Abstract
Meiotic recombination is tightly regulated by cis- and trans-acting factors. Although DNA methylation and chromatin remodeling affect chromosome structure, their impact on meiotic recombination is not well understood. To study the effect of DNA methylation on the landscape of chromosomal recombination, we analyzed meiotic recombination in the decreased DNA methylation 1 (ddm1) mutant. DDM1 is a SWI2/SNF2-like chromatin-remodeling protein necessary for DNA methylation and heterochromatin maintenance in Arabidopsis thaliana. The rate of meiotic recombination between markers located in euchromatic regions was significantly higher in both heterozygous (DDM1/ddm1) and homozygous (ddm1/ddm1) backgrounds than in WT plants. The effect on recombination was similar for both male and female meiocytes. Contrary to expectations, ddm1 had no effect on the number of crossovers between markers in heterochromatic pericentric regions that underwent demethylation. These results are surprising, because the pericentromeric regions are hypermethylated and were expected to be the regions most affected by demethylation. Thus, DDM1 loss of function may trigger changes that enhance meiotic recombination in euchromatin regions but are not sufficient to induce the same events in heterochromatic segments. This work uncovers the repressive role of methylation on meiotic recombination in euchromatic regions and suggests that additional factors may have a role in controlling the suppression of recombination in heterochromatin.
Keywords: genetic mapping, epigenetics, centromere
Meiotic recombination is a process that is tightly regulated both in its timing and in the identification of homologous partners that can exchange chromosomal segments. Recent studies have provided several details with regard to the cis and trans components regulating the rate and localization of meiotic recombination in plants and other organisms (reviewed in refs. 1 and 2). Sequence homology is one of the most critical cis requirements for determining the identity of a homolog, and sequence polymorphism negatively affects recombination (3, 4). In addition, there is evidence that DNA sequence motifs can affect recombination by acting as recombination “hotspots” along the genome (reviewed in ref. 5). An additional important cis component, whose influence on meiotic recombination is less known and which has received limited attention, is the methylation of cytosine residues.
Saccharomyces cerevisiae, the paradigm for studies on homologous recombination, has no detectable cytosine methylation and therefore is not a useful model in this context. In contrast, in a number of organisms, including Neurospora crassa and Ascobulus immersus, hypermethylation of repeats leads to gene silencing and suppression of meiotic recombination (reviewed in ref. 6). Similarly, in maize, intergenic DNA regions that are rich in transposons and other repetitive sequences are hypermethylated and represent “cold” meiotic recombination segments as compared with hypomethylated low-copy protein-coding genes (7). Overall, the degree of DNA methylation at a specific genomic region may represent an important cis factor regulating the rate of meiotic recombination; however, the effect of demethylation of these repeats has not been studied.
As with eukaryotes, plant genomes contain high percentages of 5′-methylcytosine (mC) in CG sequences and also within CHG and CHH sequences (8). DNA methylation in plants has been implicated in processes related to transposon silencing, gene transcription, development, and defense (reviewed in ref. 9). Although Arabidopsis thaliana has one of the lowest mC contents of the plant genomes analyzed so far, the machinery for regulating DNA methylation has been described thoroughly in this species (10). The methyltransferases governing methylation of cytosine nucleotides include MET1, which maintains cytosine methylation at CG sites in newly synthesized DNA (11, 12), CMT3, responsible for CHG methylation (13, 14), and DRM2, which, together with small RNAs, methylates all other sequence contexts, namely CHH (15–17). In addition to the methyltransferases, SWI2/SNF2-like chromatin-remodeling proteins, such as DDM1 and DRD1, are required for the maintenance of DNA methylation (18–20). Mutations in DDM1 are recessive and cause gradual depletion of cytosine methylation. DDM1 function is crucial for the maintenance of heterochromatin in the nucleus (21, 22) and is necessary in DNA repair (23). In ddm1 mutants, there is a significant DNA decondensation at centromeric and pericentromeric regions rich in repetitive sequences and transposons (24–27). Moreover, in ddm1 mutants, as well as in met1 mutants, some transposons become transcriptionally active (28–30) or even undergo transposition (31–35).
In addition to the depletion of DNA methylation, ddm1 mutations correlate with a gradual replacement of heterochromatin-specific histone modifications (histone H3 lysine 9 dimethylation) with marks specific for euchromatin (histone H3 lysine 4 trimethylation) (21, 25, 36, 37). A number of reports recently have linked histone modifications with meiotic recombination in several organisms. In S. cerevisiae, lysine 4 methylation of histone H3 is critical for the formation of programmed DNA double-strand breaks that initiate homologous recombination during meiosis (38, 39). Similarly, in maize, Mutator insertions as well as meiotic recombinations occur more frequently in regions with high levels of both lysine 4 methylation and lysine 9 acetylation in histone H3, together with low levels of cytosine methylation (40). In Arabidopsis, mutations of MCC1 result in hyperacetylation of histone H3 and affect the number and distribution of chiasmata, leading to abortion of approximately half of the male and female gametes (41). The interplay among histone modifications, chromatin remodeling, and DNA methylation is complex, and the direct impact of these factors on meiotic recombination in plants remains to be established.
In this work, we investigated the relation between DNA methylation and meiotic recombination. We show that the ddm1 mutation affects meiotic recombination in both male and female lineages, but the effect is not equal throughout the genome: Euchromatic regions exhibit high rates of meiotic recombination in the mutant, whereas no changes were found in heterochromatic centric and pericentric areas.
