Abstract
S100A4 is a member of the S100 family of calcium-binding proteins that is directly involved in tumor metastasis. It binds to the nonmuscle myosin IIA (NMIIA) tail near the assembly competence domain (ACD) promoting filament disassembly, which could be associated with increasing metastatic potential of tumor cells. Here, we investigate the mechanism of S100A4–NMIIA interaction based on binding studies and the crystal structure of S100A4 in complex with a 45-residue-long myosin heavy chain fragment. Interestingly, we also find that S100A4 binds as strongly to a homologous heavy chain fragment of nonmuscle myosin IIC as to NMIIA. The structure of the S100A4–NMIIA complex reveals a unique mode of interaction in the S100 family: A single, predominantly α-helical myosin chain is wrapped around the Ca2+-bound S100A4 dimer occupying both hydrophobic binding pockets. Thermal denaturation experiments of coiled-coil forming NMIIA fragments indicate that the coiled-coil partially unwinds upon S100A4 binding. Based on these results, we propose a model for NMIIA filament disassembly: Part of the random coil tailpiece and the C-terminal residues of the coiled-coil are wrapped around an S100A4 dimer disrupting the ACD and resulting in filament dissociation. The description of the complex will facilitate the design of specific drugs that interfere with the S100A4–NMIIA interaction.
Keywords: Ca2+-binding, myosin filaments, protein–protein interactions, cell migration
S100A4 is a member of the S100 family of small, dimeric, EF-hand Ca2+-binding proteins. S100 proteins are present exclusively in vertebrates. They are expressed in a tissue specific manner and they regulate specific extra- and intracellular processes (i.e., cell cycle regulation, cell growth, cell differentiation, and motility). S100 proteins form mainly homodimers stabilized by noncovalent interactions between two helices from each subunit (helices 1, 4, 1′, 4′) that form an X-type four-helix bundle dimerization motif. Each S100 subunit contains two Ca2+-binding motifs: an N-terminal pseudo- and a C-terminal canonical EF-hand (1, 2). The elevated concentration of S100A4 is observed in a number of human cancers such as breast, colorectal, ovarian, pancreatic, and others (3, 4). Overexpression of S100A4 enhanced the metastatic capability of mammary tumors (5, 6). Extracellularly, binding to annexin A2 (ANXA2) S100A4 activates matrix metalloproteinases through plasminogen activation (7), whereas in the cytoplasm it interacts with cytoskeletal elements like F-actin (8), tropomyosin (9), and nonmuscle myosin IIA (NMIIA) (10), and it has a role in cell shape remodeling. S100A4 has also been shown to interact with several other proteins: p53, CCN3, liprin β1, methionine aminopeptidase 2, and Smad3 (11–15).
Upon Ca2+ binding, S100A4 undergoes a large conformational shift in the canonical EF-hand resulting in the exposure of a hydrophobic binding pocket formed by helices 3, 4, the hinge region (loop 2), and the C-terminal coil region (3) (Fig. 1). Although the structural transition on binding Ca2+ is well characterized, atomic details of S100A4 binding to any of its partners have not been described yet. In two crystal structure of Ca2+-bound S100A4 the hydrophobic cleft of each subunit is occupied by the C terminus of an adjacent dimer (16, 17), which could explain the formation of S100A4 oligomers that were observed extracellularly (18). These clefts were also shown to interact with the calmodulin antagonist trifluoperazine (19). Interestingly, the known complex structures of the S100 family do not reveal a uniform binding mode as the orientations of the target peptides in each complex are considerably different. Nevertheless, all the structural models reveal that one S100 dimer binds symmetrically to two, predominantly α-helical peptide ligands (20–26).
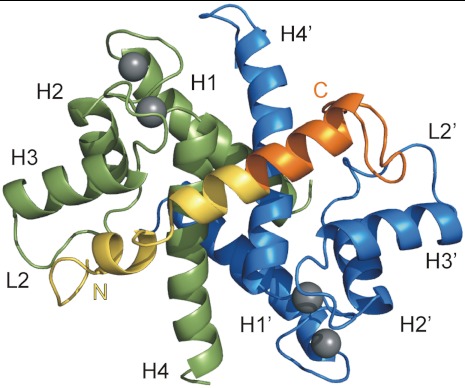
Fig. 1.
Overview of S100A4 in complex with the 45-residue-long NMIIA tail fragment along the twofold symmetry axis of the dimer. S100A4 subunit A and B are shown in green and blue, respectively, while NMIIA peptide in yellow (residues 1893–1913) and orange (residues 1914–1935), and Ca2+ ions are gray. The main secondary structural elements are indicated.
