Abstract
Nuclear translocation of cytosolic CLIC4 is an essential feature of its proapoptotic and prodifferentiation functions. Here we demonstrate that CLIC4 is induced concurrently with inducible nitric oxide synthase (iNOS) and S-nitrosylated in proinflammatory peritoneal macrophages. Chemical inhibition or genetic ablation of iNOS inhibits S-nitrosylation and nuclear translocation of CLIC4. In macrophages, iNOS-induced nuclear CLIC4 coincides with the pro- to anti-inflammatory transition of the cells because IL-1β and CXCL1 mRNA remain elevated in CLIC4 and iNOS knockout macrophages at late time points, whereas TNFα mRNA is elevated only in the iNOS knockout macrophages. Active IL-1β remains elevated in CLIC4 knockout macrophages and in macrophages in which CLIC4 nuclear translocation is prevented by the NOS inhibitor l-NAME. Moreover, overexpression of nuclear-targeted CLIC4 down-regulates IL-1β in stimulated macrophages. In mice, genetically null for CLIC4, the number of phagocytosing macrophages stimulated by LPS is reduced. Thus, iNOS-induced nuclear CLIC4 is an essential part of the macrophage deactivation program.
Keywords: protein nitrosylation, IL-1beta, phagocytosis
Chloride intracellular channel 4 (CLIC4) is the most well-characterized member of a family of channel proteins conserved from Caenorhabditis elegans to humans. Although the chloride-selective channel activity of various members has long been established by multiple groups (1–3), the signaling and adaptor functions of soluble CLIC4 and other members have only recently come to the fore. Soluble CLIC4, originally identified as a p53- and c-myc–responsive proapoptotic protein (4, 5), is important for PKC-dependent keratinocyte differentiation (6) and a modulator of TGFβ signaling in multiple cell types (7–9). Many of these proapoptotic and differentiation functions of CLIC4 are dependent on its translocation to the nucleus (6, 8, 10). CLIC4 exhibits redox sensitivity in both its soluble and membrane-associated states (11, 12). Indeed, a central mechanism for induction of nuclear CLIC4 is nitric oxide (NO)-dependent modification (S-nitrosylation) of the protein that induces a conformational change, enhancing its association with nuclear transporters and thus its nuclear levels (13).
Recently, a role for CLIC4 has been implicated in innate immunity. CLIC4 is an IRF3-dependent early response gene in LPS-stimulated macrophages, transrepressed by the anti-inflammatory activity of the glucocorticoid receptor (14). Moreover, CLIC4 knockout macrophages exhibit dysregulation of multiple inflammatory mediators during the early response to LPS, in part as a consequence of altered IRF3 activity (15). These studies suggest that CLIC4 is important in macrophage early functions in response to stimulation but do not address later aspects of macrophage biology related to deactivation and phagocytosis. Furthermore, information is not available to reveal if the CLIC4 channel or nuclear activity is involved in these functions. Therefore, we investigated the role of CLIC4 activity in inflammatory macrophages and the contribution of nuclear CLIC4 to their function.
Results
CLIC4 Is Highly Expressed in Murine Marcrophages, Is S-Nitrosylated, and Translocates to the Nucleus in Response to LPS and IFNγ.
CLIC4 is highly induced in macrophages upon stimulation of toll-like receptor 4 with LPS (14). We used a combination of LPS and IFNγ to stimulate peritoneal macrophages. LPS-induced innate immune response is modulated by Th1-dependent IFNγ to fully activate macrophages for host defense against bacterial infections (16). These peritoneally derived, primary macrophages will be referred to as macrophages through the rest of the paper. CLIC4 mRNA is induced by 10- to 15-fold by LPS/IFNγ in macrophages and the murine macrophage cell line RAW264.7, compared with unstimulated granulocytes, monocytes, B-cells, and T-cells (Fig. 1A). This robust increase is noted at 3 and 6 h post treatment, remains elevated for up to 48 h, and mirrors the early induction for inducible nitric oxide synthase (iNOS) mRNA (Fig. 1B). Both proteins show a similar temporal increase in protein levels post stimulation (Fig. 1C).
Fig. 1.