Results
DDM1 and MET1 Affect Meiotic Recombination.
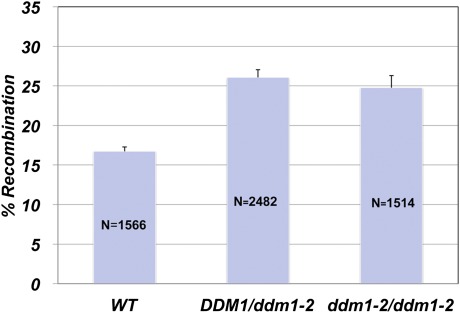
To analyze the role of DDM1 in meiotic recombination, the homozygous tester line Col3-4/20 (42) in the background of accession Columbia was crossed with WT Columbia or with the Columbia background ddm1-2 mutant (20). The tester line contains both RFP and GFP markers under the seed-specific NAPINE promoter, linked in cis, 16 cM apart. Consequently, the line enables estimation of meiotic recombination rates by counting seeds expressing none or both parental markers (red and green) and recombinant seeds expressing only red or only green markers. Seeds resulting from these crosses are heterozygous for the fluorescent markers. In the crosses with the mutant, the homolog of each chromosome pair contributed by the tester is methylated, but the homolog from the ddm1-2 mutant is not. In agreement with the results reported in ref. 43, Southern blot analysis of F1 plants performed after total genomic DNA was digested with the methylation-sensitive restriction enzyme HpaII and using the A. thaliana 180-bp centromere repeat as a probe confirmed that plants heterozygous for ddm1-2 (DDM1/ddm1-2) were more prone to HpaII digestion than WT plants. Crossover rates of progeny obtained by selfing (F2) of these plants were significantly higher than in WT plants, with a measured 26.1 cM for DDM1/ddm1-2 plants versus 16.4 cM for WT (P < 0.0001; χ2 test) (Table 1). The mutation did not affect the segregation of the two individual markers, both of which showed the expected 3:1 Mendelian ratio.
Table 1.
Rates of recombination between green and red fluorescent markers in F2 seeds of a cross between tester line Col3-4/20 × WT or Col3-4/20 × ddm1-2 mutant
| F2 seed phenotype |
||||||
| Cross | Seeds (#) | Red only | Green only | Red and green | Nonfluorescent | Genetic distance* (cM) |
| Col3-4/20 × ddm1-2 heterozygous DDM1/ddm1-2 | 312 | 35 | 44 | 204 | 29 | |
| 299 | 30 | 30 | 191 | 48 | ||
| 233 | 23 | 23 | 153 | 34 | ||
| 279 | 40 | 26 | 176 | 37 | ||
| 286 | 35 | 29 | 188 | 34 | ||
| 287 | 39 | 27 | 185 | 36 | ||
| 295 | 36 | 27 | 191 | 41 | ||
| 242 | 34 | 29 | 142 | 37 | ||
| 249 | 32 | 23 | 159 | 35 | ||
| Total | 2,482 | 304 | 258 | 1,589 | 331 | 26.1 |
| Col3-4/20 × WT | 144 | 8 | 9 | 100 | 27 | |
| 309 | 17 | 27 | 221 | 44 | ||
| 141 | 11 | 12 | 94 | 24 | ||
| 187 | 13 | 17 | 113 | 44 | ||
| 372 | 25 | 27 | 257 | 63 | ||
| 413 | 37 | 33 | 266 | 77 | ||
| Total | 1,566 | 111 | 125 | 1,051 | 279 | 16.4 |
| Col3-4/20 × ddm1-2 homozygous ddm1-2/ddm1-2 | 448 | 45 | 42 | 282 | 79 | |
| 444 | 49 | 41 | 284 | 70 | ||
| 298 | 45 | 24 | 184 | 45 | ||
| 279 | 26 | 45 | 171 | 37 | ||
| 45 | 4 | 5 | 31 | 5 | ||
| Total | 1,514 | 169 | 157 | 952 | 236 | 24.7 |
Every row represents an independent plant from the same experiment. All crosses were grown at the same time and under the same conditions.
*The rate of recombination was significantly different between WT and DDM1/ddm1-2 plants (P < 0.001; χ2 test) and between WT and ddm1-2/ddm1-2 plants (P < 0.001; χ2 test).
Fluorescent seed markers then were introgressed in plants homozygous for the ddm1 mutation (ddm1-2/ddm1-2). Crossover rates between the markers were 24.7 cM in ddm1-2/ddm1-2 and 17.3 cM in WT plants, a 43% increase (P < 0.0001; χ2 test), similar to that recorded for DDM1/ddm1-2 plants (Fig. 1 and Table 1).
Fig. 1.
Loss of DDM1 activity increases meiotic recombination rates in heterozygote DDM1/ddm1-2 and homozygote ddm1-2/ddm1-2 plants. Recombination rates of DDM1/ddm1-2 were obtained from F2 seeds of a cross between the ddm1-2 mutant and the tester line Col3-4/20, which contained green and red fluorescent markers. The seeds used to monitor recombination rates of ddm1-2/ddm1-2 were obtained from the introgression of the fluorescent seed markers from tester line Col3-4/20 into the ddm1-2 mutant background. The homozygous ddm1-2 point mutation was confirmed by sequencing. WT represents seeds from crosses between tester line Col3-4/20 and WT plants. N = number of seeds obtained from five to nine plants) scored in each background. The variation in recombination between individual plants was used to determine the SE (shown as error bars).
