Abstract
WaaA is a key enzyme in the biosynthesis of LPS, a critical component of the outer envelope of Gram-negative bacteria. Embedded in the cytoplasmic face of the inner membrane, WaaA catalyzes the transfer of 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) to the lipid A precursor of LPS. Here we present crystal structures of the free and CMP-bound forms of WaaA from Aquifex aeolicus, an ancient Gram-negative hyperthermophile. These structures reveal details of the CMP-binding site and implicate a unique sequence motif (GGS/TX5GXNXLE) in Kdo binding. In addition, a cluster of highly conserved amino acid residues was identified which represents the potential membrane-attachment and acceptor-substrate binding site of WaaA. A series of site-directed mutagenesis experiments revealed critical roles for glycine 30 and glutamate 31 in Kdo transfer. Our results provide the structural basis of a critical reaction in LPS biosynthesis and allowed the development of a detailed model of the catalytic mechanism of WaaA.
Keywords: endotoxin, GT-B, GT-30, monotopic membrane protein
Gram-negative bacteria possess an asymmetrically organized outer membrane that is composed of various glycerophospholipids in the inner leaflet and nearly exclusively of LPS in the outer leaflet. LPS is a critical component of the outer envelope, essential for viability and growth of a majority of Gram-negative bacteria. It serves as a permeability barrier, protecting the bacterial cell from potentially lethal small molecules and antimicrobials. Endotoxically active forms of LPS participate in a diverse spectrum of pathological and physiological activities associated with the human host's immune response (1).
LPS typically consists of three covalently linked sections: the membrane-anchored lipid A, a central core oligosaccharide, and a distal O-specific polysaccharide chain (2). Common to all LPS molecules is the presence of a strictly conserved 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) residue in the core oligosaccharide, which links the carbohydrate portion to the lipid A (3). The ubiquitous nature of Kdo within LPS structures and its indispensable role in maintaining outer membrane integrity and viability of the majority of Gram-negative bacteria (4, 5) have led to extensive studies of its synthesis (6–8), activation (9, 10), and incorporation into the maturating LPS molecule. The incorporation of Kdo is catalyzed by the membrane-embedded Kdo transferase WaaA, which transfers Kdo from the donor substrate CMP-Kdo to a 4′-phosphorylated lipid A precursor at the cytoplasmic face of the inner membrane. Remarkably, Kdo transferases of different bacteria are mono-, bi-, tri-, or even tetrafunctional (2, 11), typically consistent with the number of Kdo residues present in the LPS core oligosaccharide.
Structural studies aimed at unraveling the mechanism of Kdo transfer have not been reported so far. We recently characterized the strictly monofunctional Kdo transferase from the ancient hyperthermophilic, Gram-negative bacterium Aquifex aeolicus (WaaAAAE) (12), a member of the glycosyltransferase (GT) family 30 (13). Here, we report the X-ray crystal structure of WaaAAAE in its substrate-free form and in complex with one of its products, CMP. Based on the structure, we designed several protein variants to investigate the structural basis for catalysis. These data have provided experimental support for a mechanistic model of WaaA function.
Results and Discussion
Overall Structure.
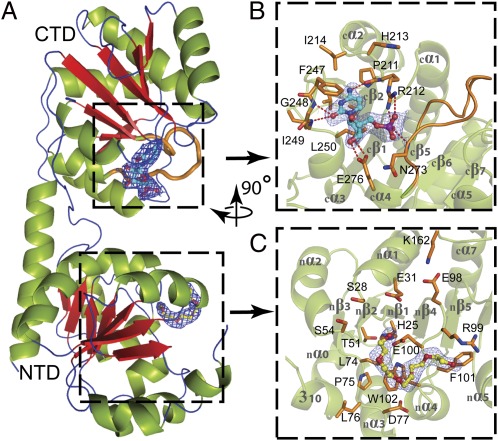
The 2.0-Å crystal structure of the Kdo transferase WaaAAAE reveals that this enzyme adopts the GT superfamily B (GT-B) fold. This fold consists of two Rossmann-like domains, each composed of a core of seven parallel β-strands sandwiched by α-helices (Fig. 1A). A structural-similarity search using the DALI server (14) identified UDP-N-acetylglucosamine-2-epimerase from Thermus thermophilus (PDB ID code 1V4V, z-score 20.7), phosphatidylinositol mannosyltransferase from Mycobacterium smegmatis (PDB ID code 2GEK, z-score 19.4) (15), MurG from Escherichia coli (PDB ID code 1NLM, z-score 17.9) (16), and WaaG from E. coli (PDB ID code 2IV7, z-score 17.2) (17) as closest homologs. The N-terminal acceptor-substrate binding domain of WaaAAAE comprises residues 1–157 and 331–352; the C-terminal donor-substrate binding domain includes residues 177–330. The two domains are joined by an extended linker (residues 158–176) between strands nβ7 and cβ1 as well as by association of the C-terminal helix cα7 (residues 331–352) with strands nβ4–nβ6 of the N-terminal domain (Fig. 1 and Fig. S1). The arrangement of the two domains results in the formation of a large interdomain groove that is about 23 Å deep and 20 Å across at its widest points (Figs. 1A and 2A), reminiscent of previously described structures of GT-B members in the open conformation.