One of the most investigated interaction partners of S100A4 is NMIIA. It has been demonstrated that metastasis-associated cellular motility is coupled to S100A4–NMIIA interaction (3, 27). The Ca2+-dependent interaction of S100A4 with NMIIA prevents filament assembly and promotes filament disassembly (8, 10, 28, 29). The increased cytoskeletal dynamics leads to the formation of few side protrusions and extensive forward protrusions in S100A4 expressing cells (29). S100A4 binds to the C-terminal end of the coiled-coil tail of NMIIA (29) overlapping the assembly competence domain (ACD) that is required for filament formation (30–34). Despite several efforts to map the S100A4 binding site on NMIIA (16, 35, 36), the observations are ambiguous, and until now there is no detailed structural model for how the S100A4 dimer dissociates NMIIA filaments. Here, we report the high-resolution crystal structure of the S100A4–NMIIA fragment complex, which reveals a unique target-recognizing mechanism in the S100 family. Based on our findings, we propose a model for the structural basis of S100A4-induced NMIIA filament disassembly.
Results
Affinity of S100A4 to NMII Fragments.
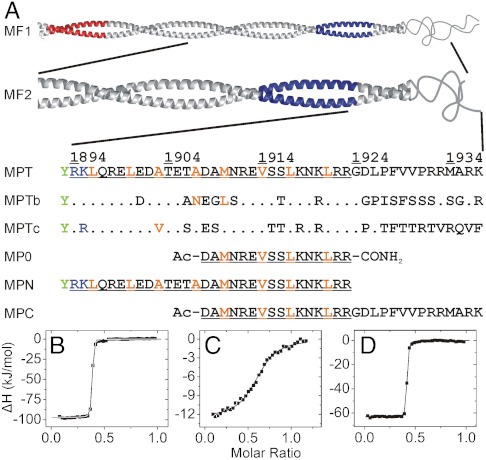
Because previous studies on the NMIIA–S100A4 interaction have not unambiguously determined the myosin sequence requirements for S100A4 binding (16, 35, 36), isothermal titration calorimetric (ITC) measurements were carried out with peptides and fragments illustrated in Fig. 2A. Our results support the findings of Badyal et al. (36) that the 16-mer peptide MP0 shows lower affinity (Kd ≈ 30 μM) to S100A4 than it was previously reported (16) (Fig. S1A and Table S1). The C-terminal extension of this peptide (MPC) binds with micromolar affinity (Kd ≈ 2 μM) and with a stoichiometry of one myosin heavy chain peptide to one S100A4 dimer (Fig. 2C). Because ITC measurements with the longer myosin fragment MF2 suggested a considerably tighter interaction (Kd ≈ 6 nM) (Fig. S1B), we added N-terminal residues to MPC, resulting in a 45-residue-long NMIIA fragment (MPT). Peptides MPT and MPN bind to S100A4 with the same stoichiometry as MPC, but with higher affinities (Kd ≈ 8 nM, Fig. 2 B–D). Although the affinities are similar, enthalpy changes are higher for MPN, whereas the entropic cost of MPT binding to S100A4 is lower, indicating different binding mechanisms for the coiled-coil and random coil regions of the myosin heavy chain (Table S1). In order to investigate the isoform specificity of S100A4 binding to the three NMII paralogs, we also measured the affinity of the homologous MPTb and MPTc fragments (Fig. 2A) to S100A4 by ITC. MPTb shows reduced affinity compared to MPT (Kd ≈ 125 nM). On the contrary, MPTc binds to S100A4 as strongly as the NMIIA peptide (Fig. S1 E and F and Table S1).
Fig. 2.
Nonmuscle myosin II heavy chain fragments and peptides used in this work and their affinity to S100A4. (A) Fragments MF1 (NMIIA Ser1712-Glu1960) and MF2 (NMIIA Gln1795-Lys1937) form coiled-coil structures, MF1 assembles into filaments (33). The mainly negatively (Glu1722-Asn1756) and positively charged (Ala1868-Lys1895) regions (the latter called ACD) that are crucial for filament formation (33, 34) colored in red and blue, respectively. Within the sequence of MPT (NMIIA Arg1894-Lys1937) the residues that are thought to form a coiled-coil in longer fragments are underlined, while orange letters show a and d positions in the heptad repeat. Note that the N-terminal basic residues of MPT overlap with ACD and MPT does not contain the full nonhelical tailpiece. Corresponding amino acid positions of S100A4 binding site that differ in NMIIB (1901-1944) and NMIIC (1918-1961) isoforms are indicated. An N-terminal Tyr (green) was added to the peptides MPT and MPN to facilitate concentration measurements. (B–D) Heat changes were recorded by ITC experiments at 25 °C, at physiological salt concentration. 75 μM S100A4 monomer was titrated with 500 μM monomeric NMIIA peptides. Peptides MPN (B) and MPT (D) show nanomolar affinity to S100A4 (Kd < 8 nM), while the binding affinity of MPC (C) is in the micromolar range (Kd ≈ 1.8 μM). In all cases, the binding stoichiometry is approximately one myosin peptide per S100A4 dimer. Thermodynamic parameters of the interactions of S100A4 with NMII heavy chain fragments are shown in Table S1.
Overall Description of the S100A4-MPT Complex.
