Abstract
Polyamines (PAs) are ubiquitous, polycationic compounds that are essential for the growth and survival of all organisms. Although the PA-uptake system plays a key role in mammalian cancer and in plant survival, the underlying molecular mechanisms are not well understood. Here, we identified an Arabidopsis L-type amino acid transporter (LAT) family transporter, named RMV1 (resistant to methyl viologen 1), responsible for uptake of PA and its analog paraquat (PQ). The natural variation in PQ tolerance was determined in 22 Arabidopsis thaliana accessions based on the polymorphic variation of RMV1. An RMV1-GFP fusion protein localized to the plasma membrane in transformed cells. The Arabidopsis rmv1 mutant was highly resistant to PQ because of the reduction of PQ uptake activity. Uptake studies indicated that RMV1 mediates proton gradient-driven PQ transport. RMV1 overexpressing plants were hypersensitive to PA and PQ and showed elevated PA/PQ uptake activity, supporting the notion that PQ enters plant cells via a carrier system that inherently functions in PA transport. Furthermore, we demonstrated that polymorphic variation in RMV1 controls PA/PQ uptake activity. Our identification of a molecular entity for PA/PQ uptake and sensitivity provides an important clue for our understanding of the mechanism and biological significance of PA uptake.
Polyamines (PAs) are widely distributed in all living organisms and are involved in various cellular processes, including transcription, RNA modification, translation, membrane stabilization, and the modulation of cell signaling (1–4). They also control the activities of various types of cation channels (5–9). Because of the multiple functions of PAs, their content must be carefully controlled for the maintenance of cellular homeostasis. The intracellular levels of PAs are tightly regulated through the coordination of PA biosynthesis, catabolism, conjugation, and transport at the cell surface (3, 10). Despite extensive studies on PA metabolism, the PA transport system is largely unknown in eukaryotes (10, 11). Nevertheless, the significance of PA uptake has been demonstrated in several studies. In mammalian cancer cells, inhibition of PA uptake enhances the antitumor effects of anticancer agents such as α-difluoromethylornithine (DFMO) (10, 12), and in plants, exogenous PA application can ameliorate the effects of environmental stresses (2, 11, 13). The PA analog methyl viologen (MV), which is commonly known as paraquat (PQ) (Fig. S1), functions as an oxidative stress inducer and is one of the most widely used herbicides, despite the fact that its ingestion can cause death by multiorgan failure (14, 15). Data showing that PA and PQ (PA/PQ) displayed similar uptake characteristics in animal and plant systems suggested that PQ uptake is mediated by PA transporters (16, 17), although little is known about the molecular components responsible for the influx of PA/PQ. Here, we identified an Arabidopsis L-type amino acid transporter (LAT) family transporter named RMV1 (resistant to methyl viologen 1) responsible for the uptake of PA and PQ by analyzing the natural variation of PQ tolerance in Arabidopsis accessions.
Results and Discussion
Natural Variation in PQ Tolerance in Arabidopsis Accessions.
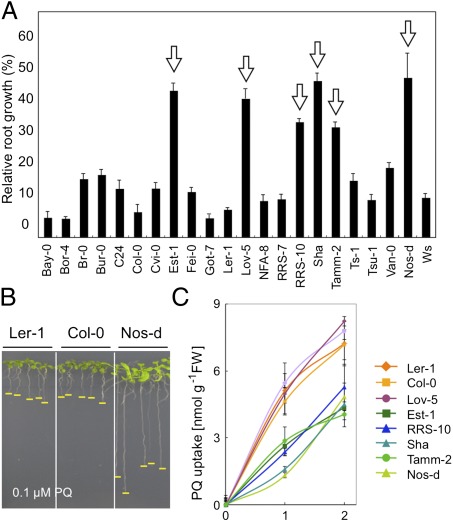
To identify the loci responsible for the natural variation in PA/PQ transport, Arabidopsis accessions showing alterations in PA/PQ tolerance were screened for primary root growth in PA- or PQ-containing media. Whereas none of the accessions examined displayed PA tolerance, several accessions, such as Nos-d, Est-1, Lov-5, RRS-10, Sha, and Tamm-2, showed significant tolerance to PQ toxicity (Fig. 1 A and B). Nos-d was more tolerant to diquat, a bipyridinium-derived PQ analog, than Col-0, whereas there was no apparent difference in tolerance to the other oxidative stress inducers, including hydroxyperoxide and Rose Bengal (Fig. S2). This suggested that the tolerance may be related to PQ structure and possibly occurs at the level of PQ uptake, rather than an oxidative stress response. Consistent with this hypothesis, the PQ uptake rate in sensitive accessions was higher than in tolerant accessions, except for accession Lov-5 (Fig. 1C).