Fig. 1.
Domain organization of WaaAAAE. (A) Overall structure. β-Strands are shown in red, α-helices in green, and loops and turns in blue. (B) Donor-substrate binding site of the C-terminal domain (CTD). Residues forming the CMP-binding site are shown in orange. Dashed lines indicate hydrogen bonds. In A and B the putative Kdo-binding loop (cβ5–cα4, 263GGTFVNIGGHNLLE276) is highlighted in orange. (C) Putative acceptor-substrate binding site of the N-terminal domain (NTD) occupied by a PEG molecule. Residues potentially involved in acceptor-substrate binding are shown in orange. The final 2|Fo|-|Fc| electron-density maps surrounding the bound CMP and PEG molecules are contoured at 1 σ above the mean. Oxygen atoms are colored red, nitrogen atoms blue, and the phosphorous atom purple. The orientation of B and C with respect to A is indicated by the rotation axis.
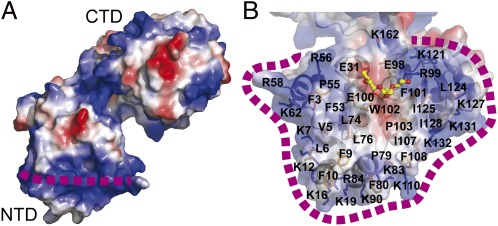
Fig. 2.
Putative membrane association of WaaAAAE. (A) Surface representation of WaaAAAE calculated with the program ABPS (41), color-coded according to the electrostatic potential (positive potential, blue; negative potential, red). The dashed purple line indicates the horseshoe of basic amino acid residues (for details see Results and Discussion). (B) Transparent surface representation as in A showing the putative membrane-association/acceptor-substrate binding site of the N-terminal domain. Lysine and arginine side chains constituting the horseshoe are shown as blue sticks. The bound PEG molecule is shown in stick representation, demonstrating that the putative membrane-association site and the acceptor-substrate binding site overlap with each other.
The N-terminal domain contains a hydrophobic, surface-exposed patch surrounded by a horseshoe-like ribbon of basic amino acid residues, which may allow the N-terminal domain to immerse into the membrane through electrostatic and hydrophobic interactions with the phospholipids (Fig. 2 A and B). This mode of membrane association would enable the lipid A precursor to access the active site (see below). Seeking support for this view, we used a computational approach that optimizes the spatial arrangement of protein structures in lipid bilayers, accounting for hydrophobic, hydrogen bonding, and electrostatic interactions with the anisotropic water–lipid environment (18). This computational approach produced a membrane-binding mode for WaaAAAE that is identical to the one described above. Sequence analyses predict that many Kdo transferases contain an N-terminal transmembrane helix. Clearly, however, the N-terminus of WaaAAAE, comprising a short α (nα0) and a 310 helix, is an integral part of the potential acceptor-substrate binding domain (Fig. 1C), again suggesting that membrane interaction is achieved solely by the distinctive arrangement of surface residues described above. Further details on the overall structure of WaaAAAE are provided in SI Results and Discussion.
Donor-Substrate Binding Site.
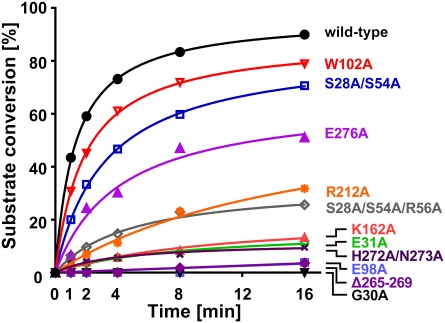
The 2.4-Å structure of WaaAAAE in complex with CMP reveals that the CMP-binding site is located in the central groove between the two domains. This site is composed mainly of residues from loops cβ2–cα2 (residues 211–214), cβ4–cα3 (residues 247–250), and cβ5–cα4 (residues 272–276) of the C-terminal domain (Fig. 1 A and B). The cytosine moiety of the nucleotide is accommodated in a hydrophobic pocket lined by residues V210, I214, V244, F247, I249, and L250. Residues F247 and L250 sandwich the cytosine base by π–π stacking and van der Waals interactions, respectively (Fig. 1B). The most prominent difference between the substrate-free and CMP-bound structure is the position of the aromatic side chain of F247, which reorients in the WaaAAAE:CMP complex to shield the base from bulk solvent. An analogous reorientation has been observed for F244 in the MurG:UDP-GlcNAc complex (16). Other notable contacts include hydrogen bonds between the cytosine N4 atom and the main-chain carbonyl of P211 and between the cytosine O2 atom and the main-chain amides of G248 and I249. The ribose hydroxyl groups form two hydrogen bonds with the carboxyl group of E276; the phosphate is engaged in three hydrogen bonds, two with the R212 side chain and one with the main-chain amide of N273 (Fig. 1B). The substitution of E276 by alanine in WaaAAAE causes a decrease in the catalytic activity of the enzyme (Fig. 3), similar to observations in the corresponding E269A variant of MurG (16).