The effect observed in the ddm1 background might be a direct effect of DNA methylation. If so, similar consequences would be expected upon mutation of other genes along the DNA methylation pathway. To this end, the Col3-4/20 tester line was crossed with the met1-3 mutant (12) and resulted in crossover rates in the F2 generation of 21.5 cM for MET1/met1-3 and 16.9 cM for WT plants (Table 2). In summary, the decreased methylation in the met1-3 mutant led to increased meiotic recombination, a result that was in line with our previous understanding.
Table 2.
Rates of recombination between green and red fluorescent markers in F2 seeds of a cross between tester line Col3-4/20 × WT or Col3-4/20 × met1-3 mutant
| F2 seed phenotype |
||||||
| Cross | Seeds (#) | Red only | Green only | Red and green | Nonfluorescent | Genetic distance* (cM) |
| Col3-4/20 × met1-3 | 446 | 49 | 39 | 284 | 74 | |
| 460 | 38 | 52 | 289 | 81 | ||
| 392 | 35 | 38 | 261 | 58 | ||
| 489 | 50 | 45 | 330 | 64 | ||
| 428 | 48 | 47 | 259 | 74 | ||
| 547 | 44 | 49 | 365 | 89 | ||
| 419 | 53 | 34 | 270 | 62 | ||
| 413 | 34 | 36 | 288 | 55 | ||
| 361 | 31 | 37 | 244 | 49 | ||
| Total | 3,955 | 382 | 377 | 2,590 | 606 | 21.5 |
| Col3-4/20 × WT | 414 | 31 | 30 | 268 | 85 | |
| 266 | 21 | 17 | 190 | 38 | ||
| 344 | 22 | 29 | 236 | 57 | ||
| 370 | 28 | 31 | 249 | 62 | ||
| 422 | 28 | 40 | 285 | 69 | ||
| 391 | 29 | 33 | 256 | 73 | ||
| 389 | 29 | 33 | 252 | 75 | ||
| Total | 2,596 | 188 | 213 | 1,736 | 459 | 16.9 |
*The rate of recombination was significantly different between the two crosses (P < 0.001; χ2 test).
Effect of ddm1-2 on Meiotic Recombination in Female and Male Meiocytes.
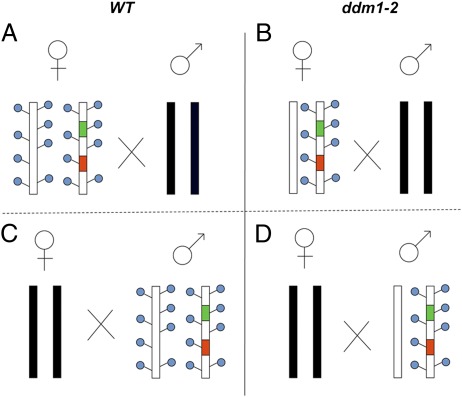
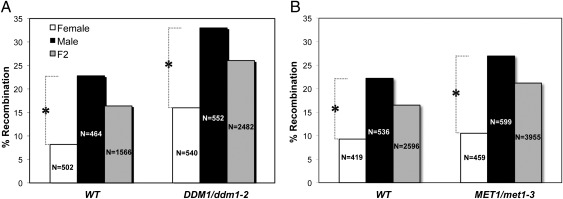
It has been well established that recombination frequencies differ significantly in female and male lineages (44), as reported for a wide range of organisms (45–47), including plants (45, 48–52). However, the genetic basis for this phenomenon remains poorly understood. To test the influence of epigenetic imprints on these sex-specific disparities, we determined the meiotic recombination rates of female and male meiocytes resulting from the crossing scheme shown in Fig. 2. Meiotic recombination in female meiocytes was determined in both WT and DDM1/ddm1-2 F1 plants heterozygous for the tester markers (Fig. 2 A and B). Similarly, meiotic recombination in male meiocytes was determined by using pollen from both WT and DDM1/ddm1-2 F1 plants heterozygous for the tester markers (Fig. 2 C and D). Note that the chromosomes used for backcrossing (shown as black chromosomes in Fig. 2) were of either WT or ddm1-2 origin and, as expected, had no influence on the recombination rates (Table S1 and Table S2). Fluorescent markers were monitored in the backcross of the F1 hybrid (BC1F1) seeds. Male meiocytes exhibited significantly higher recombination rates than female meiocytes (P < 0.001; χ2 test), regardless of the methylation state of the recombining homologs. More specifically, rates of 22.4 vs. 8.2% in WT plants and 33 vs. 14.7% in ddm1-2 mutants were recorded for male and female meiocytes, respectively (Fig. 3A). In F2 seeds the recombination rates for both male and female gametes were comparable to the average rate (Fig. 3Aand Table 1). Similar observations were seen in MET1; namely, regardless of their methylation status in the tested interval, male meiocytes exhibited significantly higher recombination rates than female meiocytes (P < 0.001; χ2 test): 22.7 vs. 8.6% in WT plants and 27% vs. 11.5% in met1-3 mutants (Fig. 3B and Table S3).
Fig. 2.