CLIC4 is highly expressed in primary macrophages (Mϕ), is S-nitrosylated, and translocates to the nucleus in response to LPS/IFNγ. (A and B) RNA from mouse hematopoietic cells (A) or mouse macrophages (B), stimulated or unstimulated, as indicated, was isolated and used for CLIC4 and iNOS real-time PCR analysis. In A, RAW and macrophages were stimulated for 6 h with LPS/IFNγ. Bars represent the mean ± SEM of three replicates. Statistical significance was determined using a two-tailed unpaired Student's t test and is indicated. (C) Whole-cell lysates from stimulated or unstimulated macrophages were used for immunoblotting with CLIC4, CLIC1, and iNOS proteins. (D and E) Primary mouse macrophages were treated with LPS and IFNγ (1 μg/mL and 10 ng/mL, respectively) for 18 h. (D) Lysates were used to perform biotin switch assays to detect S-nitrosylation. Lysate from LPS/IFNγ-treated cells was also used for a reaction that omitted ascorbate (“no Asc.”). Five percent of lysates were used as input controls. Assays were immunoblotted for CLIC4. Lane 2 of CLIC4 input is representative for both LPS/IFNγ-treated pull-down assays. The SNO-CLIC4:CLIC4 input ratio was calculated for all treatments across four independent experiments. Statistical significance was determined using a two-tailed unpaired Student's t test. Control versus LPS/IFN treatment has a P < 0.01 and LPS/IFN versus LPS/IFN (no ascorbate) has a P < 0.05. (E) Macrophage cells were immunostained for CLIC4 and visualized with confocal microscopy. Inset nuclei are visualized using DAPI.
CLIC4 nuclear translocation is regulated by NOS activity through direct modification of a CLIC4 cysteine residue by NO (13), and nuclear CLIC4 functions to enhance TGF-β signaling (8), the latter being a critical modulator of macrophage deactivation (17). Stimulation with LPS and IFNγ induces S-nitrosylation of CLIC4 in macrophages as detected by a biotin switch assay (Fig. 1D), and this is coincident with increased levels of nuclear CLIC4 (Fig. 1E) in the DAPI-stained nuclei (Insets).
S-Nitrosylation and Nuclear Translocation of CLIC4 Is Dependent on iNOS Activity in Macrophages.
NO from NOS activity is required for nitrosylation and translocation of CLIC4 in RAW264.7 cells because pretreatment with 1400W, an iNOS-specific inhibitor, reduces CLIC4 nitrosylation following LPS/IFNγ stimulation (Fig. 2A). Nuclear residence of the protein is also reduced under the same conditions (Fig. 2C) where NO levels are significantly reduced (Fig. 2B). Macrophages also show a similar dependence on NOS activity for CLIC4 nitrosylation and nuclear translocation (Fig. S1, and see Fig. 5A). Synergistic induction of iNOS protein and activity following stimulation by LPS and IFNγ is well established in macrophages (18). In macrophages genetically deleted of iNOS, CLIC4 S-nitrosylation (Fig. 2D) was significantly reduced following LPS/IFNγ stimulation. However, the modified protein migrated faster than full-length CLIC4, suggesting that it was prone to degradation. This is likely a consequence of the conformational change induced in the protein (13). iNOS knockout macrophages stimulated with LPS and IFNγ also showed reduced CLIC4 nuclear translocation (Fig. 2 E and F). These results establish that iNOS regulates nitrosylation and thus nuclear levels of CLIC4 in macrophages.
Fig. 2.