Fig. 3.
WaaAAAE activity assay. The relative conversion of lipid IVA (a lipid A precursor) to Kdo-lipid IVA is shown for wild-type WaaAAAE and various protein variants at indicated time points. For details see Materials and Methods.
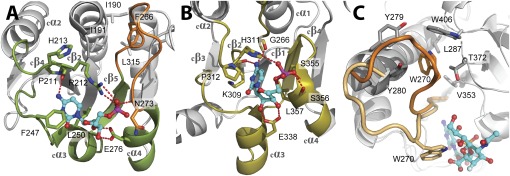
The similarities between Kdo and N-acetylneuraminic acid (Neu5Ac, a sialic acid), both of which are used as CMP-activated sugars in glycosyl transfer and contain a carboxylated anomeric carbon, prompted us to compare the CMP-binding mode of WaaAAAE with those of the sialyltransferases that adopt the GT-B fold (19–23). Superimposition of the respective C-terminal domains revealed that CMP binds to matching protein regions. Although key interactions between CMP and protein are preserved (e.g., hydrophobic residues sandwiching the cytosine base and glutamate residues contacting the ribose hydroxyl groups), the positions of the amino acid residues mediating these interactions are variable (Fig. 4 A and B and Fig. S2). Strikingly, the highly conserved amino acid triplet 211PRH213 of WaaAAAE aligns structurally with the HP motif of sialyltransferases (Fig. 4 A and B). It has been proposed that the histidine of the HP motif, which contacts the CMP-phosphate group (24), stabilizes the CMP-leaving group during the glycosyl transfer reaction (20, 22, 23). Because R212 of WaaAAAE hydrogen bonds to the CMP phosphate group in an analogous fashion, it may have a similar function. Although an equivalent arginine residue in the trifunctional Kdo transferase of Chlamydia trachomatis is crucial for the synthesis of the chlamydial family-specific LPS epitope (25), the R212A substitution in WaaAAAE clearly affected but did not completely prevent Kdo transfer (Fig. 3). Hence, we cannot define the function of this conserved arginine in WaaAAAE precisely. A more detailed discussion of the comparison of WaaAAAE with the sialyltransferases is provided in SI Results and Discussion.
Fig. 4.
Comparison of the donor-substrate binding sites of WaaAAAE (A) and a sialyltransferase of the GT-B superfamily [e. g., PM0188 (19)]. (B). Equivalent protein regions involved in the binding of the CMP moiety of the donor substrate are highlighted in green (WaaAAAE) or olive (PM0188). In both structures, an α/β/α motif, overlapping with the α/β/α motif of MurG (16), is involved in CMP binding. Hydrogen bonds are depicted as red dotted lines, and residues involved in CMP binding are shown as sticks. Note that the positions of hydrophobic residues sandwiching the cytosine base (F247/L250 for WaaAAAE and P312/L357 in PM0188) and the glutamate residue contacting the ribose hydroxyls (E276 in WaaAAAE and E338 in PM0188) vary within the equivalent protein regions. The HP motif of PM0188 (H311/P312) overlaps with the highly conserved 211PRH213 triplet of WaaAAAE. The putative Kdo-binding loop of WaaAAAE in A is highlighted in orange. For clarity, the 3F-Neu5Ac moiety of the donor substrate in B has been omitted. (C) Conformational changes upon donor-substrate binding as evident from superimposed structures of PM0188 (23). In the apo form, W270 (orange) is part of a hydrophobic cluster. Binding of the donor-substrate analog CMP-3F-Neu5Ac brings W270 (beige) into contact with the 3F-Neu5Ac moiety of the donor substrate. For clarity, the perspective of C is different from B.