Fig. 1.
Natural variation of PQ tolerance in Arabidopsis accessions. (A) Relative root growth of 22 accessions grown on MS medium containing 0.1 μM PQ. Arrows denote accessions that were resistant to PQ. Data represent median values (n = 15) for each accession. Error bars represent SD. (B) Representative sensitive (Ler-1 and Col-0) and tolerant (Nos-d) accessions grown in 1 μM PQ. Yellow lines indicate root tips. (C) PQ uptake in nine selected accessions. Error bars represent SD (n = 3).
Mapping of PQ (MV)-Resistant Genes.
To understand the genetic basis of PQ tolerance, Col-0 to Nos-d plants were crossed. The PQ sensitivity of the resultant F1 plants was intermediate to that of the parent accessions, implying that PQ tolerance is semidominant (Fig. S3A). Bulked segregant analysis (BSA) (18) of Col-0 × Nos-d or Ler-1 × Nos-d F2 plants indicated that the trait is mostly regulated by a single locus situated on the 1.4 Mb upper arm region of chromosome 5 (Fig. S3 B–D). Further BSA analyses confirmed that in all PQ-tolerant accessions, except for Lov-5, this locus mapped to the same region as in Nos-d (Fig. S4). The locus in Nos-d was therefore named resistance to methyl viologen 1 (RMV1), which is the trade name for PQ.
Map-Based Cloning and Association Analysis of the RMV1 Gene.
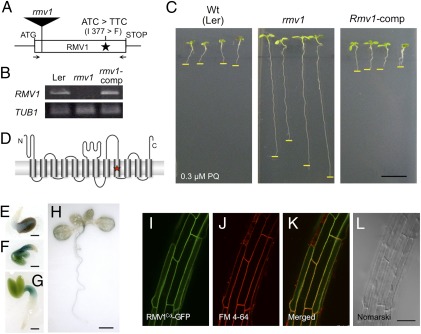
Fine mapping using 883 F2 plants narrowed down the candidate region of RMV1 to 21.1 kb of chromosome 5 (Fig. S5A). This region, which is annotated to contain nine protein-coding genes, showed a significant association between two single nucleotide polymorphisms (SNPs) in two genes (At5g05600 and At5g05630) and PQ tolerance (P < 0.0005) (Fig. S5B). To determine the gene responsible for PQ tolerance, complementation analysis was performed by reciprocal introduction of the genomic regions that contained the promoters and coding regions of these candidate genes from Col-0 and Nos-d into Nos-d and Col-0, respectively. The PQ tolerance of transgenic Nos-d was abolished by the introduction of the Col-0 At5g05630 gene, which encodes a putative amino acid permease protein, suggesting that this gene is responsible for PQ resistance (Fig. S5C). Sequence analysis of the At5g05630 gene in all accessions examined revealed that a single base substitution causing an amino acid change from isoleucine 377 to phenylalanine is linked to the loss of PQ sensitivity (Fig. 2A and Fig. S5D). To further analyze the significance of At5g05630, we obtained a T-DNA insertion mutant (designated as rmv1; Fig. 2A) and identified homozygous knockout plants with no detectable At5g05630 transcripts (Fig. 2B). The rmv1 mutant displayed higher PQ tolerance than the corresponding wild-type Ler-1, Col-0, and Nos-d lines (Fig. 2C and Fig. S6), whereas the PQ sensitivity of transgenic rmv1 plants carrying a genomic region of At5g05630 from Col-0 (designated as the complemented line, rmv1-comp) recovered to a level of PQ tolerance that was comparable to that of Col-0 (Fig. 2C). These results confirmed that At5g05630 identifies RMV1 and that the PQ tolerance phenotype of Nos-d is caused by a missense mutation at RMV1.
Fig. 2.
