Abstract
Double-stranded RNA viruses in the family Reoviridae are capable of transcribing and capping nascent mRNA within an icosahedral viral capsid that remains intact throughout repeated transcription cycles. However, how the highly coordinated mRNA transcription and capping process is facilitated by viral capsid proteins is still unknown. Cypovirus provides a good model system for studying the mRNA transcription and capping mechanism of viruses in the family Reoviridae. Here, we report a full backbone model of a transcribing cypovirus built from a near-atomic-resolution density map by cryoelectron microscopy. Compared with the structure of a nontranscribing cypovirus, the major capsid proteins of transcribing cypovirus undergo a series of conformational changes, giving rise to structural changes in the capsid shell: (i) an enlarged capsid chamber, which provides genomic RNA with more flexibility to move within the densely packed capsid, and (ii) a widened peripentonal channel in the capsid shell, which we confirmed to be a pathway for nascent mRNA. A rod-like structure attributable to a partially resolved nascent mRNA was observed in this channel. In addition, conformational change in the turret protein results in a relatively open turret at each fivefold axis. A GMP moiety, which is transferred to 5’-diphosphorylated mRNA during the mRNA capping reaction, was identified in the pocket-like guanylyltransferase domain of the turret protein.
Double-stranded RNA (dsRNA) viruses use RNA-dependent RNA polymerases (RdRps) to transcribe mRNA from the minus-strand RNA genome within virus capsids. Nascent mRNA is capped by other viral enzymes and then released into the cytoplasm to initiate viral protein translation (1). The Reoviridae family is one of the largest families of dsRNA viruses and causes disease in humans, livestock, insects, and plants (1). Viruses in the family Reoviridae have a segmented dsRNA genome enclosed by single- or double-layered icosahedral inner capsid structure (hereafter referred to as the “core”), which is usually coated by an icosahedral T = 13 (2, 3) or incomplete T = 13 (4–7) outer capsid layer. The Reoviridae family members are divided into two subfamilies according to the structural organization of their core—the Sedoreovirinae and the Spinareovirinae (8). The cores of viruses in the subfamily Sedoreovirinae, for example the Orbivirus and Rotavirus genera, have a relatively smooth proteinaceous capsid shell composed of 120 copies of capsid shell protein coated by a T = 13 layer (2, 3, 9). The cores of viruses in the subfamily Spinareovirinae, for example, the Orthoreovirus, Oryzavirus, and Cypovirus genera, have a capsid shell with a similar organization but a distinct pentameric turret formed by five copies of turret proteins, each sitting around the fivefold vertex on the capsid shell, which functions in the catalysis of mRNA 5′ cap synthesis (4–7, 10, 11). The capping enzymes in viruses of the subfamily Sedoreovirinae without turret structures are located under the fivefold vertex inside the capsid shell (2, 12). A common process adopted by all Reoviridae members, and some other dsRNA viruses, is the uncoating of the outer capsid after delivery into the cytoplasm of host cells (13, 14) prior to transcription, and the core remains intact and serves as a stable nano-scale machine for viral mRNA transcription and capping and for protecting the transcription process from the antiviral defense mechanisms of the host cell (12, 15–17).
Cryoelectron microscopy (cryo-EM) and conventional electron microscopy have been used to analyze the structure of transcribing viruses in the Reoviridae family (17–19). The structure of the transcribing rotavirus core has been determined at a resolution of 25 Å (17). It suggested that rotavirus mRNA is released from a peripentonal channel rather than the channel located at each fivefold vertex, and the structure of the transcribing rotavirus core is essentially identical to that of nontranscribing cores (17). Recently, the structure of the transcribing core of orthoreoviruses has been determined at an approximately 25-Å resolution (18). At such a resolution, it is not possible to investigate fine structural changes. How the highly coordinated mRNA transcription and capping process is facilitated by viral capsid proteins is still unknown.
In this study, we use cytoplasmic polyhedrosis virus (CPV) as a model system to study the mechanism of mRNA transcription and capping. CPV belongs to the genus Cypovirus. It is the simplest virus in the Reoviridae family in that it only has a single capsid layer (20). The single capsid layer of CPV has structurally conserved capsid shell proteins and turret proteins and has a very similar organization to the core of viruses in the subfamily Spinareovirinae (10, 16, 21, 22). Near-atomic-resolution structural analyses by X-ray crystallography and cryo-EM reveal that the structures of the capsid shell proteins and the location of RdRp in all viruses in the Reoviridae family are conserved (5, 9, 10, 16, 20, 22–24). Thus, CPV is an ideal simplified model system for studying the mRNA transcription and capping mechanism of viruses in the Reoviridae family. We have now obtained a near-atomic-resolution structure of a transcribing CPV by cryo-EM, which allows us to build backbone models for the capsid proteins. By making comparisons with the atomic model of nontranscribing CPV (10), we discovered a series of significant structural changes in CPV capsid proteins that are associated with CPV mRNA transcription and capping. The conformational changes of capsid shell protein VP1 give rise to an enlarged capsid chamber and a widened peripentonal channel connecting the inner capsid chamber and the turret cavity. Moreover, a GMP moiety structure was identified in the VP3 guanylyltransferase (GTase) domain. These observations provide a wealth of new structural information for us to understand the mechanism underlying the highly coordinated mRNA transcription and capping process which occurs in this stable nano-scale viral transcription machine.