Pharmacologic inhibition or genetic ablation of iNOS inhibits S-nitrosylation and nuclear translocation of CLIC4 in murine macrophages. (A–C) RAW 264.7 macrophages were pretreated with (N-(3-(Aminomethyl) benzyl) acetamidine dihydrochloride) 1400 W (100 μM) for 1 h in indicated cases before treatment with LPS and IFNγ (1 μg/mL and 10 ng/mL, respectively) for 24 h. (A) Lysates were used to perform biotin switch assays. Lysate from LPS/IFNγ-treated cells was also used for a reaction that omitted biotin. Lane 3 of CLIC4 input is representative for both LPS/IFNγ-treated pull-down assays. Five percent of lysates were used as input controls and immunoblotted for CLIC4. SNO-CLIC4:CLIC4 input ratio was calculated for treatments across three independent experiments. Statistical significance was determined using a two-tailed unpaired Student's t test. 1400W versus LPS/IFN treatment has a P < 0.05 and LPS/IFN versus LPS/IFN +1400W has a P < 0.05. (B) Media from treated plates was collected and assayed for nitrite + nitrate levels in control and treated cells. (C) Cells were immunostained for CLIC4, and nuclei were stained with DAPI and visualized with confocal microscopy. (D–F) Primary mouse macrophages from iNOS knockout and wild-type mice were treated with LPS and IFNγ (1 μg/mL and 10 ng/mL, respectively) for 18 h. (D) Lysates were used to perform biotin switch assays. Lysate from LPS/IFNγ-treated wild-type cells was also used for a reaction that omitted ascorbate (“no Asc.”). Five percent of lysates were used as input controls and immunoblotted for CLIC4. (E) Cells were used for subcellular fractionation, and nuclear and cytosolic lysates were immunoblotted for CLIC4 and β-actin. (F) Macrophage cells were immunostained for CLIC4 and visualized with confocal microscopy. Nuclei were counterstained with DAPI and are pseudocolored red for better visualization.
Fig. 5.
Chemical inhibition of iNOS activity enhances IL-1β and iNOS levels, whereas overexpression of CLIC4 down-regulates IL-1β and iNOS. (A–C) Primary mouse macrophages from wild-type mice were treated with LPS and IFNγ (1 μg/mL and 10 ng/mL, respectively) for 24 h (A) or for 48 h (B and C). Cells were pretreated with 2.5 mM l-NAME for 1 h where indicated. (A) Nuclear and cytosolic lysates were prepared and immunoblotted for CLIC4 and β-actin antibodies. (B) Whole-cell lysates were immunoblotted for IL-1β, iNOS, CLIC4, CLIC1, and β-actin. (C) Media from treated samples was assayed for cleaved IL-1β by ELISA. Bars represent the mean ± SEM of three replicates. (D) RAW macrophages were transduced with adenoviruses expressing nuclear-targeted CLIC4 (NUC-CLIC4), HA-tagged CLIC4 (HA-CLIC4), or GFP proteins overnight followed by treatment with LPS and IFNγ (1 μg/mL and 10ng/mL, respectively) for 24 h. Whole-cell lysates were immunoblotted for IL-1β, iNOS, CLIC4, GFP, and β-actin.
CLIC4 and iNOS Knockout Macrophages Exhibit Similar Dysregulation of Proinflammatory Mediator IL-1β.
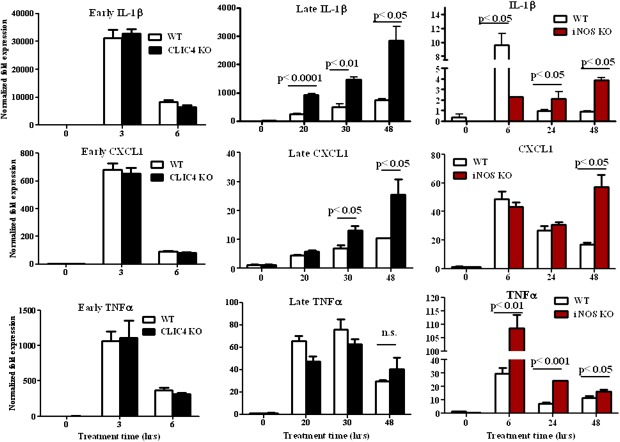
CLIC4 knockout macrophages derived from the bone marrow have decreased levels of TNFα and IL-6 cytokines immediately following LPS stimulation alone (15), a result that can be replicated with our CLIC4 knockout macrophages derived from the peritoneum (Fig. S2). In contrast, however, IP-10 mRNA levels are higher in our LPS-stimulated CLIC4 knockout macrophages (Fig. S2) and may reflect inherent differences between peritoneal and bone marrow-derived macrophage populations (19). LPS-stimulated iNOS activity is associated with transient induction of multiple cytokines followed by dissipation of the response (20). We examined transcript levels of various chemokines and cytokines at early and late time points post CLIC4 translocation. Immunokines like IL-1β, CXCL1, and TNFα mRNA are induced early by LPS/IFNγ in both CLIC4 and iNOS knockout macrophages as well as in their wild-type counterparts (Fig. 3). However, the knockout genotypes sustain elevated levels of IL-1β and CXCL1 mRNA for more than 24 h post stimulation (Fig. 3). These results suggest that iNOS-induced CLIC4 translocation may be important to dissipate the expression of certain proinflammatory mediators like IL-1β and CXCL1, but not others like TNFα, following activation of macrophages.