Characterization of RMV1. (A) Schematic representation of the structure of RMV1. The position of nucleotides and amino acid residues that differ between Ler-1 and Nos-d is indicated. The position of the T-DNA insertion of rmv1 (GT_3_3436) is shown as a filled rectangle. Arrows denote primers used for RT-PCR. (B) RMV1 transcript levels in wild-type (Ler-1), rmv1, and complementation transgenic lines (rmv1-comp). RMV1 transcripts were detected by RT-PCR using total RNA extracted from 2-d-old seedlings. (C) PQ sensitivity of the wild-type (Ler-1), rmv1, and complementation transgenic lines (rmv1-comp). (D) Schematic view of the topology of RMV1, as predicted by ConPred II (34). Transmembrane domains are indicated as cylinders. The star denotes the position of amino acid residues that differ between the RMV1 proteins derived from Ler-1 and Nos-d. (E–H) RMV1 promoter-GUS reporter gene expression in transgenic plants (E–H: 1, 2, 3, and 10 d after germination, respectively). (I–L) Subcellular localization of the RMV1c-GFP fusion protein. Roots of Arabidopsis stably transformed with RMV1Col-GFP and stained with FM4-64. (I) GFP filter. (J) FM4-64 filter. (K) merged. (L) Nomarski images. [Scale bars: 1 cm (C); 1 mm (E–G); 5 mm (H); 50 μm (L).]
RMV1 consists of one exon (Fig. 2A) with an ORF of 1,470 bp encoding a polypeptide of 490 amino acids, which is predicted to contain 12 transmembrane domains (TMs) (Fig. 2D and Fig. S7A). The amino acid substitution between RMV1Ler and RMV1Nos is located within the ninth TM (Fig. 2D). RMV1 is a putative amino acid permease that belongs to the LAT family within the amino acid, polyamine, and organic cation (APC) superfamily, which is conserved in eukaryotes (19). In Arabidopsis, five members of the LAT family are annotated (Fig. S7 A and B), among which the isoleucine 377 of RMV1 is not a conserved amino acid (Fig. S7A).
RMV1 Gene Expression Pattern and RMV1 Protein Plasma Membrane Localization.
RMV1 expression was examined in transgenic plants expressing β-glucuronidase (GUS) under the control of the 2.1 kb upstream region of RMV1-coding sequences. Transgenic plants showed strong GUS activity in the hypocotyls and petioles of cotyledons in 1- to 3-d-old seedlings (Fig. 2 E–G), and weak GUS activity in the roots and veins of leaves in 10-d-old-seedlings (Fig. 2H). PQ treatment did not affect GUS activity. The subcellular localization of GFP-tagged RMV1 was investigated by generating transgenic Arabidopsis plants (Col-0) expressing Col-0-, Nos-d-, or Ler-1-type RMV1-GFP under the control of the constitutive cauliflower mosaic virus 35S RNA (CaMV35S) promoter (Fig. S8). In these transgenic plants, the GFP signals overlapped with the FM4-64 signal, which localizes to the plasma membrane under the conditions used (Fig. 2 I–L and Fig. S8 A and B). Moreover, in onion epidermal cells, RMV1-GFP was detected in the plasma membrane after plasmolysis (Fig. S8 D–H). Together with the prediction that RMV1 has transmembrane domains, these results indicate that RMV1 is localized to the plasma membrane.
RMV1 Mediates PQ Uptake.
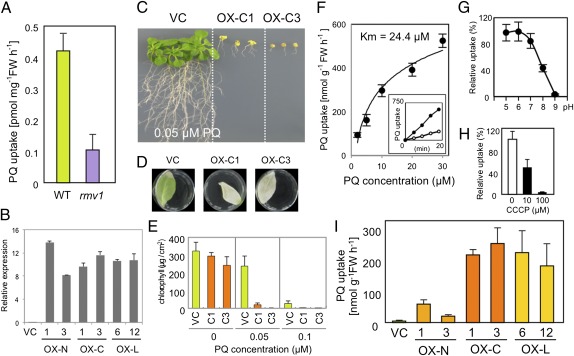
To verify that RMV1 has a role in PQ uptake, an rmv1 knockout mutant was analyzed. [14C] PQ uptake was reduced in the rmv1 mutant compared with wild-type (Ler-1) plants, suggesting that RMV1 is required for PQ uptake (Fig. 3A). To further examine the transport activity of RMV1, three types of transgenic Arabidopsis (Col-0) plants were generated expressing Nos-d-, Col-0-, or Ler-1-type RMV1 variants (designated as OX-N, OX-C, or OX-L, respectively) under the CaMV35S promoter (Fig. S9). These transgenic plants showed similar expression levels of the transgenes, as demonstrated by quantitative RT-PCR (Fig. 3B). Under normal growth conditions, there were no morphological differences between the transgenic plants and vector control plants (Fig. S9). However, when grown in a PQ-containing medium, the OX-C plants showed severe growth defects, indicating hypersensitivity to PQ (Fig. 3C). Likewise, the RMV1Col-GFP-overexpressing plants showed PQ hypersensitivity (Fig. S8C), implying that RMV1-GFP fusion proteins are functional. Furthermore, OX-C transgenic lines also displayed PQ hypersensitivity in leaves (Fig. 3 C–E), suggesting that RMV1 or the RMV1-like transporter, or both, mediates PQ entry into leaf cells as well as into root cells. Kinetic analysis further indicated that RMV1 is capable of mediating PQ transport against a concentration gradient (Fig. 3F), as shown by the Km value (24.4 μM) for PQ uptake in RMV1-overexpressing plants, which was similar to the Km for PQ influx in wild-type maize roots (17). Moreover, the PQ uptake in the OX-C transgenic plants showed pH dependency (Fig. 3G), and the reduction of the proton gradient by the protonophore carbonyl cyanidem-chlorophenylhydrazone (CCCP) significantly abolished PQ uptake in the OX-C transgenic plants (Fig. 3H). Taken together, these results suggest that RMV1-mediated PQ uptake is dependent on proton cotransport. Consistently, CCCP-dependent inhibition of PQ uptake was also observed in wild-type (Ler-1) plants (Fig. S10A).