Scheme of crosses for the analysis of meiotic crossovers in female and male meiocytes. Recombination of male and female meiocytes was determined in seeds derived from reciprocal backcrosses (BC1F1 seeds) using green and red fluorescent markers. The four panels show female meiosis in WT background (A) or ddm1-2 background (B) and male meiosis in WT background (C) or ddm1-2 background (D). F1 plants were backcrossed with plants from either WT or ddm1-2/ddm1-2 background (black chromosomes). Blue circles represent cytosine methylations.
Fig. 3.
The effect of ddm1-2 and met1-3 on meiotic recombinations in female and male meiocytes. The percentage of meiotic recombination was determined in reciprocal backcrosses of F1 plants heterozygous for the meiotic tester Col3-4/20. (A and B) The x axes refer to the genotype of the F1 plants, either WT or DDM1/ddm1-2 (A) or WT or MET1/met1-3 (B). The differences between recombination rates in male and female meiocytes was significant in both genotypes. *P < 0.001 (χ2 test). The ratio between the meiotic recombination rates in female and male meiocytes was similar in both WT vs. DDM1/ddm1-2 genotypes (P = 0.1413; χ2 test) and WT vs. MET1/met1-3 genotypes (P = 0.5911; χ2 test). Male and female meiotic recombination is shown in black and white, respectively. Gray bars represent the F2 rates of meiotic recombination. N is the number of BC1F1 or F2 seeds monitored for the fluorescent markers.
Effect of ddm1-2 on Meiotic Recombination Varies in Euchromatic and Heterochromatic Regions.
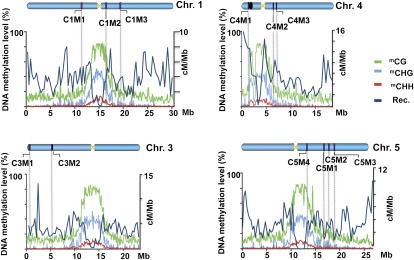
To test for the generality of the effect of ddm1 on meiotic recombination, we used genetic-marker pairs originating from various locations in the genome. Insertion/deletion (InDel) markers were chosen for mapping an F2 population derived from the cross between ecotype Columbia (WT or ddm1-2) and ecotype Landsberg. Markers for each Arabidopsis chromosome were selected (Materials and Methods and Fig. 4) using a list of polymorphisms between Landsberg and Columbia (53) and a recently described high-density InDel platform (54). All markers were designed so that the InDel polymorphism was >60 bp and the physical distance between markers was at least 1 Mb. We expected that heterochromatic regions, particularly centromeric and pericentromeric regions that are relatively hypermethylated in comparison with other regions and that are known to be cold regions of recombination (27, 52), would be influenced more than euchromatic regions by ddm1-mediated demethylation (27, 52). Exploiting the information of a DNA methylation map produced from young flower buds and leaves (55, 56) we considered the markers to be located in heterochromatin when they originated from regions that contained higher-than-average cytosine methylation (>30%) (Fig. 4). These regions include pericentric regions and the knob region on chromosome 4 and the pericentric region on chromosomes 1 and 5. For euchromatic regions, we choose markers located in subtelomeric regions (e.g., in chromosome 3) or in the center of the chromosomal arm (e.g., in chromosome 5) with average or below-average rates of cytosine methylation (Fig. 4). Physical distances for all the markers described in Table 3 were calculated using the maps of the Arabidopsis Information Resource (http://www.arabidopsis.org/servlets/mapper).
Fig. 4.
Distribution of In/Del markers along different Arabidopsis chromosomes. Eight different pairs of InDels were selected and tested in an F2 population resulting from a cross between ecotypes Columbia and Landsberg. Markers are shown as vertical lines on the chromosomes (Chr1–5) and were positioned in the diagram by physical coordinates. The percentage of methylation (y axes) along the chromosomes (taken from ref. 56) is shown as green, light blue, and red lines for mCG, mCHG, and mCHH, respectively. Markers were chosen as being in the heterochromatic region when they had an mGC level (green line) >30% in chromosome 1 (C1M1 and C1M2, around the centromere; C1M2 and C1M3 pericentric region), in chromosome 4 (C4M1 and C4M2 surrounding the knob and centromere; C4M2 and C4M3 pericentric region), and in chromosome 5 (C5M4 at a pericentric region and C5M1 at euchromatic region). Markers representing euchromatic regions are shown in chromosome 3 (C3M1 and C3M2), and two pairs of markers with mGC levels <30% are shown in chromosome 5 (C5M1, C5M2, and C5M3). Chromosome 3 contains the fluorescent markers used in this study (marked in red and green). The dark blue line represents the genome-wide crossover distribution, calculated as the average recombination rate (Rec.) in male and female meiocytes, from data of Giraut et al (52). The yellow triangles represent the centromeres of each chromosome. The black oval on chromosome 4 represents the knob. Adapted by permission from Macmillan Publishers Ltd: [Nature] (56), copyright (2008).
Table 3.