Fig. 3.
CLIC4 and iNOS knockout macrophages exhibit similar dysregulation of IL-1β and CXCL1 but not TNFα mRNA after prolonged stimulation with LPS/IFNγ. Primary mouse macrophages from CLIC4 knockout, iNOS knockout, and wild-type mice were treated with LPS and IFNγ (1 μg/mL and 10 ng/mL, respectively) for the indicated times. Lysates were used for real-time PCR analysis of IL-1β, CXCL1, and TNFα mRNA. Bars represent the mean ± SEM of three replicates. Statistical significance was determined using a two-tailed unpaired Student's t test and is indicated.
CLIC4 Knockout Macrophages Show Enhanced Levels of IL-1β and Lower F4/80 Levels with Reduced Numbers of Phagocytosing Macrophages Compared with Wild-Type Cells.
Stringent regulation of IL-1β activity is accomplished by multiple processes, including transcription, mRNA stability and translation, and protein processing and release. High levels of IL-1β precursor protein, which mirror the increased IL-1β mRNA levels, are detected in cell lysates from CLIC4 knockout macrophages at multiple time points after stimulation with LPS/IFNγ (Fig. 4A). IL-1β precursor is processed by the caspase 1-centered inflammasome to generate the active, cleaved protein that is secreted through multiple processes. CLIC4 knockout macrophages show significantly higher levels of cleaved IL-1β (Fig. 4A) on immunoblots, which are reflected by the increased levels of IL-1β activity in the media from these cells, as measured by ELISA (Fig. 4B). These results suggest that CLIC4 is required to down-regulate IL-1β activity of proinflammatory macrophages.
Fig. 4.
CLIC4 knockout macrophages show enhanced levels of IL-1β activity, lower levels of F4/80 receptor, and reduced numbers of phagocytosing macrophages in vivo. (A and B) Primary mouse macrophages from CLIC4 knockout and wild-type mice were treated with LPS and IFNγ (1 μg/mL and 10 ng/mL, respectively) for the indicated times. (A) Whole-cell lysates were immunoblotted for CLIC4, IL-1β, iNOS, and β-actin, as indicated. (B) Media from quadruplicate samples was assayed for cleaved IL-1β by ELISA. Bars represent the mean ± SEM. (C) Wild-type and CLIC4 knockout animals were injected with thioglycollate and 6 d later injected with LPS (12.5 μg/g) for 24 h. Animals were injected with pHrodo-labeled E. coli for 1 h before peritoneal macrophages were collected by lavage and stained for APC-F4/80 and with DAPI. Viable cells from each animal were analyzed for F4/80 and pHrodo fluorescence using flow cytometry. Statistical significance was determined using a two-tailed unpaired Student's t test and is indicated. Each dot represents results from a single animal.
Macrophage phagocytic activity is essential for the body's response to injury and infection, and iNOS-mediated NO production is linked with the cytotoxic and cytostatic effects of activated macrophages against pathogens (21). CLIC4 knockout macrophages were examined for phagocytic activity, in vivo, using pHrodo-labeled Escherichia coli particles followed by flow cytometric analysis of F4/80 and pHrodo-positive, viable cells. Wild-type and CLIC4 knockout macrophages show a similar level of phagocytic activity as measured by the median pHrodo fluorescence (Fig. 4C) for macrophages in the two genotypes. Strikingly, knockout cells have significantly lower levels of F4/80 macrophage receptor as measured by fluorescence intensity and significantly lower numbers of F4/80-positive, pHrodo-positive macrophages (Fig. 4C). There was no difference in cell viability between CLIC4 knockout and wild-type macrophages (Fig. S3).
Chemical Inhibition of iNOS-Induced CLIC4 Nuclear Translocation Enhances IL-1β Levels, Whereas Overexpression of Nuclear CLIC4 Down-Regulates IL-1β.