Sequence analysis reveals that loop cβ5–cα4 of WaaAAAE coincides with the conserved motif GGS/TX5GXNXLE (263GGTFVNIGGHNLLE276 in WaaAAAE) that is highly specific for Kdo transferases (Fig. S3). The position of CMP within the donor-substrate binding site suggests that residues of this motif might be involved directly in Kdo binding (Fig. 4A). Loop regions that fold over the donor substrate have been described for a number of GTs (26). In the GT-B sialyltransferase PM0188 (19, 23), W270 is located at the tip of a loop and sticks into a hydrophobic pocket, as does F266 of WaaAAAE. Binding of a donor-substrate analog to PM0188 induces a reorientation of the loop that places W270 into close contact with the 3F-Neu5Ac moiety of the donor substrate (Fig. 4C) (23). A similar rearrangement of 263GGTFVNIGGHNLLE276 in WaaAAAE could bring F266 close to the Kdo sugar. This conformation might protect the very labile CMP-Kdo substrate (27) from hydrolysis. Unfortunately, we did not observe a rearrangement of loop cβ5–cα4 in our CMP complex, most likely because of the absence of the Kdo moiety. However, either deletion of residues 265–269 or the substitution H272A/N273A drastically affected the ability of WaaAAAE to transfer Kdo (Fig. 3), indicating that loop cβ5–cα4 is indeed functionally important.
Putative Acceptor-Substrate Binding Site.
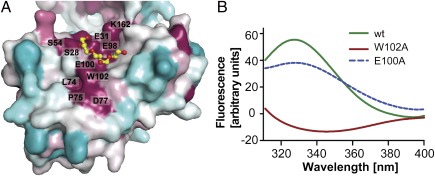
A striking feature of the WaaAAAE N-terminal domain is a depression that extends from the putative membrane-association site to the central groove between the two domains. Several lines of evidence suggest that this particular region may serve as the acceptor-substrate binding site for the lipid A precursor. First, residues at the base of the depression, S28, the potential general base E31 (see below), S54, L74, P75, D77, E98, E100, and W102, are highly conserved among Kdo transferases (Figs. 1C and 5A and Fig. S3). Second, we detected an |Fo|-|Fc| difference density in the putative acceptor-substrate binding site, which we interpreted as a polyethylene glycol (PEG) molecule after GLC/MS analysis of the protein stock solution. PEG molecules were found also to interact with the acceptor-binding site of the sialyltransferase from Neisseria meningitidis (22). Third, structures of the GT-B enzymes UGT72B1 (28), OleI (29), and UGT78G1 (30) display acceptor substrates (or analogs) bound to equivalent regions (Fig. S4). Finally, increased tryptophan fluorescence was observed upon addition of the lipid A precursor lipid IVA to solutions of wild-type protein but not to solutions of the W102A variant (Fig. 5B; see SI Results and Discussion for more details), implicating W102 in acceptor-substrate binding. For large ligands like lipid IVA, multiple interactions with WaaAAAE are expected to occur, possibly explaining why the replacement of W102 by alanine had only a limited effect on WaaAAAE activity (Fig. 3). In agreement with this interpretation, combined alanine substitutions of amino acid residues proposed to interact with the 4′-phosphate group of the lipid A precursor substrate increasingly affected WaaAAAE activity (see below).
Fig. 5.
Conservation and tryptophan-quenching experiments in the putative acceptor-substrate binding site of WaaAAAE. (A) Surface conservation plot generated with Consurf (42) and color-coded as cyan (variable) to white (average conservation) to purple (high conservation). The base of the putative acceptor-substrate binding site harbors several highly conserved amino acid residues such as W102. The PEG molecule is shown in stick representation. (B) Tryptophan-fluorescence difference spectra obtained by subtracting the spectra measured for the isolated individual protein solutions from those recorded in the presence of lipid IVA (a lipid A precursor). An increase in tryptophan fluorescence is observed for the wild-type protein and the E100A variant used as a control but not for the W102A variant, indicating that the hydrophobic part of lipid IVA interacts with W102.
Taken together, our data are consistent with a mechanistic model in which membrane association of the N-terminal domain of WaaAAAE inserts the putative acceptor-substrate binding site into the lipid bilayer in an ideal position to interact with the membrane-anchored lipid A precursor. The opening of the horseshoe-like ribbon of basic residues, defined by R56, R99, and K121 (Fig. 2B), would allow the entry of the lipid A precursor into the acceptor-substrate binding site.
Implications for the Reaction Mechanism.