Frequency of meiotic recombination throughout the genome
| WT |
DDM1/ddm1-2 |
|||||||
| Marker position | Physical distance (Mb) | No. plants | Recombinant plants | Genetic distance (cM)* | No. plants | Recombinant plants | Genetic distance (cM)* | Recombinant in ddm1-2 vs. WT (%) |
| Euchromatin | ||||||||
| Chr. 3 (Col3-4/20) Isogenic background† | 5.1 | 1,566‡ | 236 | 16.4 | 2,482‡ | 562 | 26.1 | 59 |
| Chr. 3 (Col3-4/20) nonisogenic background§ | 5.1 | 1,929‡ | 191 | 10.4 | ||||
| Chromosome 3a (C3M1-C3M2) | 5.6 | 75 | 10 | 10.7 | 80 | 26 | 16.3 | 52 |
| Chromosome 5a (C5M1-C5M2) | 1.1 | 189 | 10 | 2.65 | 167 | 16 | 4.8 | 81 |
| Chromosome 5b (C5M2-C5M3) | 1.1 | 111 | 14 | 6.3 | 141 | 27 | 9.6 | 52 |
| Heterochromatin | ||||||||
| Chromosome 1a (C1M1-C1M2) | 5.5 | 145 | 26 | 9.0 | 155 | 26 | 8.4 | −7.1 |
| Chromosome 1b (C1M2-C1M3) | 3.1 | 113 | 31 | 13.7 | 92 | 26 | 14.1 | 2.9 |
| Chromosome 4a (C4M1-C4M2) | 4.9 | 154 | 23 | 7.5 | 134 | 19 | 7.1 | −5.6 |
| Chromosome 4b (C4M2-C4M3) | 0.8 | 137 | 15 | 5.5 | 124 | 14 | 5.7 | 3.6 |
| Chromosome 5c (C5M4-C5M1) | 3.2 | 100 | 30 | 15 | 94 | 30 | 15.9 | 6 |
*Calculated as (number of recombinants/total number of gametes) × 100.
†Isogenic background: cross between the meiotic tester (in Columbia background) and Col WT.
‡Number of seeds screened by fluorescent markers.
§Nonisogenic background: cross between the meiotic tester (in Columbia background) and Ler WT.
Markers C3M1 and C3M2 were selected for chromosome 3 because they map at physical positions similar to the insertion sites of the red and green fluorescence markers in the Col3-4/20 tester. The genetic distance between C3M1 and C3M2 was 10.7 cM in the WT background and 16.3 cM in the DDM1/ddm1-2 background (see details in Table 3, chromosome 3a), demonstrating that 52% more recombination events occurred in the mutant background. This relative increase is similar to that observed in the same interval for the seed fluorescent markers (Fig. 3). However, there is a discrepancy in absolute recombination values in WT: 16 cM for the fluorescence markers in Col3-4/20 (Fig. 3) versus 10.7 cM in the same interval between C3M1 and C3M2. On the basis of previous results (3), we hypothesized that this discrepancy was the result of the genetic distance between the fluorescence markers being measured in an isogenic background (in a Col3-4/20 × Col cross) and the genetic distance between C3M1 and C3M2 being measured in a nonisogenic background (WT Col × WT Ler). To test this hypothesis, we crossed Col3-4/20 with WT Ler and determined the genetic distance in F2, in the same nonisogenic background as for C3M1 and C3M2. Under these conditions, the genetic distance obtained with the fluorescent markers was 10.4 cM (Table 3, chromosome 3 nonisogenic background), almost identical to the distance obtained with markers C3M1 and C3M2 in the WT background. Markers C5M1, C5M2, and C5M3 are in a euchromatic region of chromosome 5. The genetic distance observed between the markers was 2.65 cM in the WT and 4.8 cM in the DDM1/ddm1-2 background. Thus, ddm1-related hypomethylation correlated with an 81% increase in recombination rates (Table 3, chromosome 5a). For markers C5M2 and C5M3 the observed frequency of meiotic recombination was 6.3 cM in the WT and 9.6 cM in the DDM1/ddm1-2 background, corresponding to a 52% increase in meiotic recombination in the mutant (Table 3, chromosome 5b). C5M4, also located on chromosome 5, is positioned in a pericentric region with characteristic heterochromatic features, 3 Mb away from C5M1 (Fig. 4). The genetic distance between these two markers was 15 cM in the WT vs. 15.9 cM in the DDM1/ddm1-2 background (Table 3, chromosome 5c); a significantly lower increase in recombination rates than observed in euchromatic regions. The marker pair C1M1 and C1M2 is in a heterochromatic region flanking the centromere of chromosome 1 (Fig. 4). Surprisingly the frequency of recombination in the WT genotype (9 cM) was similar but slightly higher than in the DDM1/ddm1-2 genotype (8.4 cM) (Table 3, chromosome 1a). Distal to the centromere, but still in the pericentromeric region, an additional marker, C1M3, was selected. Recombination frequencies again were similar in WT (13.7 cM) and DDM1/ddm1-2 genotypes (14.1 cM) (Table 3, chromosome1b). Markers C4M1 and C4M2, around the heterochromatic regions of the knob and the centromere of chromosome 4, also were evaluated. These markers showed a rate of recombination of 7.5 cM in the WT background and 7.1 cM in the DDM1/ddm1-2 background (Table 3, chromosome 4a). Because C4M1 and C4M2 are markers around the knob, and there is an inverted region between the accessions Col and Ler (57), the results might have been misinterpreted. Therefore, an additional marker, C4M3, was selected; C4M3 is distal to C4M2 and the centromere but still is in the pericentric region. The genetic distance between C4M2 and C4M3 was 5.8 cM in the WT background versus 6 cM in the DDM1/ddm1-2 background (Table 3, chromosome 4b).