Fig. 3.
PQ transport by RMV1. (A) PQ uptake of the wild-type (Ler-1) and rmv1 mutant. Error bars represent SD (n = 3). (B) RMV1 transcript levels of vector control (VC) and RMV1 transgenic lines (OX-N, -C, or -L). The RMV1 transcripts were detected by real-time quantitative RT-PCR using total RNA extracted from 10-d-old seedlings. (C–E) PQ sensitivity of transgenic Arabidopsis (Col-0) plants expressing Col-type RMV1. (C) Root growth on MS plates containing 0.05 μM PQ. (D) Fully expanded leaves of transgenic plants were floated on buffer containing 3 mM MES, 0.1% Tween-20, and 0.1 mM PQ for 2 d. (E) Chlorophyll contents of leaves floated on the indicated concentrations of PQ. Error bars represent SD (n = 5). (F) Concentration-dependent kinetics of PQ influx into roots of OX-C1 plants. Seedlings were incubated with increasing concentrations of PQ for 15 min before determination of intracellular radioactivity. Inset shows a time course of PQ uptake with 2 μM (open circles) or 30 μM (closed circles) PQ. The Km value was determined from the Hanes–Woolf plot. Uptake data were fitted to Michaelis–Menten and Hanes–Woolf equations. (G) pH dependence of PQ transport in OX-C1 plants. Seedlings were incubated in MS medium at indicated pH values containing 10 μM PQ for 15 min. (H) The effect of CCCP on PQ uptake activity of OX-C plants. Seedlings were incubated in MS medium containing 10 μM PQ with indicated concentrations of CCCP for 15 min. (I) PQ uptake of overexpressing lines. Error bars represent SD (n = 3).
PQ Uptake Activity and Tolerance Is Controlled by Polymorphic Variation in RMV1.
Analysis of transgenic plants with similar levels of transgene expression revealed that the OX-N transgenic lines exhibited lower uptake activity than OX-C or OX-L (Fig. 3H), which could explain the difference in PQ sensitivity of Nos-d, Col-0, and Ler-1 lines (Fig. 1B). Recently, the crystal structure of the bacterial amino acid antiporter AdiC, which belongs to the APC superfamily, was reported (20–22). AdiC shares 20% amino acid identity with RMV1 (Fig. S7A). Structural analysis of the AdiC variant bound to Arg, a substrate of AdiC, mapped the substrate-binding sites to TM3, 6, 8, and 10 (22) (Fig. S7A). The ninth TM, which has an amino acid substitution between RMV1Ler and RMV1Nos, is predicted to locate to the molecular surface (20–22). Therefore, the substitution at position 377 may have indirect effects such as alterations in molecular stability rather than affecting the transport activity itself.
RMV1 Participates in PA Uptake.