The timing of iNOS-induced CLIC4 nuclear translocation coincides with the transition of classically activated macrophages to an anti-inflammatory program for resolution of inflammation. This suggests that nuclear CLIC4 may be important for down-regulation of proinflammatory mediators like IL-1β and iNOS. Induction and increased activity of iNOS in immune cells is also considered a hallmark of inflammation. Indeed, inhibition of nuclear translocation of CLIC4 by l-NAME in macrophages (Fig. 5A) coincides with enhanced levels of IL-1β and iNOS proteins (Fig. 5B) and IL-1β release (Fig. 5C). To establish the importance of nuclear CLIC4 in deactivation of macrophages, native (HA-CLIC4) or nuclear-targeted (NUC-CLIC4) CLIC4 was overexpressed in RAW264.7 cells by adenoviral transduction with adenoviral GFP used as a control. All adenoviral constructs have been previously characterized (10). Treatment with both CLIC4 vectors decreased IL-1β and iNOS following stimulation of cells (Fig. 5D). Taken together, these results underscore the importance of nuclear CLIC4 for deactivation of proinflammatory macrophages.
Discussion
We demonstrate that CLIC4 is significantly induced upon classical activation of peritoneal macrophages. This concurs with earlier reports of CLIC4 as an inflammatory response gene and a transcriptional target of NF-κB and IRF3 (14). The increase is detected in both the cytosolic and nuclear levels of CLIC4 and is concurrent with induction of iNOS protein. CLIC4 nuclear translocation is governed by nitric oxide through S-nitrosylation, which in turn dictates its association with nuclear import proteins (13). iNOS-induced S-nitrosylation and nuclear translocation of CLIC4 in proinflammatory macrophages seems to be essential for down-regulating that part of the proinflammatory program of macrophages that resolves inflammation. Deactivation of macrophages is essential for the transition from a microbiocidal, tissue-damaging phenotype to tissue repair in the infected or injured host. Indeed, dysregulation of this anti-inflammatory program leads to numerous autoinflammatory diseases (22).
CLIC4 knockout cells have decreased levels of proinflammatory cytokines like TNF and IL-6 at the early time points in response to LPS alone (15). We obtain similar results under those conditions; however, when cells are costimulated with IFNγ, mimicking the priming of macrophages in vivo by natural killer (NK) cells (23), CLIC4 knockout cells show only aberrant down-regulation of proinflammatory IL-1β whereas early induction seems to be comparable to wild type. CLIC4 knockout mice have less Listeria monocytogenes clearance compared with wild-type animals after 5 d and reduced levels of TNFα and IL-12 (15). Indeed, our in vivo phagocytosis results indicate that CLIC4 knockout mice have fewer phagocytically active macrophages in the peritoneum that may lead to reduced bacterial clearance. In addition, knockout macrophages have reduced levels of F4/80 receptor that may also contribute to reduced cytokine secretion, like IFNγ. F4/80 is critical for interaction of macrophages and NK cells, which in turn is essential for optimal secretion of cytokines (TNFα and IL-12) by macrophages and IFNγ by NK cells. IFNγ is a major barrier to bacterial growth during infection (24).
CLIC4 knockout macrophages have reduced expression of IRF3-dependent cytokines (15); however, dysregulation of IRF3-independent genes suggests that CLIC4 has additional roles in macrophage biology. TGF-β1 is central for deactivation of macrophages following stimulation, in particular for down-regulating proinflammatory mediators like IL-1β (25) and iNOS (26). The Smad-dependent TGF-β1 pathway, specifically Smad3, is essential for abatement of the proinflammatory macrophage phenotype, including iNOS and IL-1β (27, 28), whereas Smad4 is important for endotoxin tolerance (29). We have previously established that nuclear CLIC4 is an essential positive modulator of TGF-β1 signaling in keratinocytes through its stabilization of phospho-Smad2/3 by disrupting their interaction with the phosphatase PPM1A (8). Thus, it is likely that the prolonged proinflammatory phenotype of CLIC4 knockout macrophages stems from aberrant TGF-β1 signaling. Propagation of Smad-dependent TGF-β1 signaling may be a central function of iNOS-induced nuclear CLIC4 in facilitating deactivation of macrophages. However, we cannot rule out a function for cytosolic CLIC4 in this pathway. More recently, p38 and its phosphatase MKP-1 have been implicated in regulation of the transition from a pro- to anti-inflammatory program in macrophages (30). Chemical inhibition of phospho-p38 increases IL-1β levels at late time points, which in turn regulate anti-inflammatory molecules like IL-10 and TGF-β1 through p-p38. It remains to be determined which pathway(s) may be important for CLIC4-dependent deactivation of macrophages.