Our efforts to crystallize wild-type S100A4 in complex with NMIIA peptides failed. Therefore, we created a triple mutant S100A4 (Cys3Ser/Cys81Ser/Cys86Ser), where the solvent exposed thiol groups were eliminated in order to avoid disulfide formation, leading to the loss in binding capability of S100A4 to myosin IIA fragments. These mutations were introduced not only to the wild-type, but to the Phe45Trp mutant S100A4, which was originally designed for facilitating spectroscopic measurements. The construct with the four mutations was successfully crystallized and the crystal structure was solved by molecular replacement to 1.9 Å resolution (Table S2). According to ITC measurement, MPT binds to the mutant S100A4 with about two orders of magnitude weaker affinity than to the wild-type protein (Kd ≈ 1.5 μM), but the stoichiometry of the interaction did not change (Fig. S1D). Fig. 1 shows how a single chain of MPT (NMIIA heavy chain residues 1894–1935) wraps around the dimeric S100A4 protein in the crystal structure.
Symmetry Breakage in the S100A4 Homodimer upon MPT Binding.
Whereas MPT binds both S100A4 subunits with approximately the same area of interaction surface (1,308 Å2 and 1,251 Å2 for subunit A and B, respectively), each subunit of the symmetric homodimer has to adapt its conformation to the different character of the two termini of MPT. Based on differential distance matrix analysis, the most obvious difference between the subunits is that helix 3 (residues 52–62) and helix 4 (residues 72–92) clamp more tightly on the N-terminal part of the peptide in subunit A (by approximately 1.5–2.5 Å) (Fig. S2). In addition, loop 2 (residues 42–51) flips over to the rest of the chain in subunit B. Differences between the C termini of chain A and B are due to the crystal contacts evolved between the C-terminal tail of subunit B and a symmetry related subunit A (see SI Text for more details and Fig. S3). The most rigid portion of the structure is the N-terminal half of the S100A4 chains (residues 2–44) with no significant differences between subunit A and B. These residues are partly involved in subunit–subunit interactions and in forming the surface of the dimer on the diagonally opposite side to the MPT binding interface. Similar loss of symmetry can be observed in the atomic displacement parameters (ADPs, expressed in the form of crystallographic B-factors) between the two subunits and the two halves of MPT, respectively (see SI Text for details).
Interactions of MPT with the Two S100A4 Subunits.
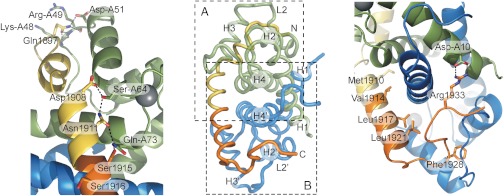
The binding interface between the N-terminal part of MPT and subunit A is abundant in polar interactions involving both side-chain and main-chain atoms. As shown in Fig. 3A, Gln1897 forms an important hub by interacting with Lys-A48, Arg-A49, and Asp-A51, simultaneously. The short α-helix interacts with a hydrophobic surface formed by Trp-A45, Leu-A58, Leu-A82, and Met-A85 via residues Leu1900 and Ala1903 (a and d positions in the coiled-coil). The central part of MPT can be characterized by a mixture of hydrophilic and hydrophobic interactions. Asp1908, Asn1911, and Ser1915 form an extended H-bond network with Ser-A64 and Gln-A73. (Fig. 3A), whereas hydrophobic residues Met1910, Val1914, Leu1917, and Leu1921 (a and d positions in the coiled-coil) in the long α-helix of MPT bridge the S100A4 dimer interface (Fig. 3B). The C-terminal part of MPT, on the other hand, is abundant with hydrophobic interactions. In this region, eight hydrophobic residues in loop 2, helix 3 and 4 form an extensive and continuous hydrophobic surface with which five hydrophobic residues on the C-terminal part of MPT interact. As a demonstration of the remarkable flexibility of loop 2 in subunit B, this loop forms a hydrophobic ring around Phe1928 (Fig. 3B). Note that in subunit A residues of this loop form an extensive hydrogen bond network with Gln1897. Finally, wrapping around the dimer Arg1933 forms an ionic interaction with Asp-A10 (Fig. 3B).
Fig. 3.
Key interactions within the S100A4-NMIIA MPT complex. (A) Polar interactions at the interface of the N-terminal part of MPT with subunit A. (B) Interactions of MPT with subunit B are dominated by hydrophobic interactions, where Met1910, Val1914, Leu1917, and Leu 1921 (a and d positions in the coiled-coil) face inward. Phe1928 is located in a hydrophobic cavity formed by loop 2 (residues 41–51) and helix 3, while Arg1933 forms ionic interaction with Asp10 of subunit A wrapping around the dimer. The overview in the middle is shown in an orientation perpendicular to the twofold symmetry axis of the S100A4 dimer.
Conformational Changes in NMIIA Peptides upon S100A4 Binding.