Discussion
The relationship between methylation and recombination might be explored by analyzing the correlation between recombination and methylation rates along the chromosome. This approach is limited by the lack of data regarding genome methylation in meiocytes. Nevertheless, we superimposed the methylome landscape from leaves (Fig. 4) (56) with the recently published distribution of crossovers along the chromosomes (52). The most striking feature of this comparison is that hypermethylation in centric and pericentric regions is correlated with reduced rates of meiotic recombination; in the remaining chromosomal regions, however, the fluctuations in recombination are not necessarily associated with changes in methylation (Fig. 4). We cannot rule out the possibility of local correlation between increased crossover rates and hypomethylation. At present, however, the resolution of recombination maps is too low, compared with the variation in methylation at the single-nucleotide level, to allow such analysis. Moreover, the level of methylation in the euchromatin is not low enough to mirror the situation observed around centromeres. Therefore, we used methylation mutants to test the hypothesis that hypomethylation could lead to hyperrecombination, as would be expected from a negative relationship between DNA methylation and recombination. Our working hypothesis was that demethylation would increase the rate of meiotic crossover, particularly in the centric and pericentric regions that would be most affected by demethylation.
By exploiting the previously described fluorescent seed meiotic tester Col3-4/20 (42), we found that the rate of meiotic recombination was 59% higher in the DDM1/ddm1-2 mutant than in WT plants. This effect was observed in both male and female lineages, suggesting that DDM1 is not involved in the higher recombination rates characteristic of the male Arabidopsis lineage. For InDel DNA markers of chromosome 3, located near the insertion site of the red and green fluorescence markers, recombination rates were 52% higher in the DDM1/ddm1-2 mutant background than in WT plants. This increase paralleled that obtained with the Col3-4/20 tester, thus substantiating the utility of the transgenic fluorescent markers as tools for the rapid analysis of factors affecting meiotic recombination. Moreover, molecular markers within the euchromatic region of chromosome 5 showed increases of 58%, 81%, or 90% in crossover rates in the DDM1/ddm1-2 background as compared with WT. This finding suggests that the DDM1/ddm1-2–related increase is not restricted to a specific chromosomal segment but instead seems to be common to different marker types from various euchromatic regions. The results, reported by Mirouze et al. (58), obtained from epigenetic recombinant inbred lines (EpiRILs) derived from a met1 mutant show a similar increase in hypomethylated euchromatic regions, suggesting that the correlation between reduced methylation and increased crossover is not restricted to ddm1 or to a specific euchromatic region.
The underlying mechanism for this increase could be interpreted in various ways. DDM1 is an SNF2 nucleosome-remodeling protein that regulates histone H3 and DNA methylation. Upon loss of DDM1 activity, a 70% reduction in DNA methylation is induced (19), promoting chromatin decondensation (27). The DNA demethylation per se or altered chromatin remodeling could make the DNA more accessible to homologous recombination enzymes, fostering DNA double-strand breaks, homology searches, strand invasion, and exchange. Alternatively, the expression of the recombination machinery might be up-regulated during meiosis of DDM1/ddm1-2, for example by activation of silent genes from hypomethylated chromosomes, thus affecting the crossover rate in trans. The finding that loss of MET1 also is associated with an increase in meiotic crossover (Fig. 3B and Table 2) supports a model whereby demethylation is responsible, at least in part, for the heightened crossover rates. The effect of DDM1/ddm1-2 was stronger than that of MET1/met1, suggesting the involvement of additional factors beyond methylation. Interestingly, the ddm1 effect was similar in the DDM1/ddm1-2 background, where each homolog pair consists of a WT and a hypomethylated chromosome, and in the homozygous ddm1-2/ddm1-2 background, where both homologs are partially hypomethylated (Fig. 1). Currently, we do not have a satisfactory explanation for this finding.
Heterochromatin demethylation and transcriptional activation of transposons are among the central phenotypes of ddm1, including in the heterozygous mutant (27, 28). Therefore we predicted that the ddm1-2 mutation would have a greater impact on meiotic recombination in heterochromatic regions than in euchromatin. However, our experimental setup demonstrated the contrary; namely, the rate of meiotic recombination in the tested heterochromatic regions was similar in WT and in the ddm1-2 mutant. In this work, two of the heterochromatic regions flanked the centromeres of chromosomes 1 and 4, and one heterochromatic region was in the pericentric area on one side of the centromere of chromosome 5. Mirouze et al. (58) report similar findings in heterochromatic regions of chromosomes 1, 2, 3, and 5 using EpiRILs derived from met1. Taken together, data from both works suggest that reducing methylation in heterochromatin regions does not affect crossover rates in all Arabidopsis chromosomes.
Thus, cytosine methylation per se does not affect meiotic recombination in heterochromatin. Note that both ddm1 and met1 mutations cause decondensation of centric heterochromatin (25), but this decondensation is not associated with increased rates of crossing over. Therefore, specific states of chromatin modification may be responsible for the low rates of meiotic recombination in heterochromatic regions. For example, in the absence of cytosine methylation, as in Saccharomyces cerevisiae, chromatin modifications such as histone methylation, acetylation, or ubiquination influence activity in recombination hotspots (reviewed in ref. 59). Similarly, trimethylation of the lysine4 residue of mammalian H3 is required for hotspot identification (39, 60–62). In plants, this type of histone modification serves as a marker for active transcription but is not known to be a signal in a meiotic context (reviewed in ref. 59).