Results and Discussion
Preparation of Transcribing CPV.
mRNA transcripts from a transcription reaction mixture were separated by 4% polyacrylamide gel electrophoresis (Fig. 1, lane 1). Radioactive mRNA transcripts were present indicating that the transcription reaction had occurred in the reaction mixture. We also designed three reactions with different concentrations of GTP and different incubation times as controls. First, we performed the reaction in the absence of GTP (the other conditions were the same as those for the transcription reaction mixture) and incubated the reaction mixture at 31 °C for 3 h. As expected, no transcribed mRNA was detected in the absence of GTP (Fig. 1, lane 2). We then decreased the concentration of GTP from 2 mM to 0.04 mM (the other conditions were the same as those for the transcription reaction mixture) and incubated the reaction mixture for 1 and 3 h, respectively. A 3-h incubation time resulted in darker RNA bands (Fig. 1, lane 3) than a 1-h incubation time (Fig. 1, lane 4), indicating that the reaction continues after 1 h. The RNA bands in lane 1 were darker than those in lane 4 indicating that the lower GTP concentration limited reaction activity. mRNA transcripts were also observed using cryo-EM. A cryo-EM image of a transcribing CPV shows that strand-like material was visible around many of the particles (Fig. 2A and Fig. S1), while it was not observed in nontranscribing CPV cryo-EM images (10).
Fig. 1.
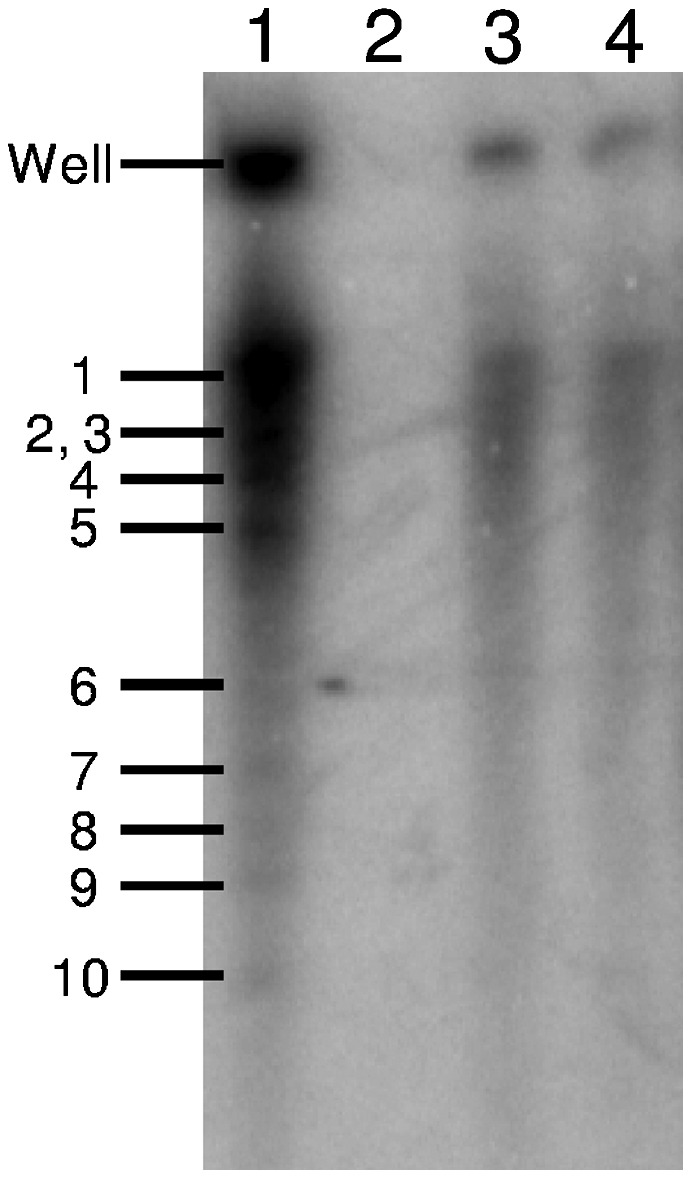
Polyacrylamide gel electrophoresis analysis of transcription reaction mixtures with different concentrations of GTP and incubation times. All 10 RNA segments are labeled. Lane 1: transcription reaction mixture consisting of 70 mM Tris-Ac (pH 8), 10 mM MgAc2, 100 mM NaAc, 4 mM ATP, 2 mM GTP, 2 mM CTP, 2 mM UTP, 20 μCi [α-32P] UTP (specific activity 3,000 Ci/mM), 1 mM SAM, 1 U/μL RNase inhibitor and purified CPV suspension was incubated at 31 °C for 3 h. Lane 2: transcription reaction mixture in the absence of GTP (other conditions are the same as those for lane 1). Lane 3 and 4: transcription reaction mixtures in which the concentration of GTP was decreased to 0.04 mM were incubated at 31 °C for 3 h and 1 h, respectively.