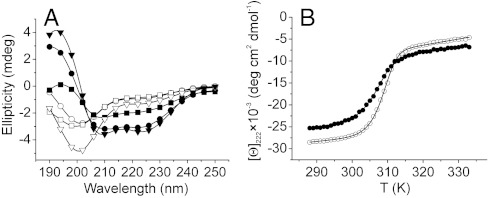
To confirm that the complex formation using wild-type S100A4 involves similar structural changes in solution, the secondary structures of the free- and S100A4-bound peptides were investigated by CD spectroscopy. These short myosin tail fragments show disordered structure in aqueous solution. In the presence of Ca2+ and a 10% excess of S100A4 dimer an increase was observed in the α-helical content in each peptide (Fig. 4A). Comparing the CD spectra of the S100A4-bound fragments indicates that the α-helical content of MPN and MPT is markedly higher than that of MPC. The helix content of the bound MPN, MPC, and MPT peptides were estimated to be 65% (19 residues), 46% (14 residues), and 42% (19 residues), respectively. These calculations are consistent with observations of the S100A4–MPT crystal structure, and suggest that the central part of MPT is bound as an α-helix, the N-terminal part is partially α-helical, while the C-terminal part assumes nonhelical conformation.
Fig. 4.
Secondary structural changes in NMIIA peptides and a coiled-coil fragment upon S100A4 binding. (A) CD spectra of 50 μM myosin heavy chain peptides MPT (▿) MPN (○), MPC (□) were measured at 25 °C, at low salt concentration. The black symbols illustrate the spectra of peptides complexed with S100A4 (110 μM). (B) Thermal denaturation of 6 μM MF1 in the absence (○) and in the presence (●) of 15 μM S100A4. The curves of the bound myosin peptides and fragments were produced as the difference between the curve of S100A4-myosin complex and of S100A4 alone (Fig. S4D).
Conformational Changes in Coiled-Coil NMIIA Fragments upon S100A4 Binding.
To assess the effect of S100A4 binding on the structure of dimeric myosin, the thermostability of free-and S100A4-bound MF1 and MF2 myosin fragments was studied by CD spectroscopy. The thermodynamic parameters of the dimer-to-monomer transition show that both the van’t Hoff enthalpy of denaturation and the melting temperature of the coiled-coil (TM) clearly decreased in MF1 (Fig. 4B and Table S3 ). In the case of MF2, which forms a relatively short coiled-coil, binding of S100A4 considerably decreases the cooperativity of thermal unfolding, preventing accurate data analysis (Fig. S4). Nevertheless, both experiments suggest that S100A4 binding partially unwinds the coiled-coil. Note that the structure of S100A4 is extremely thermostable as demonstrated in Fig. S4D. Estimating the α-helix content of free- and S100A4-bound coiled-coil fragments (at temperatures where the helix content of the free coiled-coil is maximal), approximately 25 residues could be unwound upon complex formation (Fig. S4 A and B). Of those, 10 residues are visualized as coil within the S100A4 binding pocket of our structural model, and consequently approximately 15 residues could be unwound N-terminally to the S100A4 binding site. These results clearly suggest that binding of S100A4 affects the stability of the coiled-coil ACD that could be directly related to the myosin filament disassembly.
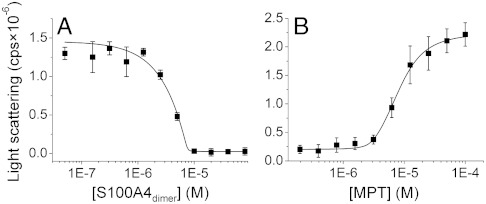
Effect of S100A4 on MF1 Filament Disassembly.
Finally, we investigated whether both myosin heavy chains have to be occupied by the wild-type S100A4 dimer for total filament dissociation. We examined the light scattering of MF1 as a function of S100A4 dimer concentration at physiological salt concentration (Fig. 5A). Note that light scattering is directly proportional to the non-S100A4-bound myosin concentration at the observed concentration range (Fig. S5). The experimental data could be fitted to a single-site binding model (apparent Kd ≈ 8 nM), notwithstanding that the binding of S100A4 to the myosin filament suggests a more complex model. Our results support that the stoichiometry is one S100A4 dimer per one myosin polypeptide chain, because at 10 μM myosin concentration (≫Kd) total solubilization of the filament was observed when 10 μM S100A4 dimer was added. We also tested whether MPT is an efficient competitor of a filament forming myosin fragment. The experimental data, derived from the titration of myosin-S100A4 complex (10 μM) with MPT, was fitted to a competitive binding model. When the affinity of MPT was fixed to Kd = 8 nM (estimated by ITC), the affinty of MF1 was calculated to be 30 nM, while addition of 8 μM peptide caused 50% increase in turbidity (Fig. 5B). These findings indicate that MPT comprises the complete S100A4 binding site.
Fig. 5.
Filament assembly properties of an NMIIA fragment in the presence of S100A4 and MPT. (A) A 10 μM MF1 fragment was titrated with S100A4 at physiological salt concentration. Light scattering was measured at 320 nm. The solid line represents the best fit to a single-site binding model. (B) Titration of the MF1-S100A4 complex with MPT. The solid line represents the best fit to a competitive binding model. The data points represent the mean ± SEM from three independent measurements.
Discussion
Unique Structure of the S100A4–NMIIA Complex.