The differential impact on the genome of nonuniform methylation and/or chromatin alterations can result from a number of mechanisms. A genetic “mismatch antirecombination” model could explain the observed phenomenon. In Arabidopsis, the centromeric regions are rich in repeats, whereas the pericentric regions are rich in transposons (63). Transposon regions are, almost by definition, more polymorphic between the two parental types than are gene-rich regions. Moreover, repeats tend to accumulate more mutations than single-copy genes. It has been well established that such polymorphisms can recruit the mismatch-repair machinery during pairing and can lay the foundation for the dissociation of divergent recombination substrates (3, 64). In this case, the putative boosting effect of demethylation on recombination would be masked by the inhibitory effect of sequence divergence. This model could be tested using transgenic markers that flank heterochromatic regions in a purely isogenic background between the recombining homologs.
Alternatively, distinctive regulators of condensation may control different chromatin regions. For example, Perrella et al. (41) showed that histone H3 hyperacetylation in Arabidopsis pollen mother cells affects the distribution of meiotic crossover in a nonuniform manner throughout the genome. In centromeres heterochromatin can be distinguished from the rest of the genome by the presence of histone H3 variants, such as CENH3 (reviewed in refs. 65 and 66), which contains less H3 Lys9 methylation than does H3 (67).
An epigenetic “double-lock” model also may be the basis of the observed differential impact of methylation on genome subsections. It is possible that, in addition to methylation-related condensation, recombination is inhibited by an additional factor controlling chromatin structures in heterochromatic regions. This second lock might inhibit recombination even though the first lock had been opened. For example, it has been shown in Arabidopsis that the silencing of the heterochromatic 5S rDNA repeats array is under the dual control of both DDM1 and the siRNA machinery (68). In the present model, reduction of DDM1 activity might not be sufficient to enable recombination in heterochromatin. Such a multiple-lock protective mechanism might have provided the advantage of positive selection during evolution, because loose control of genomic recombination in repetitive regions can have highly deleterious effects.
Our results show the repressive role of methylation on meiotic recombination in euchromatin. Hypomethylation may be sufficient to trigger the changes in euchromatin necessary to allow enhanced meiotic recombination. In heterochromatin, however, loss of DDM1 function did not affect recombination despite the observed demethylation in these regions. This lack of effect may be the result of the high sequence divergence in these regions or of specific chromatin modifications that are not altered in the ddm1-2 mutant.
Materials and Methods
Plant Material.
The WT A. thaliana plants described were from the Columbia and Landsberg ecotypes. The meiotic tester Col3-4/20 was as described by Melamed-Bessudo et al. (42). The mutant ddm1-2 was kindly provided by Eric Richards (Boyce Thompson Institute, Cornell University, Ithaca, NY). The mutant met1-3 was kindly provided by Jerzy Paszkowski (University of Geneva, Switzerland).
Seeds were selected by fluorescence detection with an SMZ1500 stereomicroscope (Nikon Instruments, Inc.) adapted to the X-CITE 120PC Q light source system (Lumen Dynamics Group Inc), equipped with filter sets for Texas red (Ex 560/40, DM595 DCLP, Em 630/60) and narrow-band GFP (Ex 480/20, DM 495 LP, Em HQ 510/20).
In every experiment WT plants were grown together with mutants to avoid variability in environmental conditions.
DNA Isolation and PCR.
DNA was extracted by the CTAB method as described by Melamed-Bessudo et al. (42). For screening of the InDel markers, the primers used for PCR amplification were synthesized by Sigma-Aldrich and are described in Table S4. The products were separated on 2–3% agarose gels (Agarose I; Amresco Inc.).
Supplementary Material
Acknowledgments
We thank Prof. Ortrun Mittelsten Scheid for critical comments on the manuscript, Hadar Fuchs and Naomi Avivi-Ragolsky for technical help, and members of the A.A.L. and Paszkowski laboratoriess for useful discussions. This work was supported by a grant from the European Union Seventh Framework Programme (FP7) RECBREED Project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 5932 (volume 109, number 16).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120742109/-/DCSupplemental.