Substrate-induced closing of the interdomain groove of GT-B enzymes places the donor and acceptor substrates as well as catalytically important amino acid residues into close contact (16, 28, 31). The required rigid-body motion has been attributed to rotation around pivot points located in the interdomain linker and the C-terminal regions (16, 32). DynDom (33) analysis reveals equivalently positioned hinges (residues 165–166 and 322–332) in WaaAAAE. Following closure of the central groove, the transfer reaction takes place at the newly formed interface between the N- and C-terminal domains (Fig. 6A). Kdo transfer requires a general base to deprotonate the 6′-hydroxyl group of the lipid A precursor. The strictly conserved amino acid residues E31 and E98, both part of the putative acceptor-substrate binding site and located at the interdomain groove, seem to be in a position to fulfill this function in WaaAAAE. Independent substitutions of E31 and E98 with alanine point to crucial catalytic roles for both residues (Fig. 3) but do not allow the unambiguous identification of the general base. However, structural alignment of respective N-terminal domains reveals that E31 of WaaAAAE, but not E98, aligns with the general bases of WaaC (34) (rmsd 2.9 Å for 102 aligned Cα atoms), UGT72B1 (28) (rmsd 3.1 Å for 120 aligned Cα atoms), and OleI (29) (rmsd 2.8 Å for 107 aligned Cα -atoms) (Fig. S5). In addition, the general base of many GT-B members is preceded by a glycine (28, 29, 34, 35), and a glycine corresponding to G30 in WaaAAAE is highly conserved across Kdo transferases (Fig. S3). Careful inspection of the ternary enzyme–substrate complexes of UGT72B1 and OleI revealed that a side chain other than glycine preceding the general base would cause a steric clash with the donor substrate (28, 29). We indeed found the G30A variant of WaaAAAE to be completely inactive (Fig. 3), in agreement with this hypothesis. This result underscores the importance of the conserved glycine 30 and supports our assertion that E31 is indeed the general base (Fig. 6B).
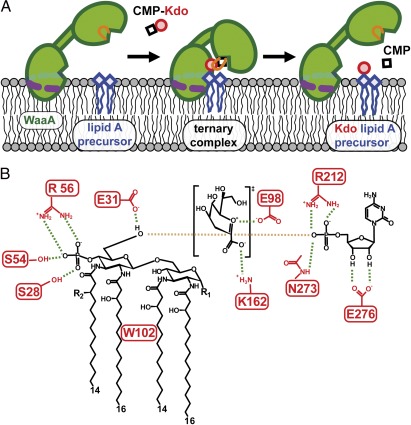
Fig. 6.
Schematic representation of the model for Kdo transfer in WaaAAAE. (A) Membrane association and large-scale conformational changes upon substrate binding. The dashed purple line marks the horseshoe-like arrangement of basic residues interacting with the negatively charged groups of membrane components. Upon substrate binding, reorientation of the Kdo-binding loop (orange) and domain closure takes place, followed by the Kdo transfer step. (B) Interactions proposed to play a role in substrate binding and catalysis. Tetraacyl-4′-phosphate lipid A of A. aeolicus is shown as the acceptor for Kdo glycosylation. Galacturonic acid or phosphate (R1) and ester-bound octadecanoic acid (R2) could make up the natural acceptor. Residue E31 serves as the catalytic base, E98 and K162 interact with the oxocarbenium ion-like intermediate of Kdo, and R212 and N273 are involved in phosphate binding. The crucial 4′-phosphate group of the acceptor substrate interacts with R56 and the highly conserved S28 and S54. Hydrogen bonds and electrostatic contacts are drawn as green and brown dashed lines, respectively.
To be deprotonated, the 6′-hydroxyl group of the lipid A precursor must be in close proximity to E31. This orientation of the lipid A precursor with respect to the N-terminal domain would place the 4′-phosphate of the acceptor substrate in the vicinity of R56 and the highly conserved S28 and S54 (Fig. 6B). Thus, together with the dipole of the nα1 helix (Fig. 1C), a suitable environment for efficient binding of the 4′-phosphate exists at this position. Consistent with this model of multiple interactions, the WaaAAAE variant S28A/S54A/R56A displayed more severe catalytic defects than did the S28A/S54A variant (Fig. 3).
With E31 acting as the general base, E98 may play a role in stabilizing the transition state by electrostatic pairing with the positively charged oxocarbenium ion-like transition state (Fig. 6B). Such an interaction has been proposed also for E249 of the fucosyltransferase FucT of Helicobacter pylori (36). The highly conserved linker residue K162 located in close proximity to E31 and E98 is another important residue that might facilitate further transition-state formation in WaaAAAE (Fig. 6B). Similar roles have been proposed for R373 in influenza-B virus neuraminidase (37) and R282 in the lipooligosaccharide sialyltransferase of N. meningitidis (22). We replaced WaaAAAE residue K162 with alanine, and, as previously observed for the R282A variant of the latter sialyltransferase, the replacement severely affected the enzymatic activity (Fig. 3).