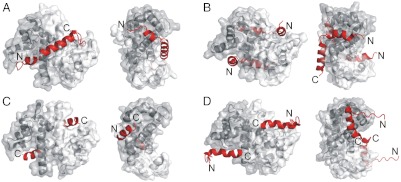
Our efforts to refine the native interaction footprint of S100A4 on the myosin heavy chain resulted in the 45-residue-long fragment that showed nanomolar affinity to S100A4. We determined the stoichiometry to be one myosin peptide per S100A4 dimer as also suggested in very recent studies (36, 37). This peptide (MPT) was successfully crystallized complexed to a quadruple mutant of S100A4. Although the Phe45Trp mutation does not affect the interaction of S100A4 with NMIIA fragments (Fig. S1 B and C), according to recent results the Cys81Ser mutation could be responsible for the differences between the wild-type and the crystallized mutant S100A4 in respect to the interaction with NMIIA (38). Using the crystal structure, we rationalize the reduced binding properties of the Cys81Ser mutation with the smaller van der Waals radius of the hydroxyl group and a potential competition with water (one such buried water molecule can be observed hydrogen bonded to Ser-A81, whereas in subunit B Ser-B81 is buried in a hydrophobic environment). Nevertheless, the 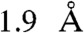 resolution structure of the S100A4–NMIIA complex reveals an alternate mechanism of target recognition in the S100 family. The crystal structure contains one myosin polypeptide chain that is wrapped around an S100A4 dimer forming a particularly large interface (2,559 Å2) compared to other S100 complexes, where both hydrophobic binding pockets and the surface formed by α-helices 4-4′ are buried. Our structural model revealed a unique interacting interface, hitherto unseen in any S100 protein complexes, formed by helices 3-3′ and 4-4′, to which the central α-helical part of the myosin peptide binds. This groove (the “waist” of the dimer) exists on other S100 proteins as well, but its width and depth vary considerably (Fig. 6).
resolution structure of the S100A4–NMIIA complex reveals an alternate mechanism of target recognition in the S100 family. The crystal structure contains one myosin polypeptide chain that is wrapped around an S100A4 dimer forming a particularly large interface (2,559 Å2) compared to other S100 complexes, where both hydrophobic binding pockets and the surface formed by α-helices 4-4′ are buried. Our structural model revealed a unique interacting interface, hitherto unseen in any S100 protein complexes, formed by helices 3-3′ and 4-4′, to which the central α-helical part of the myosin peptide binds. This groove (the “waist” of the dimer) exists on other S100 proteins as well, but its width and depth vary considerably (Fig. 6).
Fig. 6.
Comparison of four peptide-bound S100 proteins. The structure of (A) S100A4-NMIIA (3ZWH), (B) S100A6-SIP (2JTT) (25), (C) S100A10-ANXA2 (1BT6) (21), and (D) S100B-p53 (1DT7) (22) complexes are shown in two orientations. The left figures illustrate the two canonical hydrophobic pockets, while in the right ones the dimers are rotated to show the groove (the waist) formed by helices 3-3′ and 4-4′ that is occupied in the S100A4 complex by the central helical part of the myosin binding peptide. Peptide ligands are shown in red.
There are only a few cases when a homodimeric protein binds to a single asymmetric peptide. A notable example is the dimeric protein kinase A (PKA) regulatory domain that binds an A-kinase anchoring protein (AKAP) peptide in a similar manner (39, 40). The PKA regulatory domain dimer also forms an X-type four-helix bundle interface, and the bound peptide is α-helical. Helices 1 and 2 in the PKA regulatory domain are in approximately equivalent positions as helices 3 and 4 in S100A4, though the interactions with the nonhelical NMIIA peptide segments beyond the waist region makes this complex markedly more asymmetric. The ligand-induced asymmetry in the homodimeric S100A4, as well as the existence of multiple binding partners, required the evolution of multifunctional structural elements in S100A4. An example of such a structural element is loop 2, which is able to bind both the hydrophilic N terminus and the hydrophobic C terminus of MPT. Conformational adaptation is also extensive when we compare the peptide-free, Ca2+-bound wild-type S100A4 (16, 17) with the mutant S100A4 protein in complex with MPT (Fig. S6). Interestingly, it has been recently demonstrated that p53 and MDM2 bind to their S100 partners in solution with the same stoichiometry (i.e., one chain to one S100 dimer) (41, 42), strongly suggesting that interactions similar to the S100A4–NMIIA complex potentially exist in other S100 proteins as well.
Phosphorylation of NMIIA at Ser1916 by protein kinase C (43) appears to have little interference with binding the S100A4 dimer, as previously demonstrated experimentally (44), because the Ser1916 side chain points away from the binding interface (Fig. 3A). Regarding the phosphorylation of Ser1943 by casein kinase 2, because residues located C-terminally to Ala1935 apparently do not interact directly with S100A4, we support the hypothesis by Dulyaninova et al. in that the dianionic phosphate on Ser1943 may facilitate the formation of intramolecular ionic interactions between the nonhelical tailpiece and the ACD, which inhibits filament assembly and the binding of S100A4 (45).
We also studied the selectivity of S100A4 binding to NMII isoforms and confirmed the lower affinity of S100A4 to NMIIB (29). However, strikingly strong binding to NMIIC was found. Sequence comparison of MPT in the three isoforms (Fig. 2A) reveals a few key residues that could contribute to the observed selectivity (see SI Discussion for details).