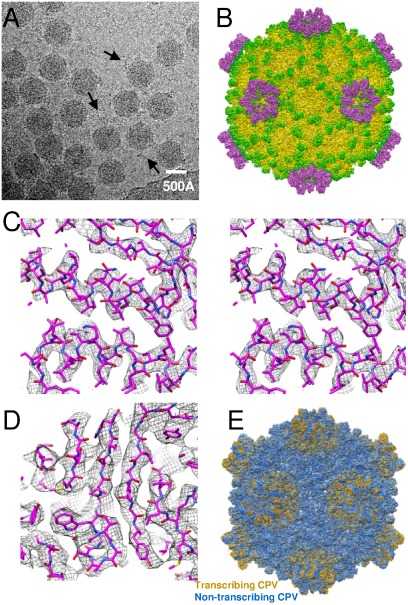
Fig. 2.
Overall structure of the CPV capsid. (A) A cryo-EM image of CPV. Strand-like material was visible around many of the particles (arrows). (B) A radially colored shaded surface representation of CPV viewed along a twofold axis. (C) Wall-eye stereo view of part of the capsid shell protein α-helix density map (mesh) superimposed on the atomic model. (D) Part of the capsid shell protein β-strands density map (mesh) superimposed on the atomic model. (E) Structure of transcribing CPV (yellow) superimposed on nontranscribing CPV (blue). The spike-like structures of nontranscribing CPV were removed computationally for clarity.
Structure Determination.
We obtained a 3D structure of the transcribing CPV by cryo-EM and single-particle reconstruction (Fig. 2 A and B). About 8,400 particle images from 845 micrographs collected by CCD with an FEI Titan Krios microscope were selected for final 3D reconstruction. Based on the clarity of the resolved features (Fig. 2 C and D), and by comparing these features with those of nontranscribing CPV (Fig. S2) (10), we estimated that the transcribing CPV is resolved to a resolution of about 4.1 Å. The structural features, including separated β-strands and turns of α-helices were clearly resolved. More than 60% of the side chains were clearly visible, allowing us to build a full backbone model for all major capsid proteins (Fig. 2 C and D and Fig. S2).
The transcribing CPV has a similar overall capsid structure to that of nontranscribing CPV except that the transcribing CPV does not have the spike-like protein complex on the top of each fivefold turret that is present in nontranscribing CPV (Fig. 2B). In order to study the structural changes in capsid proteins associated with mRNA transcription, we superimposed the density map of transcribing CPV on a structure of nontranscribing CPV that was filtered to the same resolution (Fig. 2E). The two superimposed maps show that the major differences are located in regions around the fivefold axis (Fig. 2E). By closely examining the difference maps, we identified the proteins that are involved in the conformational changes—the capsid shell proteins VP1A, VP1B, and the turret protein VP3. No structural changes in protrusion protein VP5 were detected.
Conformational Changes in Capsid Shell Protein VP1 and Related Structural Changes in the Capsid Shell.
According to the domain nomenclature used for the capsid shell protein VP3 counterpart in bluetongue virus (23), the two CPV VP1 conformers, VP1A and B, each have four domains: the apical (residues 439–775), carapace (residues 134–438, 790–825, 963–1070, and 1237–1333), and dimerization (residues 1071–1236) domains, and a distinct protrusion domain (residues 825–962). A structural comparison of transcribing and nontranscribing CPV VP1A and B revealed conformational changes in VP1A and B that are localized in the apical domain (Fig. 3A and Movies S1 and S2). In transcribing CPV VP1A, the apical domain appears to tilt toward the turret region. The tilting of the apical domain is like the movement of a hinge (Movie S1). Structural changes in VP1B of transcribing CPV are smaller than those in VP1A. The major conformational change in VP1B occurs at the tip region of its apical domain (Fig. S3 and Movie S2).