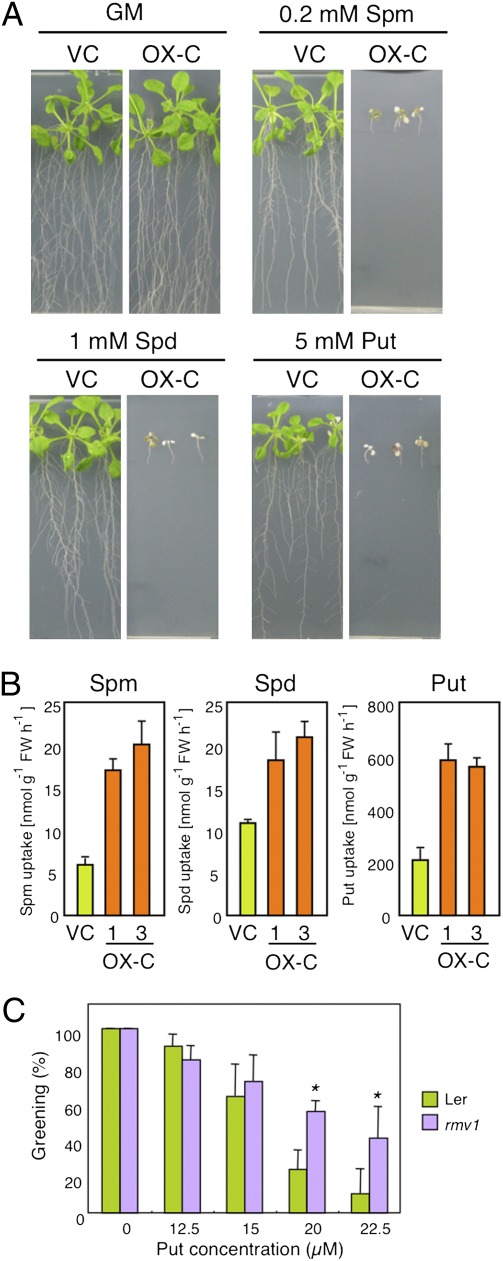
The PA transport activity of RMV1 was examined using OX-C and control transgenic plants. OX-C transgenic lines were significantly more sensitive to PAs such as spermine, spermidine, and putrescine (Fig. 4A), and exhibited higher PA uptake activity than the control lines, as demonstrated with 14C-labeled PAs and HPLC (Fig. 4B and Fig. S10 B and C). RMV1 showed high affinity for spermine (Km = 0.6 μM) and spermidine (Km = 2.2 μM), and lower affinity for putrescine (Km = 56.5 μM) (Fig. S10D). Although there was no significant difference in PA uptake between the rmv1 knockout mutant and the wild type (Ler-1) under the conditions used, possibly attributable to genetic redundancy, the rmv1 knockout mutant showed significantly enhanced tolerance to putrescine in the early stage of seedling development (Fig. 4C). These results support the notion that RMV1 is also involved in PA uptake, which is consistent with previous findings (23, 24) and our observations that the exogenous application of PAs counteract PQ toxicity (Fig. S10 E and F). In fact, the simultaneous application of PA rather than amino acids appeared to inhibit PQ uptake in roots (Fig. S10A).
Fig. 4.
RMV1 is a PA transporter. (A) PA sensitivity of OX-C plants. Four-day-old plants were transferred to MS medium with the indicated concentration of PAs and grown for 6 d. (B) PA uptake of OX-C plants. Put, putrescine; Spm, spermine; Spd, spermidine. Error bars represent SD (n = 3). (C) Putrescine sensitivity of wild-type and rmv1 mutant plants. The wild type (Ler-1) and rmv1 mutant were germinated on MS medium containing the indicated concentrations of Put and grown for 9 d. The average frequencies of expanded green cotyledons are expressed as the average percentage and SD of plants grown on three plates containing 30 seeds each (*P < 0.01).
Collectively, these results provide evidence that the LAT family transporter RMV1 is engaged in the proton-dependent transport of PA and PQ (MV) in Arabidopsis, consistent with the recent report of a rice gene mediating spermidine uptake (25). Moreover, we demonstrated that natural variation in RMV1 defines PQ uptake in A. thaliana. These data support the hypothesis that PQ enters plant cells via a transport system that inherently functions in the transport of PAs, which have structural similarities to PQ (17). Our data imply that other transporters in addition to RMV1 may participate in PA uptake in Arabidopsis, suggesting the existence of diverse transport systems, which might serve to increase the repertoire of PAs that plants can absorb from the environment. Our identification provides insight into the molecular basis of the PA/PQ uptake system in eukaryotes and may contribute to the development of genetically engineered crops, phytoremediation strategies, and anticancer drugs.
Materials and Methods
Plant Materials and Growth Conditions.