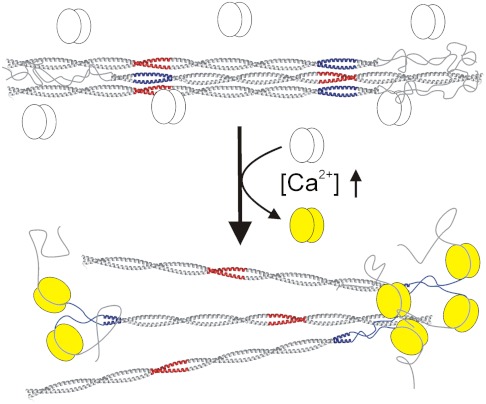
Mechanism of Myosin Filament Disassembly by S100A4.
We have also examined the effect of S100A4 binding on the coiled-coil forming NMIIA fragments: A drop in melting temperature and in cooperativity of the monomer–dimer transition was observed (Fig. 4B, Fig. S4, and Table S3). The data demonstrate partial unwinding of the coiled-coil upon S100A4 binding (that affects approximately 15 residues), strongly suggesting at least partial disruption of the nearby ACD that is required for myosin II filament assembly (32–34). Disassembly of the ordered rods of MF1 upon S100A4 binding was also investigated, and the same stoichiometry was obtained as with the shorter peptides. Our results support previous findings that a twofold excess of S100A4 monomer is able to disassemble completely the NMIIA filaments (36). Based on our results, we propose the following hypothetical model for S100A4–NMIIA heavy chain interaction (Fig. 7). Binding of S100A4 consists of a minimum of two steps. Firstly, S100A4 is supposed to catch the nonhelical tailpiece, then partially unwind the C-terminal end of the coiled-coil (which overlaps with the ACD) and bind it to the waist region. To achieve this, it has to possess a two-partite binding site, as we indeed observed in our structural model. Besides disrupting the ACD, binding of two S100A4 dimers to one myosin tail (one dimer to a single heavy chain) could result in serious steric constraints between neighboring myosins, which could also contribute to the filament disassembly or inhibit filament formation. Confirmation of the above hypothesis and the elucidation of the molecular details of the filament disassembly require further kinetic and structural studies.
Fig. 7.
Schematic model of S100A4-induced NMIIA filament disassembly. The Ca2+-loaded S100A4 (yellow) binds to NMIIA at the nonhelical tailpiece, partially unwinds the ACD (blue) and sterically blocks myosin-myosin interactions causing filament disassembly.
Materials and Methods
Expression, Synthesis, and Purification of Proteins and Peptides.
The human wild-type and mutant S100A4, nonmuscle myosin II fragments and peptides were obtained by heterologous expression in Escherichia coli, with the exception of peptide MP0, which was synthesized in-house. For detailed cloning, protein expression, peptide synthesis and purification procedures, see SI Text.
Isothermal Titration Calorimetry (ITC).
Titrations were carried out at 298 or 320 K in 20 mM Hepes pH 7.5, 150 mM or 500 mM NaCl, 1 mM CaCl2, and 1 mM TCEP using a Microcal VP-ITC apparatus. S100A4 variants were titrated up to 2-fold molar excess of ligands. Depending on the Kd and ligand concentration, up to 40 injections were performed with 400-s time intervals between injections. The Origin for ITC 5.0 (OriginLab) software package was used for data processing, and the model “One Set of Sites” was fitted.
Circular Dichroism (CD) Spectroscopy.
CD measurements were carried out on a Jasco J-715 spectropolarimeter (Jasco) The relative α-helix content of each peptide was estimated based on the molar residual ellipticity measured at 222 nm (46) and corrected with the amount of free peptides calculated from the dissociation constant of each complex (Table S1). The CD spectra of myosin peptides were measured in 10 mM Hepes pH 7.5, 20 mM NaCl, 1 mM CaCl2, and 1 mM TCEP. Thermostability studies were made in a buffer containing 500 mM NaCl, with a temperature ramp of 1 °C/ min, and the ellipticity was monitored at 222 nm. With the assumption that the unfolding is a two-state transition between the folded dimer and the unfolded monomers, the experimental data were fitted using the equations previously published (47) complemented with the parameters of the linear pre- and posttransitional phases (Eq. S1). F-test was used to estimate the statistical uncertainties in the fitted parameters (SI Text and Eq. S2).
Filament Disassembly Assays.
Light scattering measurements of NMIIA fragment MF1 were carried out on Spex Fluoromax-2 spectrofluorimeter (Jobin Yvon) at 320 nm, and at 298 K. The buffer was the same as in the ITC measurements (150 mM NaCl) complemented with 2 mM MgCl2. The experimental data of three parallels derived from the filament titration with S100A4 were fitted using Eq. S3. In the case of the competitive assay the experimental data were analyzed using a modified competitive binding model (48).
X-Ray Structure Determination and Analysis.