CLIC4 knockout macrophages, with high IL-1β and iNOS proinflammatory activities, exhibit a more M1 tumoricidal phenotype. Overexpression of CLIC4 inhibits the proinflammatory program and may shift the cells to a more M2 anti-inflammatory phenotype that may have implications for both tumor biology and wound healing. Indeed, we have previously reported the reciprocal changes in expression of CLIC4 in multiple human cancers, with decreased levels in the tumor epithelium and a marked up-regulation in the tumor stroma (31). It remains to be determined if any of the stromal up-regulation occurs in tumor-associated macrophages and what significance CLIC4 may have in the immunosuppressive tumor microenvironment.
Materials and Methods
Reagents.
Lactacystin, 1400W, and l-NAME were purchased from Alexis; Salmonella LPS from Sigma-Aldrich; and mouse IFNγ from Peprotech. MG132 was purchased from Calbiochem. All reagents used for the biotin switch assay were purchased from Pierce. pHrodo-labeled E. coli particles for phagocytosis were from Invitrogen. Antibodies were from the following sources: β-actin (Abcam), lamin A/C (Santa Cruz Biotechnology), α-tubulin (Invitrogen), GFP (Roche), GAPDH (Chemicon), FITC-labeled secondary anti-rabbit (Vector Labs), rabbit polyclonal CLIC4 N-terminal (Covance), and mouse monoclonal CLIC1, CLIC4, iNOS, CD3ε, CD11b, and CD19 (BD Biosciences). CD16/32 and APC-F4/80 antibodies were from Biolegend, anti-CD45 from eBioscienceand anti–IL-1β from R&D Systems. Results are representative of at least two independent experiments. Prism was used for all graphs, and data analysis and bars represent the mean ± SEM of at least three replicates.
Mice, Cell Culture, Transfections, and Adenoviral Infections.
Mouse studies were performed under a protocol approved by the National Cancer Institute and the National Institutes of Health Animal Care and Use Committee. iNOS knockout mice, kindly provided by Perwez Hussein (National Cancer Institute, Bethesda, MD) (32), were backcrossed to an FVB/N background for eight generations, and CLIC4 knockout mice were generated in our laboratory. Isolation and culture of cells is detailed in SI Materials and Methods.
Immunofluorescent Cell Staining.
Cell staining was performed as described previously (13), except cells were fixed with 4% (vol/vol) paraformaldehyde for 20 min.
Protein Lysates and Subcellular Fractionation.
Whole-cell lysates and subcellular fractionations were done as previously outlined (13). Lysates were separated by SDS/PAGE and immunoblotted with the relevant antibodies.
IL-1β ELISA and NO Assays.
IL-1β activity in supernatants of wild-type or CLIC4 knockout macrophages was quantified with an ELISA (BD Biosciences) using the manufacturer's protocol and normalized to total protein in the respective wells. Cell supernatant was used to detect nitrite + nitrate concentrations using a colorimetric assay kit (Oxford Biomedical Research) or fluorometric assay kit (Cayman Chemical).
Real-Time PCR.
RNA was extracted using TRIzol (Invitrogen), following the manufacturer's instructions. cDNA was prepared from 0.5 μg of total RNA, and transcript expression levels were determined and analyzed as previously described (33). Primers for mouse CLIC4 (catalog no. PPM04113E-200) and iNOS (catalog no. PPM02928B-200) were purchased from SABiosciences. IL-1β, IL-6, CXCL1 (KC), CXCL2 (MIP2), IL-18, Myc, and TNFα were predesigned Quantitect primers from Qiagen. The GAPDH sequence was forward (5′-CATGGCCTTCCGTGTTCCTA-3′) and reverse (5′-GCGGCACGTCAGATC CA-3′).
Biotin Switch Assay for S-Nitrosylation.
The assay was performed as outlined in the original protocol (34) with some modifications (13). Cells were lysed in HEN buffer (250 mM Hepes-NaOH pH 7.7, 1 mM EDTA, 0.1 mM neocuproine). Protease inhibitors, PMSF, and lactacystin were included in all buffers throughout the assay. Lysate from LPS/IFNγ-treated cells was also used for a reaction that omitted ascorbate as a specificity control.