The following A. thaliana accessions used in this study were provided by the RIKEN BioResource Center, Japan (http://www.brc.riken.go.jp/lab/epd/Eng/), and Arabidopsis Biological Resource Center (ABRC): Bay-0 (CS22676), Bor-4 (CS22677), Br-0 (CS22678), Bur-0 (CS22679), C24 (CS22680), Col-0 (CS22681), Cvi-0 (CS22682), Est-1 (CS22683), Fei-0 (CS22684), Got-7 (CS22685), Ler-1 (CS22686), Lov-5 (CS22695), NFA-8 (CS22687), RRS-7 (CS22688), RRS-10 (CS22689), Sha (CS22690), Tamm-2 (CS22691), Ts-1 (CS22692), Tsu-1 (CS22693), Van-0 (CS22694), and Ws (CS28823). The Nossen line used here is the parent line of the RIKEN Ds transposon line (CS8522) that contains different polymorphisms from a generally distributed Nossen (No-0) ecotype (26), which was temporary named “Nos-d” in this report. The rmv1 T-DNA insertion mutant (GT_3_3436) was obtained from the ABRC. Plants were grown on agar plates containing half-strength Murashige and Skoog (MS) salt (pH 7.5), a vitamin mixture, 1% sucrose, and 0.8% or 1.2% (wt/vol) agar under long-day conditions (16-h light/8-h dark cycle; 60 ± 10 μmol m−2 s−1) at 22 °C. For the analysis of stress tolerance, 4-d-old plants grown on MS medium were transferred to MS medium containing 1.2% (wt/vol) agar and the indicated concentrations of chemicals and grown vertically for 6 d. Tolerance was analyzed by monitoring the primary root growth of seedlings.
Mapping.
An F2 population derived from a Col × Nos-d cross showed a 1:2:1 (sensitive:intermediate:tolerant) segregation ratio based on the root growth assay (Fig. S3B), suggesting that the PQ-resistant phenotype in Nos-d is controlled either by a single locus or by several closely-linked loci. To obtain a rough map position, a bulked segregant analysis (BSA) experiment was performed in combination with PCR using fluorescently-labeled primers. More details will be published elsewhere. Briefly, genomic DNA from 32 plants with the highest MV tolerance (Fig. S3B) was pooled and used for PCR analysis with fluorescently-labeled primers flanking the simple sequence length polymorphism (SSLP) marker site (Fig. S3D) generated using the INDEL database, Monsanto SNP, and Ler Collections (www.arabidopsis.org/browse/Cereon/index.jsp). The resulting SSLP-containing PCR products were separated by size using a capillary sequencer (ABI PRISM3100 DNA Sequencer, Applied Biosystems) and the amount of PCR product for each genotype was calculated based on the fluorescence intensity. Data are presented as linkage, which represents the ratio of the amount of PCR product from Nos-d/Col-0 or Nos-d/Ler-1 normalized to that of the F1 line from the Nos-d × Col-0 or Nos-d × Ler-1 cross. A strong peak was detected for two SSLP markers located at 1,372,518 bp (CER457340) and 5,386,803 bp (CER456831) in the upper arm region of chromosome 5 (Figs. S2C and S3A), indicating that the PQ-resistant trait is mostly regulated by a single locus between these markers. To further narrow down the map position of the Rmv locus, genotyping of individual F2 plants was performed using SSLP and SNP markers. Analysis of 883 F2 progeny mapped the RMV1 locus to a 26.1 kb region between the newly generated SNP markers at positions 1670281 kb (S8) and 1691394 kb (S9) on chromosome 5 (Fig. S3A).
Association Analysis.
The association analysis was carried out by a general linear model (GLM) function of TASSEL using the haploid setting, as described previously (27). The analysis was carried out based on the Q (population structure), while the principle component (PC) was used to replace Q in TASSEL according to Zhao et al. (28). The PC analysis datasets were created with 250K genome-wide SNPs obtained from the laboratory of Magnus Nordborg (University of Southern California, Los Angeles).
Uptake Assay.
Fourteen-day-old seedlings grown on MS agar plates were transferred to MS liquid medium. After overnight incubation, plants were soaked into 4.5 mL of fresh medium and the reaction was started by the addition of 0.5 mL of MS medium containing 10 μM [14C] PQ (American Radiolabeled Chemicals), 10 μM [14C]Spm, 100 μM [14C]spermidine, or 10 mM [14C]Put (Amersham Biosciences). The total substrate concentration was adjusted by adding unlabeled substrate. For competition analysis, competitors or inhibitors were added 5 min before the start of the reaction. After incubation, seedlings were washed three times with 40 mL of buffer (MS containing a 10-fold concentration of substrates) and roots were excised. Roots from six seedlings were blotted and weighed in bulk and soaked into 2 mL of scintillation fluid (Clear-sol II; Nacalai Tesque), and the radioactivity was determined using a liquid scintillation spectrophotometer (TriCarb2800-TR; Perkin-Elmer). Fifteen-minute uptake periods were used in all concentration-dependent kinetic studies because uptake was linear up to at least 20 min.