X-ray diffraction data were collected at 100 K with an X-ray wavelength of 0.873 Å on the beamline ID23-2 at the European Synchrotron Radiation Facilities (ESRF, Grenoble, France). X-ray diffraction data were processed and scaled in XDS (49), and a molecular replacement solution was found with Phaser (50) using human Ca2+-bound S100A4 (PDB ID code 3C1V) as search model. The structural model and crystallographic data are deposited in the Protein Data Bank under PDB ID code 3ZWH. Details are provided in SI Text and Table S2. Difference distance matrices were calculated using the program ESCET in order to compare the S100A4 structures (51) (see SI Text).
Note Added in Proof.
While the present manuscript was under revision, we became aware that Elliott et al. (52) determined the NMR structure of wild-type S100A4 in complex with an NMIIA fragment and reached very similar conclusions.
Supplementary Material
Acknowledgments.
We thank Dr. Didier Nurizzo for his expert assistance with data collection at ESRF beamline ID23-2. We also thank Drs. Zsuzsa Majer, Ferenc Tölgyesi, and András Patthy for their help in CD and ITC measurements, as well as in-house peptide synthesis, respectively. We are grateful to Dr. József Kardos and Dr. Erik Malmerberg for their continuous advice and Philip Smith for careful reading of the manuscript. The work was supported by the Hungarian Scientific Research Fund (OTKA K81784, NK81950) and the European Union and the European Social Fund (TAMOP 4.2.1./B-09/KMR-2010-0003). A.D. and G.K. were supported by the Swedish Research Council and the Harald and Greta Jeanssons Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3ZWH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114732109/-/DCSupplemental.
References
- 1.Donato R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 2.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 3.Garrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J Biol Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- 4.Boye K, Maelandsmo GM. S100A4 and metastasis: A small actor playing many roles. Am J Pathol. 2010;176:528–535. doi: 10.2353/ajpath.2010.090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambartsumian NS, et al. Metastasis of mammary carcinomas in GRS/A hybrid mice transgenic for the mts1 gene. Oncogene. 1996;13:1621–1630. [PubMed] [Google Scholar]
- 6.Davies MP, et al. Expression of the calcium-binding protein S100A4 (p9Ka) in MMTV-neu transgenic mice induces metastasis of mammary tumours. Oncogene. 1996;13:1631–1637. [PubMed] [Google Scholar]
- 7.Semov A, et al. Metastasis-associated protein S100A4 induces angiogenesis through interactionwith Annexin II and accelerated plasmin formation. J Biol Chem. 2005;280:20833–20841. doi: 10.1074/jbc.M412653200. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe Y, et al. Calvasculin, as a factor affecting the microfilament assemblies in rat fibroblasts transfected by src gene. FEBS Lett. 1993;324:51–55. doi: 10.1016/0014-5793(93)81530-d. [DOI] [PubMed] [Google Scholar]
- 9.Takenaga K, et al. Binding of pEL98 protein, an S100-related calcium-binding protein, to nonmuscle tropomyosin. J Cell Biol. 1994;124:757–768. doi: 10.1083/jcb.124.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriajevska MV, et al. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J Biol Chem. 1994;269:19679–19682. [PubMed] [Google Scholar]
- 11.Grigorian M, et al. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: Functional consequences of their interaction. J Biol Chem. 2001;276:22699–22708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- 12.Li CL, Martinez V, He B, Lombet A, Perbal B. A role for CCN3 (NOV) in calcium signalling. Mol Pathol. 2002;55:250–261. doi: 10.1136/mp.55.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriajevska M, et al. Liprin β1, a member of the family of LAR transmembrane tyrosine phosphatase-interacting proteins, is a new target for the metastasis-associated protein S100A4 (Mts1) J Biol Chem. 2002;277:5229–5235. doi: 10.1074/jbc.M110976200. [DOI] [PubMed] [Google Scholar]
- 14.Endo H, Takenaga K, Kanno T, Satoh H, Mori S. Methionine aminopeptidase 2 is a new target for the metastasis-associated protein, S100A4. J Biol Chem. 2002;277:26396–26402. doi: 10.1074/jbc.M202244200. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura I, Lai CY, Chiang KN. Functional interaction between Smad3 and S100A4 (metastatin-1) for TGF-β-mediated cancer cell invasiveness. Biochem J. 2010;426:327–335. doi: 10.1042/BJ20090990. [DOI] [PubMed] [Google Scholar]
- 16.Malashkevich VN, et al. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingras AR, et al. Crystal structure of the Ca2+-form and Ca2+-binding kinetics of metastasis-associated protein, S100A4. FEBS Lett. 2008;582:1651–1656. doi: 10.1016/j.febslet.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 18.Novitskaya V, et al. Oligomeric forms of the metastasis-related Mts1 (S100A4) protein stimulate neuronal differentiation in cultures of rat hippocampal neurons. J Biol Chem. 2000;275:41278–41286. doi: 10.1074/jbc.M007058200. [DOI] [PubMed] [Google Scholar]