Recent biochemical studies on the bifunctional Kdo transferase of E. coli (WaaAECO) have provided experimental evidence that amino acid residues 20–30 and 91–165 of the N-terminal acceptor-substrate binding domain play a critical role in the ability of this enzyme to add two Kdo molecules to the lipid IVA acceptor substrate (38). There is no equivalent for WaaAECO residues 20–30 in the monofunctional WaaAAAE. WaaAECO segment 91–165, on the other hand, corresponds to WaaAAAE region 59–121 (Fig. S3). The acceptor-substrate promiscuity of the WaaA enzymes suggests there can be sufficient flexibility in this region of the N-terminal domain to accommodate various acceptor substrates, including Kdo-glycosylated lipid A precursors. Because residues corresponding to WaaAAAE E31, E98, and K162 are highly conserved across Kdo transferases of different functionality, these residues might perform identical functions (deprotonation of the acceptor substrate and transition-state stabilization) upon transfer of two or more Kdo residues to the acceptor molecules. However, a complete understanding of the determinants conferring different functionality to Kdo transferases awaits further investigations.
In conclusion, the determination and analysis of the WaaAAAE Kdo transferase structure suggests a plausible model for membrane association of the enzyme and, along with extensive site-directed mutagenesis data, provides insight into the binding of the substrates CMP-Kdo and lipid A precursor. Our results provide a structural framework for future research addressing further details of the Kdo transfer reaction that holds promise for the development of unique antimicrobials targeting a critical step of LPS biosynthesis.
Materials and Methods
Recombinant Protein Production and Purification.
Wild-type WaaA of A. aeolicus was overproduced and purified to homogeneity essentially as reported (12), except that 0.1% (vol/vol) Triton X-100 in the WaaAAAE buffer was replaced by 2 mM 6-cyclohexyl-1-hexyl-β-d-maltoside (Cymal-6) in the final HiTrap Heparin-Sepharose HP purification and dialysis steps. The heparin resin was washed with at least 50 column-volumes of the Cymal-6–containing buffer before elution of WaaAAAE.
Crystallization, Data Collection, and Structure Determination.
WaaAAAE was crystallized using the hanging-drop vapor-diffusion technique by mixing equal volumes of protein stock solution and reservoir [100 mM Tris⋅HCl (pH 8.5), 35–40% (vol/vol) PEG 400, 200 mM Na-citrate, 50 mM β-mercaptoethanol]. Crystal growth at 19 °C took up to 2 wk. Crystals of WaaAAAE in complex with CMP were obtained by soaking crystals in 100 mM Tris⋅HCl (pH 8.5), 36% (vol/vol) PEG 400, 200 mM Na-citrate, and 10 mM CMP for 4 d. X-ray diffraction data were collected at Berlin Electron Storage Ring Society for Synchrotron Radiation II (BESSY) (Berlin, Germany), Deutsches Elektronen-Synchrotron (DESY) (Hamburg, Germany), and MAX-lab National Laboratory for Synchrotron Radiation (Lund, Sweden). The structure was determined using a single isomorphous replacement with anomalous scattering (SIRAS) approach. Details of methods used for crystallization, data collection, structure determination and refinement are provided in SI Materials and Methods. Refinement statistics are summarized in Table S1. Figures were made with PyMOL (Schrödinger LLC).
Mutagenesis.
Amino acid substitutions in WaaAAAE and construction of the WaaAΔ265–269 deletion variant were performed as detailed in SI Materials and Methods. Primers used for site-directed mutagenesis are listed in Table S2. The conditions used to purify the WaaAAAE variants were basically as described for the wild-type protein (12), except that the final detergent-replacement step was omitted, and the temperature of the heating step was reduced to 70 °C during purification of WaaAAAE W102A.
Analysis of WaaAAAE Activity.
The WaaAAAE-catalyzed transfer of Kdo from CMP-Kdo to the synthetic tetraacyl-1,4′-bisphosphate lipid A precursor 406 was assayed as previously reported (12). To identify catalytically important residues, each WaaAAAE variant was assayed at 60 °C for 1, 2, 4, 8, and 16 min. The reaction products were analyzed by electrospray-ionization Fourier-transformed ion cyclotron mass spectrometry in the negative ion mode using a hybrid Apex Qe Instrument (Bruker Daltonics) equipped with a 7-Tesla actively shielded magnet. Details on sample preparation for MS analysis have been reported (12, 39). Compound 406 has been synthesized as described previously (40). To obtain the percentage of conversion at different time points provided in Fig. 3, the absolute intensity value of the Kdo–Compound 406 peak was divided by the sum of the absolute intensity values of the Kdo–Compound 406 and the Compound 406 peaks.