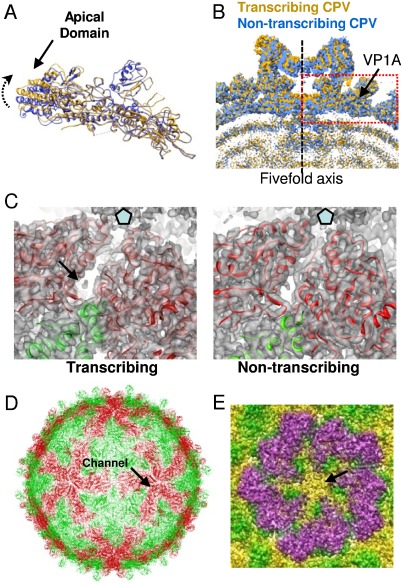
Fig. 3.
Capsid shell proteins. (A) VP1A of the transcribing CPV model (yellow) superimposed on VP1A of nontranscribing CPV (blue). The direction of the apical domain tilt is indicated by an arrow. (B) A slab view of the turret of transcribing CPV (yellow) superimposed on nontranscribing CPV (blue). The structure in the red box is a copy of the VP1A of transcribing CPV (yellow) and a copy of the VP1A of nontranscribing CPV (blue), in the same orientation as that in A, indicating that the tilt of the VP1A apical domain gives rise to an enlarged capsid chamber. (C) Left: two copies of VP1A (red) and one of VP1B (blue) are superimposed on the density map of transcribing CPV. A peripentonal RNA channel and a rod-like density are indicated by an arrow. Right: two copies of VP1A (red) and a copy of VP1B (blue) are superimposed on the density map of nontranscribing CPV. These proteins have the same orientation as those on the left for comparison. The peripentonal channel on the left (transcribing) is wider than that on the right (nontranscribing). The small protrusion in the channel on the right is the side chain of VP1 Arg 653. (D) Location (arrow) of one of the peripentonal channels in the capsid shell formed by VP1A (red) and B (green). (E) The turret of transcribing CPV showing that a nodule-like structure blocks the pentameric channel at the fivefold axis.
To investigate the functional implications of these conformational changes in VP1A and B, we need first to investigate the organization of the capsid shell proteins as an ensemble. The capsid shell of CPV is formed by 120 copies of VP1: 60 copies of VP1A are located around the fivefold axes and 60 copies of VP1B are located around the threefold axes. An RNA-dependent RNA polymerase (RdRp) is located close to the inner surface of the capsid shell at each fivefold axis and is embedded in the surrounding dsRNA (20, 25) so that mRNA can be transcribed in the capsid shell. Comparing transcribing CPV with nontranscribing CPV, we observed two important structural changes in the capsid shell that are functionally associated with mRNA transcription. The first structural change in the capsid shell is that the tilt of the VP1A apical domain gives rise to an enlarged capsid chamber where the mRNA transcription process occurs (Fig. 3B and Movies S3 and S4). The enlarged volume under the 12 fivefold vertices is about 9.4 × 105 Å3 in total and accounts for 1.4% of the total volume of the capsid chamber. Although the transcription mechanism of the Reoviridae family has not been fully elucidated, a generally accepted transcription model is that RdRp is anchored to the capsid shell while a template RNA slides through RdRp for mRNA transcription (4, 26, 27). The enlarged capsid chamber enables genomic RNA to have greater flexibility to slide within the densely packed capsid.
Another structural change in the capsid shell accompanying conformational changes in VP1A and B is that a peripentonal channel formed by two adjacent copies of VP1A and their neighboring copy of VP1B becomes wider (Fig. 3 C and D and Movie S5). This peripentonal channel is not perpendicular to the surface of the capsid shell but at an angle of approximately 30 degrees to the shell surface (Fig. S4). The entrance of the peripentonal channel facing the RNA genome is at a distance of about 35 Å from the fivefold axis and is located above the region where the RdRp is located (Fig. 3C), and its exit opens into the turret cavity (Fig. S4). The peripentonal channel is irregular in shape and has a diameter of about 9–15 Å, while the peripentonal channel in nontranscribing CPV has a diameter of about 6–10 Å. Moreover, the peripentonal channel in nontranscribing CPV is blocked by the side chain of VP1 Arg 653 (Fig. 3C). It is worth noting that the nodule-like structure that was observed to block the pentameric channel at the fivefold axis in the capsid shell of nontranscribing CPV (10) was also observed in transcribing CPV (Fig. 3E). Thus, it is likely that this nodule-like structure belongs to the RdRp, presumably serving to anchor the RdRp to the capsid shell. The presence of the nodule-like structure in transcribing CPV also excludes the possibility that the pentameric channel is the pathway for nascent mRNA release. So the peripentonal channel is the only channel connecting the inner capsid chamber and the turret cavity. In addition, a small rod-like structure, which was not observed in nontranscribing CPV, was observed in the peripentonal channel (Fig. 3C). This rod-like structure may represent partially resolved nascent mRNA. The mRNA structure was not well resolved because mRNA is flexible and the icosahedrally symmetrical channels are not all occupied by mRNA. Taken together, these observations strongly suggest that the peripentonal channel is the pathway where the nascent mRNA exits through the capsid shell to the turret cavity. This location for the nascent mRNA pathway is in agreement with the pathway of orthoreoviruses proposed by Zhang et al. (4). Based on the position and orientation of the RdRp crystal structure fitted into the cryo-EM structure of the orthoreovirus virion, Zhang et al. hypothesized that the channel could become wider during transcription to allow passage of the nascent mRNA into the turret cavity. Our structure of transcribing CPV provides direct structural evidence for this hypothesis. A previous reconstruction of the transcribing rotavirus also suggested that its transcribed mRNA exits through a peripentonal channel, not through the pentameric channel at the fivefold axis (17). It appears that at least a subset of viruses in the Reoviridae family take the peripentonal pathway for mRNA exit.