- 19.Malashkevich VN, et al. Phenothiazines inhibit S100A4 function by inducing protein oligomerization. Proc Natl Acad Sci USA. 2010;107:8605–8610. doi: 10.1073/pnas.0913660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rety S, et al. Structural basis of the Ca2+-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure. 2000;8:175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 21.Rety S, et al. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 22.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(ββ) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 23.Inman KG, et al. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol. 2002;324:1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: Insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 25.Lee YT, et al. Structure of the S100A6 complex with a fragment from the C-terminal domain of Siah-1 interacting protein: A novel mode for S100 protein target recognition. Biochemistry. 2008;47:10921–10932. doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright NT, et al. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J Biol Chem. 2008;283:26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- 28.Ford HL, Silver DL, Kachar B, Sellers JR, Zain SB. Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry. 1997;36:16321–16327. doi: 10.1021/bi971182l. [DOI] [PubMed] [Google Scholar]
- 29.Li ZH, Spektor A, Varlamova O, Bresnick AR. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42:14258–14266. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- 30.Sohn RL, et al. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J Mol Biol. 1997;266:317–330. doi: 10.1006/jmbi.1996.0790. [DOI] [PubMed] [Google Scholar]
- 31.Cohen C, Parry DA. A conserved C-terminal assembly region in paramyosin and myosin rods. J Struct Biol. 1998;122:180–187. doi: 10.1006/jsbi.1998.3983. [DOI] [PubMed] [Google Scholar]
- 32.Ikebe M, et al. The tip of the coiled-coil rod determines the filament formation of smooth muscle and nonmuscle myosin. J Biol Chem. 2001;276:30293–30300. doi: 10.1074/jbc.M101969200. [DOI] [PubMed] [Google Scholar]
- 33.Nakasawa T, et al. Critical regions for assembly of vertebrate nonmuscle myosin II. Biochemistry. 2005;44:174–183. doi: 10.1021/bi048807h. [DOI] [PubMed] [Google Scholar]
- 34.Ricketson D, Johnston CA, Prehoda KE. Multiple tail domain interactions stabilize nonmuscle myosin II bipolar filaments. Proc Natl Acad Sci USA. 2010;107:20964–20969. doi: 10.1073/pnas.1007025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriajevska M, et al. Metastasis-associated Mts1 (S100A4) protein modulates protein kinase C phosphorylation of the heavy chain of nonmuscle myosin. J Biol Chem. 1998;273:9852–9856. doi: 10.1074/jbc.273.16.9852. [DOI] [PubMed] [Google Scholar]
- 36.Badyal SK, et al. Mechanism of the Ca2+-dependent interaction between S100A4 and tail fragments of nonmuscle myosin heavy chain IIA. J Mol Biol. 2011;405:1004–1026. doi: 10.1016/j.jmb.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.House RP, et al. Two Functional S100A4 Monomers Are Necessary for Regulating Nonmuscle Myosin-IIA and HCT116 Cell Invasion. Biochemistry. 2011;50:6920–6932. doi: 10.1021/bi200498q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dulyaninova NG, et al. Cysteine 81 Is Critical for the Interaction of S100A4 and Myosin-IIA. Biochemistry. 2011;50:7218–7227. doi: 10.1021/bi200853y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newlon MG, et al. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarma GN, et al. Structure of D-AKAP2:PKA RI complex: Insights into AKAP specificity and selectivity. Structure. 2010;18:155–166. doi: 10.1016/j.str.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem. 2009;284:13804–13811. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Dieck J, Lum JK, Teufel DP, Fersht AR. S100 proteins interact with the N-terminal domain of MDM2. FEBS Lett. 2010;584:3269–3274. doi: 10.1016/j.febslet.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Conti MA, Sellers JR, Adelstein RS, Elzinga M. Identification of the serine residue phosphorylated by protein kinase C in vertebrate nonmuscle myosin heavy chains. Biochemistry. 1991;30:966–970. doi: 10.1021/bi00218a012. [DOI] [PubMed] [Google Scholar]
- 44.Dulyaninova NG, Malashkevich VN, Almo SC, Bresnick AR. Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–6876. doi: 10.1021/bi0500776. [DOI] [PubMed] [Google Scholar]
- 45.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohl CA, Baldwin RL. Comparison of NH exchange and circular dichroism as techniques for measuring the parameters of the helix-coil transition in peptides. Biochemistry. 1997;36:8435–8442. doi: 10.1021/bi9706677. [DOI] [PubMed] [Google Scholar]
- 47.Greenfield NJ, Montelione GT, Farid RS, Hitchcock-DeGregori SE. The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: Nuclear magnetic resonance structure and circular dichroism studies. Biochemistry. 1998;37:7834–7843. doi: 10.1021/bi973167m. [DOI] [PubMed] [Google Scholar]
- 48.Wang ZX. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- 49.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider TR. A genetic algorithm for the identification of conformationally invariant regions in protein molecules. Acta Crystallogr D Biol Crystallogr. 2002;58:195–208. doi: 10.1107/s0907444901019291. [DOI] [PubMed] [Google Scholar]
- 52.Elliott PR, et al. Asymmetric mode of Ca2+-S100A4 interaction with non-muscle myosin IIA heavy chain generates nanomolar affinity required for filament remodelling. Structure. 2012 doi: 10.1016/j.str.2012.02.002. 10.1016/j.str.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.