Fig. 4A and Table S3). Compound 5 weakly inhibited virus growth, having an ED50 >100 μM, whereas compounds 10, 18, and 31 had no significant effect on FluA virus replication at the tested concentrations (Fig. 4A). The AL5 compound, active against the HCMV polymerase, and compound 3 showed no activity, whereas the PB11–15–Tat peptide exhibited inhibitory activity, as expected (Fig. 4A and Table S3). No cytotoxic effect was observed at the tested concentrations for any of the compounds.
Fig. 4.
Antiviral activity of selected compounds. (A) Effects of the indicated compounds on plaque formation by PR8 virus were determined in MDCK cells. (B) Effects of compounds 1, 3, 5, and RBV on the yield of PR8 virus following low MOI infections of MDCK cells at 12 and 48 h p.i. (C) Effects of compounds 1, 3, and 5 on intracellular localization of PA in PR8-infected MDCK cells were analyzed at 6 h p.i. by immunofluorescence with an anti-PA antibody. (Scale bar, 10 μm.) (D) Effects of compounds 1, 3, and 5 on viral protein synthesis. MDCK cells were infected with PR8 virus and treated with test compounds. At 12 h p.i., [35S]-methionine was added to the cell medium and labeled proteins were analyzed by SDS/PAGE and autoradiography. (E) Effects of the indicated compounds on plaque formation by B/Lee/40 virus.
Next, we tested the effects of selected compounds in viral yield assays at 12 and 48 h postinfection (p.i.). Compound 1 inhibited virus yield with an ED50 of 1.5 ± 0.9 μM at 12 h p.i. and of 19.4 ± 3.6 μM at 48 h (Fig. 4B and Table 1). Similarly, compound 5 showed higher activity at 12 h p.i. (ED50 of 30.7 ± 4.1 μM) than at 48 h p.i. (ED50 >100 μM). As expected, compound 3 showed no significant antiviral activity at 12 or 48 h p.i., whereas RBV exhibited ED50 of ∼9 μM at both times p.i. The analysis of the intracellular localization of PA in infected cells in the presence of test compounds (Fig. 4C) demonstrated that the antiviral effects of compounds 1 and 5 were indeed due to the block of PA intranuclear translocation, likely reflecting inhibition of PA–PB1 interactions in the cytosol.
Then, we analyzed inhibition of viral protein synthesis. MDCK cells were infected with influenza PR8 virus at an multiplicity of infection (MOI) of 5 and treated with compounds 1, 3, and 5. At 12 h p.i., [35S]-Met was added for 1 h in the cell medium to label de novo viral protein synthesis, which was then analyzed by SDS/PAGE and autoradiography. Cells treated with compound 1 showed reduced expression of viral proteins (Fig. 4D), but as expected, compound 3 had no effect. No significant activity of compound 5 was observed, perhaps due to the high MOI used in these experiments.
We also tested the activity of compounds 1 and 5 against a number of clinical isolates of FluA virus. As with PR8, compound 5 showed weak antiviral activity against several strains in plaque reduction assays (Table S3). However, compound 1 effectively inhibited all FluA viruses tested, including pandemic swine-originated influenza virus (S-OIV) strains, with ED50 ranging from 12.2 to 22.5 μM. Importantly, this included potent activity against an oseltamivir-resistant clinical isolate (A/Parma/24/09) (ED50 of 22.5 ± 3.7 μM; Table S3). Thus, compound 1 displays broad-spectrum antiviral activity against FluA virus.
Compound 1 also Blocks FluB Virus Replication.
We next investigated whether compounds active against FluA virus polymerase could also inhibit FluB viruses. Thus, compounds 1 and 5 were tested against several FluB virus strains by plaque reduction assays. In these experiments, RBV blocked FluB virus replication with an ED50 of 12.4 ± 3.4 μM, whereas compound 3 and the unrelated small molecule AL5 had no effect. Compound 1 inhibited the replication of FluB viruses with ED50 values ranging from 12.5 to 21.0 μM, whereas compound 5 exhibited ED50 values >100 μM against all virus tested (Fig. 4E and Table S4).