Superimposing CPV VP1A on VP1B reveals that two major structural changes occur in the apical and protrusion domains (10). It is intriguing that a systematic comparison between the two capsid shell protein conformers of rotavirus, bluetongue virus, and orthoreovirus reveals that the major structural difference between the two conformers of all these viruses is a conformational change in the apical domain (9). Except for the unique protrusion domain present in CPV VP1, the structure of VP1 and its counterparts in other Reoviridae family members are highly conserved (10). Furthermore, the organization of VP1A and B in the CPV capsid shell is shared by all other Reoviridae family members and even some other dsRNA viruses (1, 28). It is likely that the flexibility of the apical domain is a common property of this group of viruses.
Conformational Changes in Turret Protein VP3.
After exiting through the channel in the capsid shell, the nascent mRNA moves directly to the cavity of the turret to undergo mRNA capping (16, 29, 30). The hollow pentameric turret on each icosahedral fivefold vertex is formed by five copies of VP3. A structural comparison of transcribing and nontranscribing CPV VP3 reveals that the major structural change is a pivotal movement around residue 355 (Fig. 4A and Movie S6). This pivotal movement drives the movement of the domains of 7-N-methyltransferase (7-N-MTase), 2′-O-methyltransferase (2′-O-MTase), and the bridge. Another conformational change in VP3 occurs at residues 279–309 in the GTase domain, which includes a loop and an α-helix (Fig. 4A). No other significant structural changes were observed. Maintaining a rigid structure is presumably important for preserving enzymatic activity.
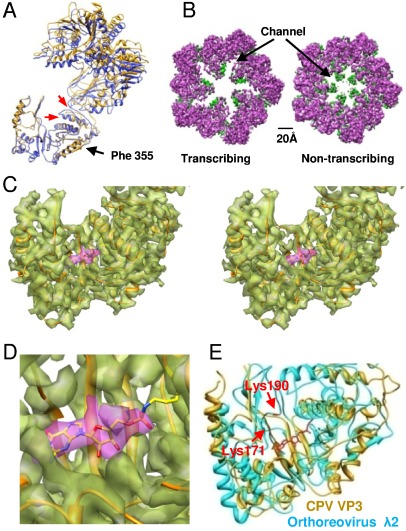
Fig. 4.
Turret protein VP3. (A) The VP3 of transcribing CPV (yellow) superimposed on the VP3 of nontranscribing CPV (blue). Two red arrows point to conformational changes of a loop and an α-helix. (B) A comparison of the turret of transcribing and nontranscribing CPV. The color scheme is the same as that in Fig. 2B. (C) A wall-eye stereo view of the density map (transparent) of the VP3 GTase domain with its atomic model superimposed. The density map of the GMP moiety is in purple. (D) Zoom-in view of the GMP in C. (E) The CPV VP3 GTase domain is superimposed on that of orthoreovirus λ2. The locations of Lys 190 and Lys 171 in λ2 are marked. The GMP moiety in VP3 is in red.
The role of VP3 is to catalyze the last three of the four reactions in mRNA capping: mRNA GTase, 7-N-MTase, and 2′-O-MTase (16, 29, 30). Accordingly, VP3 has a series of four domains: the GTase domain, the 7-N-MTase domain, the 2′-O-MTase domain, the brace domain, and the bridge domain that bridges the GTase domain and the two MTase domains (10). Conformational change in VP3 transforms the turret from a relatively closed state to a relatively open state, a process that is like the opening of a flower (Fig. 4B and Movie S4). This structural change makes the channel connecting the GTase domain and the 7-N-MTase domain, hypothesized to be the mRNA pathway (10), become wider (Fig. 4B). These structural changes are probably important in mRNA capping; however, no mRNA density was observed in the turret cavity. This may be because only one mRNA transcript can be processed in the turret because only one copy of RdRp is bound per fivefold axis (4), and because the mRNA transcript has considerable flexibility to meander in the relatively large hollow turret. The spike-like protein complexes held by brace domains in nontranscribing CPV (10) are absent in transcribing CPV probably because the brace domains of transcribing CPV are not able to hold the spike-like protein complexes due to the conformational changes in VP3 (Fig. 4B).