Phagocytosis Assays.
Phagocytosis assays were carried out essentially as outlined in ref. 35, using pHrodo-labeled E. coli. Thioglycollate-stimulated (6 d) mice were injected with a low dose of LPS (12.5 mg/kg animal) for 24 h, followed by an intraperitoneal injection of pHrodo-labeled E. coli particles (0.4 mL) for 1 h. Macrophages were collected by peritoneal lavage and prepared as detailed above. Two million cells per mouse were blocked with CD16/32 antibody (2 μg) for 20 min on ice, followed by incubation with APC-F4/80 antibody (1.4 μg) for 30 min. Cells were washed once with PBS and labeled with DAPI. Viable APC-F4/80-labeled cells were analyzed for pHrodo fluorescence by FACS analysis (BD LSR II).
Supplementary Material
Acknowledgments
We thank Kathleen Noer and Roberta Matthai of the Cancer and Inflammation Program Fluorescence-Activated Cell Sorter Core for technical assistance in immune cell sorting; Susan Garfield and the National Cancer Institute Confocal Core for microscopy assistance; and Marta Custer for maintenance of animal colonies. We also thank Anjali Shukla and Francesca Mascia for helpful suggestions. This work was supported by the intramural program of the National Cancer Institute and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201351109/-/DCSupplemental.
References
- 1.Qian Z, Okuhara D, Abe MK, Rosner MR. Molecular cloning and characterization of a mitogen-activated protein kinase-associated intracellular chloride channel. J Biol Chem. 1999;274:1621–1627. doi: 10.1074/jbc.274.3.1621. [DOI] [PubMed] [Google Scholar]
- 2.Tulk BM, Kapadia S, Edwards JC. CLIC1 inserts from the aqueous phase into phospholipid membranes, where it functions as an anion channel. Am J Physiol Cell Physiol. 2002;282:C1103–C1112. doi: 10.1152/ajpcell.00402.2001. [DOI] [PubMed] [Google Scholar]
- 3.Berryman M, Bruno J, Price J, Edwards JC. CLIC-5A functions as a chloride channel in vitro and associates with the cortical actin cytoskeleton in vitro and in vivo. J Biol Chem. 2004;279:34794–34801. doi: 10.1074/jbc.M402835200. [DOI] [PubMed] [Google Scholar]
- 4.Fernández-Salas E, et al. mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53. Mol Cell Biol. 2002;22:3610–3620. doi: 10.1128/MCB.22.11.3610-3620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiio Y, et al. Quantitative proteomic analysis of myc-induced apoptosis: A direct role for Myc induction of the mitochondrial chloride ion channel, mtCLIC/CLIC4. J Biol Chem. 2006;281:2750–2756. doi: 10.1074/jbc.M509349200. [DOI] [PubMed] [Google Scholar]
- 6.Suh KS, et al. CLIC4 mediates and is required for Ca2+-induced keratinocyte differentiation. J Cell Sci. 2007;120:2631–2640. doi: 10.1242/jcs.002741. [DOI] [PubMed] [Google Scholar]
- 7.Rønnov-Jessen L, Villadsen R, Edwards JC, Petersen OW. Differential expression of a chloride intracellular channel gene, CLIC4, in transforming growth factor-beta1-mediated conversion of fibroblasts to myofibroblasts. Am J Pathol. 2002;161:471–480. doi: 10.1016/s0002-9440(10)64203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla A, et al. TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3. Nat Cell Biol. 2009;11:777–784. doi: 10.1038/ncb1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao Q, Qu X, Yang Q, Wei M, Kong B. CLIC4 mediates TGF-beta1-induced fibroblast-to-myofibroblast transdifferentiation in ovarian cancer. Oncol Rep. 2009;22:541–548. doi: 10.3892/or_00000469. [DOI] [PubMed] [Google Scholar]
- 10.Suh KS, et al. The organellular chloride channel protein CLIC4/mtCLIC translocates to the nucleus in response to cellular stress and accelerates apoptosis. J Biol Chem. 2004;279:4632–4641. doi: 10.1074/jbc.M311632200. [DOI] [PubMed] [Google Scholar]