References
- 1.Lichten M. Meiotic chromatin: The substrate for recombination initiation. In: Egel R, Lankenau DH, editors. Recombination and Meiosis Models, Means, and Evolution. Vol 3. New York: Springer; 2008. pp. 165–193. [Google Scholar]
- 2.Lieberman-Lazarovich M, Levy AA. Homologous recombination in plants: An antireview. Methods Mol Biol. 2011;701:51–65. doi: 10.1007/978-1-61737-957-4_3. [DOI] [PubMed] [Google Scholar]
- 3.Emmanuel E, Yehuda E, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep. 2006;7:100–105. doi: 10.1038/sj.embor.7400577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Surtees JA, Argueso JL, Alani E. Mismatch repair proteins: Key regulators of genetic recombination. Cytogenet Genome Res. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- 5.Wahls WP, Davidson MK. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 2010;26:202–208. doi: 10.1016/j.tig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender J. Cytosine methylation of repeated sequences in eukaryotes: The role of DNA pairing. Trends Biochem Sci. 1998;23:252–256. doi: 10.1016/s0968-0004(98)01225-0. [DOI] [PubMed] [Google Scholar]
- 7.Dooner HK, He LM. Maize genome structure variation: Interplay between retrotransposon polymorphisms and genic recombination. Plant Cell. 2008;20:249–258. doi: 10.1105/tpc.107.057596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- 9.Gehring M, Henikoff S. DNA methylation dynamics in plant genomes. Biochim Biophys Acta. 2007;1769:276–286. doi: 10.1016/j.bbaexp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 11.Kankel MW, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 13.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindroth AM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 15.Cao XF, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 16.Chan SWL, et al. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 17.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJM. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- 19.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 20.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 21.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 22.Mathieu O, et al. Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell. 2003;15:2929–2939. doi: 10.1105/tpc.017467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaked H, Avivi-Ragolsky N, Levy AA. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mittelsten Scheid O, Probst AV, Afsar K, Paszkowski J. Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:13659–13662. doi: 10.1073/pnas.202380499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soppe WJ, et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fransz P, Soppe W, Schubert I. Heterochromatin in interphase nuclei of Arabidopsis thaliana. Chromosome Res. 2003;11:227–240. doi: 10.1023/a:1022835825899. [DOI] [PubMed] [Google Scholar]
- 27.Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–749. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 28.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 30.Tsukahara S, et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- 31.Hirochika H, Okamoto H, Kakutani T. Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell. 2000;12:357–369. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura A, et al. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- 33.Singer T, Yordan C, Martienssen RA. Robertson's mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1) Genes Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinders J, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirouze M, et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 36.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 37.Johnson LM, Cao XF, Jacobsen SE. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 38.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu SZ, et al. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 2009;5:e1000733. doi: 10.1371/journal.pgen.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrella G, et al. Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J. 2010;62:796–806. doi: 10.1111/j.1365-313X.2010.04191.x. [DOI] [PubMed] [Google Scholar]
- 42.Melamed-Bessudo C, Yehuda E, Stuitje AR, Levy AA. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J. 2005;43:458–466. doi: 10.1111/j.1365-313X.2005.02466.x. [DOI] [PubMed] [Google Scholar]
- 43.Kakutani T, Munakata K, Richards EJ, Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151:831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan TH. Special Articles. Science. 1912;36:718–720. doi: 10.1126/science.36.934.718. [DOI] [PubMed] [Google Scholar]
- 45.Lenormand T, Dutheil J. Recombination difference between sexes: A role for haploid selection. PLoS Biol. 2005;3:e63. doi: 10.1371/journal.pbio.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford AM, et al. An autosomal genetic linkage map of the sheep genome. Genetics. 1995;140:703–724. doi: 10.1093/genetics/140.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zenger KR, McKenzie LM, Cooper DW. The first comprehensive genetic linkage map of a marsupial: The tammar wallaby (Macropus eugenii) Genetics. 2002;162:321–330. doi: 10.1093/genetics/162.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moran GF, Bell JC, Hilliker AJ. Greater meiotic recombination in male vs female gametes in Pinus-Radiata. J Hered. 1983;74:62. [Google Scholar]
- 49.Robertson DS. different frequency in the recovery of crossover products from male and female gametes of plants hypoploid for B-a translocations in maize. Genetics. 1984;107:117–130. doi: 10.1093/genetics/107.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vizir I, Korol A. Sex difference in recombination frequency in Arabidopsis. Heredity. 1990;65:379–383. [Google Scholar]
- 51.Drouaud J, et al. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 2007;3:e106. doi: 10.1371/journal.pgen.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7:e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jander G, et al. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou XH, et al. A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. Plant J. 2010;63:880–888. doi: 10.1111/j.1365-313X.2010.04277.x. [DOI] [PubMed] [Google Scholar]
- 55.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fransz PF, et al. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell. 2000;100:367–376. doi: 10.1016/s0092-8674(00)80672-8. [DOI] [PubMed] [Google Scholar]
- 58.Mirouze M, et al. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:5880–5885. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edlinger B, Schlögelhofer P. Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J Exp Bot. 2011;62:1545–1563. doi: 10.1093/jxb/erq421. [DOI] [PubMed] [Google Scholar]
- 60.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 61.Grey C, Baudat F, de Massy B. Genome-wide control of the distribution of meiotic recombination. PLoS Biol. 2009;7:e35. doi: 10.1371/journal.pbio.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parvanov ED, Ng SH, Petkov PM, Paigen K. Trans-regulation of mouse meiotic recombination hotspots by Rcr1. PLoS Biol. 2009;7:e36. doi: 10.1371/journal.pbio.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 64.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birchler JA, Han F. Maize centromeres: Structure, function, epigenetics. Annu Rev Genet. 2009;43:287–303. doi: 10.1146/annurev-genet-102108-134834. [DOI] [PubMed] [Google Scholar]
- 66.Talbert PB, Henikoff S. Centromeres convert but don't cross. PLoS Biol. 2010;8:e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang W, Lee HR, Koo DH, Jiang J. Epigenetic modification of centromeric chromatin: Hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell. 2008;20:25–34. doi: 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blevins T, Pontes O, Pikaard CS, Meins F., Jr Heterochromatic siRNAs and DDM1 independently silence aberrant 5S rDNA transcripts in Arabidopsis. PLoS ONE. 2009;4:e5932. doi: 10.1371/journal.pone.0005932. [DOI] [PMC free article] [PubMed] [Google Scholar]