Direct Observation of a GMP Moiety in the GTase Domain of the Turret Protein VP3.
We found an extra density, which could not be assigned to any side chain or backbone, located in the center of the pocket-like VP3 GTase domain of transcribing CPV. The density map surrounding this extra density fits well with the atomic model (Fig. 4C and Movie S7). Moreover, this density was not observed in the VP3 of nontranscribing CPV. Biochemical data has shown that the GTase domain of the orthoreovirus turret protein functions as an RNA GTase that transfers a GMP moiety to the 5′ end of the 5′-diphosphorylated nascent mRNA (31). This transfer reaction occurs through a covalent intermediate, a phosphoamide bond between the GMP and a lysine of the turret protein (32). This evidence leads us to conclude that the extra density is a GMP moiety. Indeed, a GMP atomic model fits very well to this density (Fig. 4C) and the distance between the GMP structure and an adjacent Lys234 coincides with the length of a phosphoamide bond (Fig. 4D and Movie S7). Lys234 was the only lysine found near the GMP moiety. The shape of the GTase domain is like a pocket and the GMP moiety is located right in its center. We did not find an obvious groove for guiding the mRNA 5′ terminus from the exit of the peripentonal channel to the GTase pocket, but the presence of five copies of the GMP moiety in the five GTase domains of the pentameric turret definitely increases the chances that the mRNA 5′ terminus would fall into any one of the five pockets of the GTase domain to undergo GMP transfer.
Biochemical and mutational analyses revealed that the Lys190 of the orthoreovirus turret protein λ2 is necessary for the generation of the phosphoamide bond, and that Lys171 makes an important contribution to the phosphoamide bond (30). A previous study has shown that Lys190 in the orthoreovirus λ2 protein is in the sequence KDLS, and sequences similar to KDLS are found to be widely conserved among the RNA GTases of the Reoviridae family within the genera Rotavirus, Orbivirus, and Phytoreovirus (30). However, the sequence KILE in CPV VP3 is only partially conserved when compared with the sequence KDLS in orthoreovirus λ2. In addition, the sequence KILE is located in an α-helix in VP3 while the sequence KDLS is located in a loop in λ2 (Fig. 4E and Movie S7). This implies that the location of lysine in the GTase domain that contributes to the phosphoamide bond differs among members of the Reoviridae family. Recently, we have demonstrated that the turret protein VP3 of CPV is essentially structurally conserved with its counterpart proteins in orthoreoviruses despite their lack of significant amino acid sequence identity (10). By superimposing the VP3 of transcribing CPV on orthoreovirus λ2, we found that the two lysines of orthoreovirus λ2 are located right in the vicinity of the GMP moiety in the CPV GTase domain (Fig. 4E).
A previous study by Qiu and Luongo has shown that protonation of histidines 223 and 232 in the orthoreovirus GTase domain is necessary for GTase activity (33). These two histidines are also conserved in aquareovirus (33). CPV VP3 lacks detectable sequence identity with orthoreovirus λ2; however, a tertiary-structure-based amino acid sequence alignment between CPV VP3 and orthoreovirus λ2 (10) showed that the two histidines are actually conserved in these viruses (Fig. S5). In our CPV GTase domain, the corresponding two histidines (His 208 and 217) are located adjacent to the GMP moiety (Fig. S6). The GTase domain in our CPV VP3 is rigid, contradicting the hypothesis that histidine protonation causes the loop connecting the two histidines to move (33). Based on the relative positions of the GMP moiety and the two histidines in our CPV structure, a possible mechanism proposed by Qiu and Luongo, which could result in increased GTase activity is that the two solvent-exposed positive charges resulting from protonation of histidines 208 and 217 in CPV VP3 (histidines 223 and 232 in orthoreovirus λ2) may increase the affinity of GTP for GTase by neutralizing the negative charge of the GTP phosphate groups (33).
These results further confirm that the mechanism of GTase activity is conserved among the Reoviridae family. The diversity of the sequence and the conservation of structural topology and key residues among capsid proteins of viruses in the Reoviridae family may reflect the selection pressure that the viruses have been facing during their long evolution.
Materials and Methods
CPV Purification and mRNA Transcription Assay.