Constructs and Real-Time Quantitative RT-PCR Analysis.
For complementation analysis, the At5g05600 or At5g05630 genome regions including the 1.32 or 1.16 kb promoter region upstream of the ATG start codon, respectively, and the 0.84 or 0.55 kb region downstream of the stop codon, respectively, were amplified from Col-0, Ler-1 or Nos-d genomic DNA by PCR with SfiI linker primers (5′-gcttggcctgcttggccGATCCGATTTCTAGAAACTTTCAC-3′ and 5′-aattcggccgcgacggccCTATTCAGGATCGAAGTGGGG-3′ for At5g05600; and 5′-gcttggcctgcttggccTGATGTTTAGTGCAACCCACCC-3′ and 5′-aattcggccgcgacggccGAGAGGGAGTGACGACTTAG-3′ for At5g05630) and cloned into the SfiI site of the binary vector pBIG-CD. The RMV1 promoter-GUS plasmid was constructed by amplifying a 1.99 kb DNA fragment upstream of the RMV1 coding region by PCR with an upstream XbaI linker primer (5′-gcgtctagaACTGTGCTGGATGTTATGAGG-3′) and a downstream EcoRI linker primer (5′-cctgaattcCGAGAATTGGAACTTAAAATAATTG-3′), and cloned into the XbaI/EcoRI site of the pGH-GUS plasmid, a hygromycin-version of pGK-GUS (29). For construction of the 35S-RMV1-GFP, the RMV1 coding region was amplified by PCR with the XbaI linker primers 5′-gcgtctagaATGACTGAGCTTAGCTCTCCG-3′ and 5′-gcgtctagaCTCCATCAGGTTTGGTAGGTG-3′ and cloned into the XbaI site of pGHX-CsGFP, a hygromycin-version of pGHX-CsGFP (29). This construct was introduced into Agrobacterium tumefaciens strain GV3101 and used for plant transformation. To analyze the expression level of the transgene, quantitative RT-PCR analysis was performed as described previously (30) using gene-specific primers, 5′-CACATAAGCATTGGTGTCACCAC-3′ and 5′-TTCTTGATTTGGTTGACGGTTCT-3′.
Histochemical GUS Staining and Analysis of Subcellular Localization.
Histochemical analysis was performed as described previously (31). For transient expression analysis of the GFP fusion protein in onion epidermal cells, the 35S-RMV1-GFP or pGHX-CsGFP plasmid was introduced into onion epidermal cells as described previously (32). After incubation at 22 °C for 16 h, cells were plasmolyzed by incubating onion epidermal strips in a 0.8 M sucrose solution for 10 min. The roots of transgenic plants expressing the RMV1-GFP fusion protein were stained with FM4-64 for 1 min, according to the plasma membrane labeling protocol in the instructions of the manufacturer. Fluorescence was observed in whole mounts using a confocal laser scanning microscope (FV1000, Olympus).
Leaf Floating Test.
Fully expanded third or fourth rosette leaves detached from soil-grown transgenic plants were floated in a buffer containing 3 mM 2-(N-morpholino) ethanesulfonic acid monohydrate (Mes) and 0.05% Tween-20, containing appropriate concentrations of PQ (Sigma) at 22 °C for 24 h under continuous light (130 μmol m−2 s−1). Chlorophyll was extracted overnight at 4 °C with 1 mL of dimethylformamide. Chlorophyll contents (Chl a + b = 17.67A646.8 + 7.12A663.8) were calculated according to Porra et al. (33) and divided by leaf area, which was measured using ImageJ software (National Institutes of Health).
Supplementary Material
Acknowledgments
We thank Dr. Yusuke Saijo for critical reading of this manuscript and Dr. Toru Fujiwara for helpful suggestions. This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121406109/-/DCSupplemental.