To confirm that the antiviral activity of compound 1 against FluB virus was due to inhibition of the viral RNA polymerase, we investigated whether the compound could inhibit a FluB virus minireplicon system. Compound 1 efficiently reduced expression of the GFP reporter gene, whereas compound 3 showed no effect (Fig. S3). Thus, compound 1 also inhibits the FluB virus RNA polymerase.
Activity of Compound 1 Against NonInfluenza Viruses.
Finally, to further evaluate the therapeutic potential and selectivity of compound 1, we tested its effects on the replication of RNA viruses other than influenza virus: vesicular stomatitis virus, respiratory syncytial virus, measles virus, and coxsackie virus B1. We also tested it against DNA viruses: herpes simplex virus type 1, HCMV, and human adenovirus. Compound 1 did not inhibit the replication of any of the tested viruses (Table S5). Thus, compound 1 possesses specific antiviral activity against FluA and FluB viruses.
Discussion
Here we report the identification of small molecules that disrupt the interactions between the PA and PB1 subunits of influenza virus RNA polymerase and block virus growth in cell culture. Several of these compounds showed no cytotoxicity at concentrations up to 1 mM and one molecule—compound 1—blocked the formation of virus progeny with low micromolar potency, thus having a selectivity index >500. Additionally, the most active compound was effective not only against FluA but also against FluB. Conversely, in contrast to other antiviral agents that act on viral RNA polymerases such as RBV (27) and favipiravir (T-705) (23), this compound does not possess broad-spectrum antiviral activity against RNA and DNA viruses of other families.
Our work provides a proof-of-principle that the PA–PB1 interaction can be disrupted by a small molecular-weight compound. A major obstacle to the inhibition of protein–protein interactions is that these interactions often involve a large, flat surface area and multiple contacts (28, 29) and therefore they cannot be easily disrupted through the binding and competition of a small molecule. Thus, whereas there are numerous reports of the use of dominant negative proteins, antibodies, or peptides to inhibit protein–protein interactions (e.g., refs. 30, 31), there are few examples of small “drug-like” molecules that selectively disrupt these interactions. In the case of the FluA virus polymerase complex, a 25-amino-acid peptide corresponding to the PA-binding domain of PB1 has been shown to block the polymerase activity of FluA virus and inhibit viral spread (19). However, although these results demonstrate the feasibility of targeting the protein–protein interaction domains of the influenza virus polymerase complex, one should keep in mind that the in vivo use of peptides is often thwarted by a number of problems, particularly in relation to their cell permeation, intracellular localization, and stability. Thus, synthetic peptides rarely reach the clinics. Recent crystal structures and mutational studies showed that only a small subset of amino acid residues in PA and PB1 contribute to most of the free energy of binding (16, 17). In addition, it has been shown that alteration of these conserved residues abrogates subunit interactions accompanied with restricted assembly of polymerase heterotrimers, resulting in decreased polymerase activity (32, 33). These observations prompted us to search for nonpeptide molecules, with low toxicity, that efficiently penetrate the plasma membrane and bind with high affinity to their targets. Our successful efforts have led to the discovery of structurally diverse small molecules that can provide the basis for developing new antiinfluenza drugs.
The inhibitors that we have identified have a number of advantages compared with other classes of antiinfluenza compounds. First, because protein–protein interactions are highly specific, their inhibitors are likely to be highly specific. In line with this specificity, our compounds did not inhibit the replication of DNA viruses or of RNA viruses other than influenza virus. Further supporting their specificity, the active compounds did not exhibit significant cytotoxicity in cell culture, although their toxicities in vivo remain to be investigated.
A major concern in the use of antiinfluenza drugs is the development of resistance (34). Our antiviral agents have a different mode of action to the current antiinfluenza drugs and thus are unlikely to suffer from cross-resistance. In fact, here we show that the most active of our molecules—compound 1—inhibited the replication of an oseltamivir-resistant virus strain. In addition, the possibility of targeting other interaction sites in the polymerase complex, e.g., those between PB1 and PB2 (35), may allow the generation of an antiviral mixture that could reduce the probability of generating escape mutants.