Fluorescence Measurements and GLC/MS Analysis of WaaAAAE.
GLC/MS analyses of WaaAAAE and tryptophan-fluorescence experiments were performed according to standard protocols. For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Brigitte Kunz and Regina Engel (Research Center Borstel) for technical assistance; M. S. Weiss (European Molecular Biology Laboratory Outstation at Deutsches Elektronen-Synchrotron), and T. Ursby (MAX-lab National Laboratory for Synchrotron Radiation) for assistance with synchrotron data collection; and Tod Holler at the College of Pharmacy of the University of Michigan for critical reading of the manuscript. Access to beamline BL14.2 of the Berlin Electron Storage Ring Society for Synchrotron Radiation storage ring was provided by the Joint Berlin MX-Laboratory. Optimization of WaaAAAE crystals was performed within the OptiCryst project of the European Commission (Contract LSH-2005-037793; http://www.opticryst.org/). This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants MA 1408/2 (to U.M.) and ME 2741/1 (to J.R.M. and R.H.); by the Transnational Access to Research Infrastructures program of the European Commission; and by National Institutes of Health Grant AI61531 (to R.W.W.). R.H. was supported by the DFG Cluster of Excellence “Inflammation at Interfaces” EXC 306, by the Fonds der Chemischen Industrie, and by the Chinese Academy of Sciences through Visiting Professorship for Senior International Scientists Grant 2010T1S6.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates described in this article have been deposited in the Protein Data Bank, http://www.pdb.org (PDB ID codes 2XCI and 2XCU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119894109/-/DCSupplemental.
References
- 1.Wiese A, Brandenburg K, Ulmer AJ, Seydel U, Müller-Loennies S. The dual role of lipopolysaccharide as effector and target molecule. Biol Chem. 1999;380:767–784. doi: 10.1515/BC.1999.097. [DOI] [PubMed] [Google Scholar]
- 2.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holst O. The structures of core regions from enterobacterial lipopolysaccharides - an update. FEMS Microbiol Lett. 2007;271:3–11. doi: 10.1111/j.1574-6968.2007.00708.x. [DOI] [PubMed] [Google Scholar]
- 4.Belunis CJ, Clementz T, Carty SM, Raetz CR. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J Biol Chem. 1995;270:27646–27652. doi: 10.1074/jbc.270.46.27646. [DOI] [PubMed] [Google Scholar]
- 5.Rick PD, Young DA. Isolation and characterization of a temperature-sensitive lethal mutant of Salmonella typhimurium that is conditionally defective in 3-deoxy-d-manno-octulosonate-8-phosphate synthesis. J Bacteriol. 1982;150:447–455. doi: 10.1128/jb.150.2.447-455.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meredith TC, Woodard RW. Escherichia coli YrbH is a d-arabinose 5-phosphate isomerase. J Biol Chem. 2003;278:32771–32777. doi: 10.1074/jbc.M303661200. [DOI] [PubMed] [Google Scholar]
- 7.Radaev S, Dastidar P, Patel M, Woodard RW, Gatti DL. Structure and mechanism of 3-deoxy-d-manno-octulosonate 8-phosphate synthase. J Biol Chem. 2000;275:9476–9484. doi: 10.1074/jbc.275.13.9476. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Woodard RW. Escherichia coli YrbI is 3-deoxy-d-manno-octulosonate 8-phosphate phosphatase. J Biol Chem. 2003;278:18117–18123. doi: 10.1074/jbc.M301983200. [DOI] [PubMed] [Google Scholar]
- 9.Ray PH, Benedict CD. CTP:CMP-3-deoxy-d-manno-octulosonate cytidylyltransferase (CMP-KDO synthetase) Methods Enzymol. 1982;83:535–540. doi: 10.1016/0076-6879(82)83049-8. [DOI] [PubMed] [Google Scholar]
- 10.Heyes DJ, et al. Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases. J Biol Chem. 2009;284:35514–35523. doi: 10.1074/jbc.M109.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brabetz W, Lindner B, Brade H. Comparative analyses of secondary gene products of 3-deoxy-d-manno-oct-2-ulosonic acid transferases from Chlamydiaceae in Escherichia coli K-12. Eur J Biochem. 2000;267:5458–5465. doi: 10.1046/j.1432-1327.2000.01619.x. [DOI] [PubMed] [Google Scholar]