References
- 1.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 2.Kusano T, Berberich T, Tateda C, Takahashi Y. Polyamines: Essential factors for growth and survival. Planta. 2008;228:367–381. doi: 10.1007/s00425-008-0772-7. [DOI] [PubMed] [Google Scholar]
- 3.Cohen SS. A Guide to the Polyamines. New York: Oxford University Press; 1998. [Google Scholar]
- 4.Igarashi K, Kashiwagi K. Polyamines: Mysterious modulators of cellular functions. Biochem Biophys Res Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 5.Lopatin AN, Makhina EN, Nichols CG. Potassium channel block by cytoplasmic polyamines as the mechanism of intrinsic rectification. Nature. 1994;372:366–369. doi: 10.1038/372366a0. [DOI] [PubMed] [Google Scholar]
- 6.Gomez M, Hellstrand P. Endogenous polyamines modulate Ca2+ channel activity in guinea-pig intestinal smooth muscle. Pflugers Arch. 1999;438:445–451. doi: 10.1007/s004249900068. [DOI] [PubMed] [Google Scholar]
- 7.Lu Z, Ding L. Blockade of a retinal cGMP-gated channel by polyamines. J Gen Physiol. 1999;113:35–43. doi: 10.1085/jgp.113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CJ, Moczydlowski E. Cytoplasmic polyamines as permeant blockers and modulators of the voltage-gated sodium channel. Biophys J. 2001;80:1262–1279. doi: 10.1016/S0006-3495(01)76102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandolfi C, Pottosin I, Cuin T, Mancuso S, Shabala S. Specificity of polyamine effects on NaCl-induced ion flux kinetics and salt stress amelioration in plants. Plant Cell Physiol. 2010;51:422–434. doi: 10.1093/pcp/pcq007. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi K, Kashiwagi K. Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol Biochem. 2010;48:506–512. doi: 10.1016/j.plaphy.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi T, Kakehi J. Polyamines: Ubiquitous polycations with unique roles in growth and stress responses. Ann Bot. 2010;105:1–6. doi: 10.1093/aob/mcp259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer AJ, Wallace HM. The polyamine transport system as a target for anticancer drug development. Amino Acids. 2010;38:415–422. doi: 10.1007/s00726-009-0400-2. [DOI] [PubMed] [Google Scholar]
- 13.Groppa MD, Benavides MP. Polyamines and abiotic stress: Recent advances. Amino Acids. 2008;34:35–45. doi: 10.1007/s00726-007-0501-8. [DOI] [PubMed] [Google Scholar]
- 14.Bus JS, Aust SD, Gibson JE. Superoxide- and singlet oxygen-catalyzed lipid peroxidation as a possible mechanism for paraquat (methyl viologen) toxicity. Biochem Biophys Res Commun. 1974;58:749–755. doi: 10.1016/s0006-291x(74)80481-x. [DOI] [PubMed] [Google Scholar]
- 15.Winterbourn CC. Production of hydroxyl radicals from paraquat radicals and H2O2. FEBS Lett. 1981;128:339–342. doi: 10.1016/0014-5793(81)80112-3. [DOI] [PubMed] [Google Scholar]
- 16.Chen N, Bowles MR, Pond SM. Competition between paraquat and putrescine for uptake by suspensions of rat alveolar type II cells. Biochem Pharmacol. 1992;44:1029–1036. doi: 10.1016/0006-2952(92)90364-o. [DOI] [PubMed] [Google Scholar]
- 17.Hart JJ, Ditomaso JM, Linscott DL, Kochian LV. Transport Interactions between Paraquat and Polyamines in Roots of Intact Maize Seedlings. Plant Physiol. 1992;99:1400–1405. doi: 10.1104/pp.99.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460:1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, et al. Structure and mechanism of an amino acid antiporter. Science. 2009;324:1565–1568. doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010;463:828–832. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 23.Kurepa J, Smalle J, Van Montagu M, Inzé D. Oxidative stress tolerance and longevity in Arabidopsis: The late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14:759–764. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- 24.Minton KW, Tabor H, Tabor CW. Paraquat toxicity is increased in Escherichia coli defective in the synthesis of polyamines. Proc Natl Acad Sci USA. 1990;87:2851–2855. doi: 10.1073/pnas.87.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulangi V, Phuntumart V, Aouida M, Ramotar D, Morris P. Functional analysis of OsPUT1, a rice polyamine uptake transporter. Planta. 2012;235:1–11. doi: 10.1007/s00425-011-1486-9. [DOI] [PubMed] [Google Scholar]
- 26.Kuromori T, et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- 27.Tazib T, et al. Association mapping of cadmium, copper and hydrogen peroxide tolerance of roots and translocation capacities of cadmium and copper in Arabidopsis thaliana. Physiol Plant. 2009;137:235–248. doi: 10.1111/j.1399-3054.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin F, et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita M, et al. Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem Biophys Res Commun. 2007;364:250–257. doi: 10.1016/j.bbrc.2007.09.124. [DOI] [PubMed] [Google Scholar]
- 31.Fujita Y, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita M, et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 33.Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta. 1989;975:384–394. [Google Scholar]
- 34.Arai M, et al. ConPred II: A consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.