Finally, because the amino acids of both PB1 and PA that are essential for polymerase subunit interaction are highly conserved among all known FluA virus strains (19, 36), inhibitory molecules will likely have broad efficacy against FluA viruses of both human and animal origin. Consistent with this prediction, here we show that compound 1 inhibited the replication of a number of FluA virus strains, including the new pandemic S-OIV. Furthermore, our identification of a compound that inhibits the replication of both FluA and FluB viruses suggests the possibility of developing drugs that are active against both major subtypes of human orthomyxoviruses. This compares favorably with adamantanes, which are only effective against FluA virus (2). It is also noteworthy that compound 1 exhibited similar potencies against FluA and FluB viruses, whereas oseltamivir is less active against FluB than FluA virus (37).
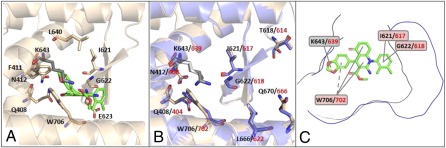
By analyzing FLAP virtual screening docked poses of compound 1 with the PA subunit of FluA virus RNA polymerase, possible molecular explanations for the broad-spectrum activity of compound 1 can be proposed. The similarity score between compound 1 and the PA cavity is mainly driven by hydrophobic interactions, in agreement with previous data showing the cavity's hydrophobic nature (36). The best pose for compound 1 in the PA structure obtained by optimizing the hydrophobic interactions is reported in Fig. 5A. Intriguingly, compound 1 appears to be docked in a region of the PA cavity where PB1-interacting residues (16, 17, 36) that are highly conserved residues among all FluA virus strains are present (i.e., Q408, F411, N412, I621, G622, E623, L640, and W706). In particular, it is notable that the 1,3-benzodioxole moiety of compound 1 is oriented toward W706, likely involving π–π interactions and H-bond interactions. Another conserved residue, I621, is also linked to compound 1 via hydrophobic interactions. The compound 1–PA interaction pattern is completed by K643; this residue is not reported to interact with PB1, but it is located in the PA cavity in proximity to W706. These observations could account for the broad efficacy of compound 1 against FluA virus strains. To explore the ability of compound 1 to also inhibit interactions between the PA and PB1 proteins of FluB virus, a homology model for FluB PA was built (SI Materials and Methods). Residues corresponding to Q408, N412, T618, I621, K643, L666, Q670, and W706 of FluA PA are conserved in FluB and match well upon structural alignment (Fig. 5B). Despite their role in PB1 binding, T618 and Q670 of FluA PA are not involved in compound 1 binding, being located in the outer part of the cavity. An analysis of the binding site of FluA and FluB PA for compound 1 and of the interaction energies showed that, despite the cavities of FluA and FluB PA having different shapes (Fig. 5C), the I621/617, G622/618, K643/639, and W706/702 residues of FluA and FluB PA are most likely crucial for the interaction with compound 1 (Fig. 5C).
Fig. 5.
Molecular basis of the interaction of compound 1 with PA. (A) FLAP best pose for compound 1 in FluA PA. Certain residues highly conserved in FluA viruses are reported in sticks mode. Among them, residues that are known to interact with PB1 are reported in ivory color, whereas K643 is reported in gray color. (B) Alignment of the homology model of FluB PA (in blue) on the crystal structure of the PA subunit of FluA virus (in ivory). Certain residues conserved in FluA and FluB viruses are reported in sticks mode. Black and red numbering refer to FluA and FluB, respectively. (C) FluA and FluB PA cavities (in black and blue lines, respectively) projected in the compound 1 2D-structure plane. Labels report the residues (for FluA PA in black, for FluB PA in red) showing higher interaction energies with compound 1.