CPV particles were isolated and purified as previously described (10, 34), and transcription was assayed according to a previously published method (35). In order to confirm whether CPV transcription had occurred, we used [α-32P] UTP in the transcription reaction mixture. The transcription reaction mixture (50 μL) consisted of 70 mM Tris-Ac (pH 8), 10 mM MgAc2, 100 mM NaAc, 4 mM ATP, 2 mM GTP, 2 mM CTP, 2 mM UTP, 20 μCi [α-32P] UTP (specific activity 3,000 Ci/mmol), 1 mM SAM, 1 U/μL RNase inhibitor and purified CPV suspension. The transcription reaction mixture was incubated at 31 °C for 3 h. The reaction was terminated by chilling the sample to 0 °C, a condition under which RNA synthesis is discontinued. Radioactive mRNA transcripts were then detected by polyacrylamide gel electrophoresis and storage phosphor technology as described below.
Detection of mRNA Transcripts.
32P-labeled CPV mRNA transcripts in the transcription reaction mixture were separated by 4% polyacrylamide gel electrophoresis. The 32P-labeled mRNA transcripts were visualized by storage phosphor technology using a Typhoon Trio+ Variable Mode Imager (Amersham Biosciences Inc).
Cryo-EM Imaging, 3D Reconstruction and Structure Analysis.
An aliquot of 3.5 μL transcription reaction mixture (70 mM Tris-Ac (pH 8), 10 mM MgAc2, 100 mM NaAc, 4 mM ATP, 2 mM GTP, 2 mM CTP, 2 mM UTP, 1 mM SAM, 1 U/μL RNase inhibitor and purified CPV suspension) was applied to a holey grid and blotted for 4 s in a chamber at 100% humidity using an FEI Vitrobot Mark IV. Viruses were imaged with an FEI 300 kV Titan Krios cryoelectron microscope equipped with a Gatan UltraScan4000 (model 895) 16-megapixel CCD. Viruses were imaged at 300 kV at an absolute magnification of 125,390×, corresponding to a pixel size of 1.19 Å. The dose for each micrograph was about 20–25 e-/Å2. The microscope was carefully aligned before image collection so that a maximum visible contrast transfer function ring of a dry portion of the carbon film image (20–25 e-/Å2 dose) at approximately 1/3 Å-1 spatial frequency was observed. The defocus values of all micrographs were set around 1.5–3.0 μm. The contrast transfer function correction for each micrograph was determined using the CTFIT program from the EMAN package (36) based on incoherently averaged Fourier transforms of each image. The orientations and centers of all virus particles were determined using the IMIRS package (37) based on the common-line strategy (38). About 8,400 selected particles (80% of all particles) were combined and reconstructed using a reconstruction program based on icosahedral symmetry-adapted functions (39). Protein subunit densities were segmented from the maps and visualized using UCSF Chimera (40).
Backbone Model Building.
The models of nontranscribing CPV VP3, VP1A and B were used as initial models. They were modified based on the density map of transcribing CPV using Coot (41). The models were then refined in a pseudocrystallographic manner using the Crystallography and NMR System (42), by a process that included simulated annealing refinement, crystallographic conjugate gradient minimization, and REFMAC5 idealization (43).
Supplementary Material
Acknowledgments.
We thank Professor Jingqiang Zhang and Professor Jingchen Sun for providing CPV polyhedra, Dr. Joy Fleming for help with manuscript editing and critical discussion, and Dr. Xiaoxing Huang and Dr. Yanxia Jia for assisting with electron microscopy and discussion on virus transcription. This research was supported by the National Basic Research Program of China (2010CB912403, 2009CB825503), the National Natural Science Foundation of China (31170697, 31170706, 31070663, 31000333, U0832604, 90919041), and the Scientific Foundation of Education Department of Hunan Province, China (11A124).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron density maps and atomic models have been deposited in the EBI, www.ebi.ac.uk (accession no. EMD-5376), and Protein Data Bank, www.pdb.org (PDB ID code 3J17).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200206109/-/DCSupplemental.