- 12.Mamat U, et al. WaaA of the hyperthermophilic bacterium Aquifex aeolicus is a monofunctional 3-deoxy-d-manno-oct-2-ulosonic acid transferase involved in lipopolysaccharide biosynthesis. J Biol Chem. 2009;284:22248–22262. doi: 10.1074/jbc.M109.033308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltranferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 14.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerin ME, et al. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J Biol Chem. 2007;282:20705–20714. doi: 10.1074/jbc.M702087200. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, et al. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci USA. 2003;100:845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Fleites C, et al. Insights into the synthesis of lipopolysaccharide and antibiotics through the structures of two retaining glycosyltransferases from family GT4. Chem Biol. 2006;13:1143–1152. doi: 10.1016/j.chembiol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: A computational approach. Protein Sci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DU, Yoo JH, Lee YJ, Kim KS, Cho HS. Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar. BMB Rep. 2008;41:48–54. doi: 10.5483/bmbrep.2008.41.1.048. [DOI] [PubMed] [Google Scholar]
- 20.Kakuta Y, et al. Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 alpha2,6-sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: Catalytic mechanism and substrate recognition. Glycobiology. 2008;18:66–73. doi: 10.1093/glycob/cwm119. [DOI] [PubMed] [Google Scholar]
- 21.Iwatani T, et al. Crystal structure of α/β-galactoside α2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum. FEBS Lett. 2009;583:2083–2087. doi: 10.1016/j.febslet.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 22.Lin LYC, et al. Structure and mechanism of the lipooligosaccharide sialyltransferase from Neisseria meningitidis. J Biol Chem. 2011;286:37237–37248. doi: 10.1074/jbc.M111.249920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni L, et al. Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry. 2007;46:6288–6298. doi: 10.1021/bi700346w. [DOI] [PubMed] [Google Scholar]
- 24.Audry M, et al. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology. 2011;21:716–726. doi: 10.1093/glycob/cwq189. [DOI] [PubMed] [Google Scholar]
- 25.Ekpo P, Nano FE. Arg276 of GseA, a Chlamydia trachomatis Kdo transferase, is required for the synthesis of the chlamydial genus-specific epitope in Escherichia coli. FEMS Microbiol Lett. 1992;75:49–53. doi: 10.1016/0378-1097(92)90455-w. [DOI] [PubMed] [Google Scholar]
- 26.Qasba PK, Ramakrishnan B, Boeggeman E. Substrate-induced conformational changes in glycosyltransferases. Trends Biochem Sci. 2005;30:53–62. doi: 10.1016/j.tibs.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Lin CH, Murray BW, Ollmann IR, Wong CH. Why is CMP-ketodeoxyoctonate highly unstable? Biochemistry. 1997;36:780–785. doi: 10.1021/bi962055c. [DOI] [PubMed] [Google Scholar]
- 28.Brazier-Hicks M, et al. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolam DN, et al. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc Natl Acad Sci USA. 2007;104:5336–5341. doi: 10.1073/pnas.0607897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modolo LV, et al. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids. J Mol Biol. 2009;392:1292–1302. doi: 10.1016/j.jmb.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Ni L, et al. Cytidine 5′-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry. 2006;45:2139–2148. doi: 10.1021/bi0524013. [DOI] [PubMed] [Google Scholar]
- 32.Mulichak AM, et al. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci USA. 2003;100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 34.Grizot S, et al. Structure of the Escherichia coli heptosyltransferase WaaC: Binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose. J Mol Biol. 2006;363:383–394. doi: 10.1016/j.jmb.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 35.Shao H, et al. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17:3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun HY, et al. Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design. J Biol Chem. 2007;282:9973–9982. doi: 10.1074/jbc.M610285200. [DOI] [PubMed] [Google Scholar]
- 37.Burmeister WP, Henrissat B, Bosso C, Cusack S, Ruigrok RW. Influenza B virus neuraminidase can synthesize its own inhibitor. Structure. 1993;1:19–26. doi: 10.1016/0969-2126(93)90005-2. [DOI] [PubMed] [Google Scholar]
- 38.Chung HS, Raetz CR. Interchangeable domains in the Kdo transferases of Escherichia coli and Haemophilus influenzae. Biochemistry. 2010;49:4126–4137. doi: 10.1021/bi100343e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondakova AN, Lindner B. Structural characterization of complex bacterial glycolipids by Fourier transform mass spectrometry. Eur J Mass Spectrom (Chichester, Eng) 2005;11:535–546. doi: 10.1255/ejms.721. [DOI] [PubMed] [Google Scholar]
- 40.Imoto M, et al. Chemical synthesis of phosphorylated tetraacyl disaccharide corresponding to a biosynthetic precursor of lipid A. Tetrahedron Lett. 1984;25:2667–2670. [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33(Web Server issue):W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.