In conclusion, our screen has resulted in the identification of a new class of influenza virus inhibitors that act by targeting the PA–PB1 interaction. Optimization of these compounds, in particular compounds 1 and 5, could result in the development of a new generation of therapeutic agents exhibiting high specificity and low resistance to influenza virus.
Materials and Methods
For a complete description of the source of materials and our methods, see SI Materials and Methods. It includes detailed procedures for the virtual screening and homology model creation, expression and purification of proteins, PA–PB1 interaction ELISA, and His-pulldown assay. It also includes description of PA–PB1 nuclear import assays, luciferase-based reporter assays, the cytotoxicity assay, plaque assays, virus yield reduction assays, analysis of viral protein synthesis, and antiviral assays with noninfluenza viruses.
Supplementary Material
Acknowledgments
We thank R. Cusinato, A. Calistri, C. Salata, I. Donatelli, and W. S. Barclay for FluA and FluB viruses; G. Gribaudo for VSV; E. Fodor, L. Tiley, and W. S. Barclay for plasmids; H. Wise and E. Sinigalia for experimental help; and E. Carosati and F. Sirci for help in virtual screening and sequence/structure alignment. This work was supported by Italian Ministry of Health and Istituto Superiore Sanità, Progetto Finalizzato 2009 “Studio e Sviluppo di Nuovi Farmaci Antivirali Contro Infezioni da Virus Influenzale A-H1N1” (to A.L. and G.P.), by MURST EX60% and PRIN 2008 Grant 20085FF4J4 (to A.L.); by Regione Veneto and Progetto Strategico di Ateneo 2008 (to G.P. and A.L.); by United Kingdom Medical Research Council Grant G0801931 (to P.D.); a Wellcome Trust Studentship (to Á.F.); and by a grant from Molecular Discovery, London (to G.C.).
Footnotes
Conflict of interest statement: A provisional patent application has been filed covering some of the results described in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119817109/-/DCSupplemental.
References
- 1.Palese P, Shaw ML. Orthomyxoviridae: The viruses and their replication. Fields’ Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 2.De Clercq E. Antiviral agents active against influenza A viruses. Nat Rev Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden FG, Hay AJ. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr Top Microbiol Immunol. 1992;176:119–130. doi: 10.1007/978-3-642-77011-1_8. [DOI] [PubMed] [Google Scholar]
- 4.Colman PM, Varghese JN, Laver WG. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 5.de Jong MD, et al. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med. 2005;353:2667–2672. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- 6.Biswas SK, Nayak DP. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Digard P, Blok VC, Inglis SC. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology. 1989;171:162–169. doi: 10.1016/0042-6822(89)90523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilligay D, et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol. 2008;15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 9.Yuan P, et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 10.Dias A, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 11.Torreira E, et al. Three-dimensional model for the isolated recombinant influenza virus polymerase heterotrimer. Nucleic Acids Res. 2007;35:3774–3783. doi: 10.1093/nar/gkm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez DR, Donis RO. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J Virol. 1995;69:6932–6939. doi: 10.1128/jvi.69.11.6932-6939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohtsu Y, Honda Y, Sakata Y, Kato H, Toyoda T. Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol Immunol. 2002;46:167–175. doi: 10.1111/j.1348-0421.2002.tb02682.x. [DOI] [PubMed] [Google Scholar]
- 14.González S, Zürcher T, Ortín J. Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: A model for the viral RNA polymerase structure. Nucleic Acids Res. 1996;24:4456–4463. doi: 10.1093/nar/24.22.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poole EL, Medcalf L, Elton D, Digard P. Evidence that the C-terminal PB2-binding region of the influenza A virus PB1 protein is a discrete alpha-helical domain. FEBS Lett. 2007;581:5300–5306. doi: 10.1016/j.febslet.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 16.Obayashi E, et al. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature. 2008;454:1127–1131. doi: 10.1038/nature07225. [DOI] [PubMed] [Google Scholar]