- 11.Littler DR, et al. Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4. FEBS J. 2005;272:4996–5007. doi: 10.1111/j.1742-4658.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- 12.Greco TM, et al. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik M, et al. S-nitrosylation regulates nuclear translocation of chloride intracellular channel protein CLIC4. J Biol Chem. 2010;285:23818–23828. doi: 10.1074/jbc.M109.091611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogawa S, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He G, et al. Role of CLIC4 in the host innate responses to bacterial lipopolysaccharide. Eur J Immunol. 2011;41:1221–1230. doi: 10.1002/eji.201041266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nacy CA, Meltzer MS. T-cell-mediated activation of macrophages. Curr Opin Immunol. 1991;3:330–335. doi: 10.1016/0952-7915(91)90033-w. [DOI] [PubMed] [Google Scholar]
- 17.Tsunawaki S, Sporn M, Ding A, Nathan C. Deactivation of macrophages by transforming growth factor-beta. Nature. 1988;334:260–262. doi: 10.1038/334260a0. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Morrison DC, Parmely TJ, Russell SW, Murphy WJ. An interferon-gamma-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-gamma and lipopolysaccharide. J Biol Chem. 1997;272:1226–1230. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 19.Berton G, Gordon S. Superoxide release by peritoneal and bone marrow-derived mouse macrophages. Modulation by adherence and cell activation. Immunology. 1983;49:693–704. [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto T, et al. Multiple contributing roles for NOS2 in LPS-induced acute airway inflammation in mice. Am J Physiol Lung Cell Mol Physiol. 2004;286:L198–L209. doi: 10.1152/ajplung.00136.2003. [DOI] [PubMed] [Google Scholar]
- 21.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 22.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon S, Hamann J, Lin HH, Stacey M. F4/80 and the related adhesion-GPCRs. Eur J Immunol. 2011;41:2472–2476. doi: 10.1002/eji.201141715. [DOI] [PubMed] [Google Scholar]
- 25.Imai K, Takeshita A, Hanazawa S. Transforming growth factor-beta inhibits lipopolysaccharide-stimulated expression of inflammatory cytokines in mouse macrophages through downregulation of activation protein 1 and CD14 receptor expression. Infect Immun. 2000;68:2418–2423. doi: 10.1128/iai.68.5.2418-2423.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vodovotz Y, Bogdan C, Paik J, Xie QW, Nathan C. Mechanisms of suppression of macrophage nitric oxide release by transforming growth factor beta. J Exp Med. 1993;178:605–613. doi: 10.1084/jem.178.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner F, et al. Transforming growth factor-beta 1 inhibition of macrophage activation is mediated via Smad3. J Biol Chem. 2000;275:36653–36658. doi: 10.1074/jbc.M004536200. [DOI] [PubMed] [Google Scholar]
- 28.McCartney-Francis N, Jin W, Wahl SM. Aberrant Toll receptor expression and endotoxin hypersensitivity in mice lacking a functional TGF-beta 1 signaling pathway. J Immunol. 2004;172:3814–3821. doi: 10.4049/jimmunol.172.6.3814. [DOI] [PubMed] [Google Scholar]
- 29.Pan H, et al. SMAD4 is required for development of maximal endotoxin tolerance. J Immunol. 2010;184:5502–5509. doi: 10.4049/jimmunol.0901601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perdiguero E, et al. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J Cell Biol. 2011;195:307–322. doi: 10.1083/jcb.201104053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh KS, et al. Reciprocal modifications of CLIC4 in tumor epithelium and stroma mark malignant progression of multiple human cancers. Clin Cancer Res. 2007;13:121–131. doi: 10.1158/1078-0432.CCR-06-1562. [DOI] [PubMed] [Google Scholar]
- 32.Hussain SP, et al. Nitric oxide, a mediator of inflammation, suppresses tumorigenesis. Cancer Res. 2004;64:6849–6853. doi: 10.1158/0008-5472.CAN-04-2201. [DOI] [PubMed] [Google Scholar]
- 33.Cataisson C, et al. CXCR2 ligands and G-CSF mediate PKCalpha-induced intraepidermal inflammation. J Clin Invest. 2006;116:2757–2766. doi: 10.1172/JCI27514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 35.Medina E, Goldmann O. In vivo and ex vivo protocols for measuring the killing of extracellular pathogens by macrophages. Current Protoc Immunol. 2011;Chapter 14:Unit 14.19.1–17. doi: 10.1002/0471142735.im1419s92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.