References
- 1.Mertens P. The dsRNA viruses. Virus Res. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Li ZL, Baker ML, Jiang W, Estes MK, Prasad BVV. Rotavirus architecture at subnanometer resolution. J Virol. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, et al. Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins. Proc Natl Acad Sci USA. 2010;107:6292–6297. doi: 10.1073/pnas.0913403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 angstrom. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng LP, et al. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazaki N, et al. Structural evolution of reoviridae revealed by oryzavirus in acquiring the second capsid shell. J Virol. 2008;82:11344–11353. doi: 10.1128/JVI.02375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan XD, et al. Virion structure of baboon reovirus, a fusogenic orthoreovirus that lacks an adhesion fiber. J Virol. 2011;85:7483–7495. doi: 10.1128/JVI.00729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses 2009. Arch Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClain B, Settembre E, Temple BRS, Bellamy AR, Harrison SC. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 angstrom resolution. J Mol Biol. 2010;397:587–599. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng L, et al. Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping. Proc Natl Acad Sci USA. 2011;108:1373–1378. doi: 10.1073/pnas.1014995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology. 2005;343:25–35. doi: 10.1016/j.virol.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mertens PPC, Diprose J. The bluetongue virus core: A nano-scale transcription machine. Virus Res. 2004;101:29–43. doi: 10.1016/j.virusres.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Huismans H, Vandijk AA, Els HJ. Uncoating of parental bluetongue virus to core and subcore particles in infected L-Cells. Virology. 1987;157:180–188. doi: 10.1016/0042-6822(87)90327-8. [DOI] [PubMed] [Google Scholar]
- 14.Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. Structure of the reovirus membrane-penetration protein, mu 1, in a complex with its protector protein, sigma 3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nibert ML, Baker TS. CPV, a stable and symmetrical machine for mRNA synthesis. Structure. 2003;11:605–607. doi: 10.1016/s0969-2126(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 16.Reinisch KM, Nibert M, Harrison SC. Structure of the reovirus core at 3.6 angstrom resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 17.Lawton JA, Estes MK, Prasad BVV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 18.Mendez II, Weiner SG, She YM, Yeager M, Coombs KM. Conformational changes accompany activation of reovirus RNA-dependent RNA transcription. J Struct Biol. 2008;162:277–289. doi: 10.1016/j.jsb.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartlett NM, Gillies SC, Bullivan S, Bellamy AR. Electron-microscopy study of reovirus reaction cores. J Virol. 1974;14:315–326. doi: 10.1128/jvi.14.2.315-326.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Visualization of protein-RNA interactions in cytoplasmic polyhedrosis virus. J Virol. 1999;73:1624–1629. doi: 10.1128/jvi.73.2.1624-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng LP, Fang Q, Shah S, Atanasov IC, Zhou ZH. Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol. 2008;382:213–222. doi: 10.1016/j.jmb.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu XK, Ge P, Jiang JS, Atanasov I, Zhou ZH. Atomic model of CPV reveals the mechanism used by this single-shelled virus to economically carry out functions conserved in multishelled reoviruses. Structure. 2011;19:652–661. doi: 10.1016/j.str.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 24.Prasad BVV, et al. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 25.Xia Q, Jakana J, Zhang JQ, Zhou ZH. Structural comparisons of empty and full cytoplasmic polyhedrosis virus—protein-RNA interactions and implications for endogenous RNA transcription mechanism. J Biol Chem. 2003;278:1094–1100. doi: 10.1074/jbc.M205964200. [DOI] [PubMed] [Google Scholar]
- 26.Tao YZ, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage—structural studies of reovirus polymerase lambda 3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 27.Gouet P, et al. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 28.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struc Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Luongo CL, Contreras CM, Farsetta DL, Nibert ML. Binding site for S-adenosyl-L-methionine in a central region of mammalian reovirus lambda 2 protein—evidence for activities in mRNA CAP methylation. J Biol Chem. 1998;273:23773–23780. doi: 10.1074/jbc.273.37.23773. [DOI] [PubMed] [Google Scholar]
- 30.Luongo CL, Reinisch KM, Harrison SC, Nibert ML. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein lambda 2. J Biol Chem. 2000;275:2804–2810. doi: 10.1074/jbc.275.4.2804. [DOI] [PubMed] [Google Scholar]
- 31.Cleveland DR, Zarbl H, Millward S. Reovirus guanylyltransferase is L2-gene product lambda-2. J Virol. 1986;60:307–311. doi: 10.1128/jvi.60.1.307-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fausnaugh J, Shatkin AJ. Active site localization in a viral mRNA capping enzyme. J Biol Chem. 1990;265:7669–7672. [PubMed] [Google Scholar]
- 33.Qiu T, Luongo CL. Identification of two histidines necessary for reovirus mRNA guanylyltransferase activity. Virology. 2003;316:313–324. doi: 10.1016/j.virol.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 34.Zhou ZH, Zhang H, Jakana J, Lu XY, Zhang JQ. Cytoplasmic polyhedrosis virus structure at 8 angstrom by electron cryomicroscopy: Structural basis of capsid stability and mRNA processing regulation. Structure. 2003;11:651–663. doi: 10.1016/s0969-2126(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 35.Smith RE, Furuichi Y. Gene-mapping of cytoplasmic polyhedrosis-virus of silkworm by the full-length messenger-RNA prepared under optimized conditions of transcription invitro. Virology. 1980;103:279–290. doi: 10.1016/0042-6822(80)90187-7. [DOI] [PubMed] [Google Scholar]
- 36.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 37.Liang YY, Ke EY, Zhou ZH. IMIRS: A high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 38.Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 39.Liu HR, et al. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J Struct Biol. 2008;161:64–73. doi: 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Pettersen EF, et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D . 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.