- 17.He X, et al. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature. 2008;454:1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 18.Baroni M, Cruciani G, Sciabola S, Perruccio F, Mason JS. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): Theory and application. J Chem Inf Model. 2007;47:279–294. doi: 10.1021/ci600253e. [DOI] [PubMed] [Google Scholar]
- 19.Ghanem A, et al. Peptide-mediated interference with influenza A virus polymerase. J Virol. 2007;81:7801–7804. doi: 10.1128/JVI.00724-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wunderlich K, et al. Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication. PLoS ONE. 2009;4:e7517. doi: 10.1371/journal.pone.0007517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loregian A, et al. Inhibition of human cytomegalovirus DNA polymerase by C-terminal peptides from the UL54 subunit. J Virol. 2003;77:8336–8344. doi: 10.1128/JVI.77.15.8336-8344.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loregian A, Coen DM. Selective anti-cytomegalovirus compounds discovered by screening for inhibitors of subunit interactions of the viral polymerase. Chem Biol. 2006;13:191–200. doi: 10.1016/j.chembiol.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Furuta Y, et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu AL, Wang HD, Lee SM, Wang YT, Du GH. Structure-activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities. Bioorg Med Chem. 2008;16:7141–7147. doi: 10.1016/j.bmc.2008.06.049. [DOI] [PubMed] [Google Scholar]
- 25.Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J Virol. 2004;78:9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullin AE, Dalton RM, Amorim MJ, Elton D, Digard P. Increased amounts of the influenza virus nucleoprotein do not promote higher levels of viral genome replication. J Gen Virol. 2004;85:3689–3698. doi: 10.1099/vir.0.80518-0. [DOI] [PubMed] [Google Scholar]
- 27.Sidwell RW, et al. Broad-spectrum antiviral activity of Virazole: 1-Beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science. 1972;177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 28.Tsai CJ, Nussinov R. Hydrophobic folding units at protein-protein interfaces: Implications to protein folding and to protein-protein association. Protein Sci. 1997;6:1426–1437. doi: 10.1002/pro.5560060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loregian A, Palù G. Disruption of protein-protein interactions: towards new targets for chemotherapy. J Cell Physiol. 2005;204:750–762. doi: 10.1002/jcp.20356. [DOI] [PubMed] [Google Scholar]
- 30.Marcello A, et al. Specific inhibition of herpes virus replication by receptor-mediated entry of an antiviral peptide linked to Escherichia coli enterotoxin B subunit. Proc Natl Acad Sci USA. 1994;91:8994–8998. doi: 10.1073/pnas.91.19.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loregian A, et al. Intranuclear delivery of an antiviral peptide mediated by the B subunit of Escherichia coli heat-labile enterotoxin. Proc Natl Acad Sci USA. 1999;96:5221–5226. doi: 10.1073/pnas.96.9.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wunderlich K, et al. Limited compatibility of polymerase subunit interactions in influenza A and B viruses. J Biol Chem. 2010;285:16704–16712. doi: 10.1074/jbc.M110.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez DR, Donis RO. Functional analysis of PA binding by influenza a virus PB1: Effects on polymerase activity and viral infectivity. J Virol. 2001;75:8127–8136. doi: 10.1128/JVI.75.17.8127-8136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayden FG, de Jong MD. Emerging influenza antiviral resistance threats. J Infect Dis. 2011;203:6–10. doi: 10.1093/infdis/jiq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reuther P, Mänz B, Brunotte L, Schwemmle M, Wunderlich K. Targeting of the influenza A virus polymerase PB1-PB2 interface indicates strain-specific assembly differences. J Virol. 2011;85:13298–13309. doi: 10.1128/JVI.00868-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Yao X. Molecular basis of the interaction for an essential subunit PA-PB1 in influenza virus RNA polymerase: Insights from molecular dynamics simulation and free energy calculation. Mol Pharm. 2010;7:75–85. doi: 10.1021/mp900131p. [DOI] [PubMed] [Google Scholar]
- 37.Hatakeyama S, et al. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA. 2007;297:1435–1442. doi: 10.1001/jama.297.13.1435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.