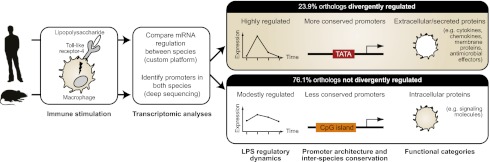
Abstract
Evolutionary change in gene expression is generally considered to be a major driver of phenotypic differences between species. We investigated innate immune diversification by analyzing interspecies differences in the transcriptional responses of primary human and mouse macrophages to the Toll-like receptor (TLR)–4 agonist lipopolysaccharide (LPS). By using a custom platform permitting cross-species interrogation coupled with deep sequencing of mRNA 5′ ends, we identified extensive divergence in LPS-regulated orthologous gene expression between humans and mice (24% of orthologues were identified as “divergently regulated”). We further demonstrate concordant regulation of human-specific LPS target genes in primary pig macrophages. Divergently regulated orthologues were enriched for genes encoding cellular “inputs” such as cell surface receptors (e.g., TLR6, IL-7Rα) and functional “outputs” such as inflammatory cytokines/chemokines (e.g., CCL20, CXCL13). Conversely, intracellular signaling components linking inputs to outputs were typically concordantly regulated. Functional consequences of divergent gene regulation were confirmed by showing LPS pretreatment boosts subsequent TLR6 responses in mouse but not human macrophages, in keeping with mouse-specific TLR6 induction. Divergently regulated genes were associated with a large dynamic range of gene expression, and specific promoter architectural features (TATA box enrichment, CpG island depletion). Surprisingly, regulatory divergence was also associated with enhanced interspecies promoter conservation. Thus, the genes controlled by complex, highly conserved promoters that facilitate dynamic regulation are also the most susceptible to evolutionary change.
Keywords: evolution, inflammation, innate immunity, pattern-recognition receptor, transcriptional regulation
Rodents are widely used as models for humans in experimental pathology. The assumption that mice provide a reliable model for understanding and treating human disease is a particular concern for immunological studies, as humans and mice diverged 65 to 75 Mya (1), and pathogens evolve quickly and exert strong pressure on the immune system to coevolve. Accordingly, many aspects of innate and acquired immunity are different between mouse and human (reviewed in ref. 2). High-profile commentaries have highlighted limitations of mice for immunological studies and emphasized the urgent need to identify critical points of divergence between the species (3, 4).
Macrophages and other cells of the innate immune system recognize distinct microbial products through families of pattern recognition receptors such as the Toll-like receptors (TLRs) (5). Although the TLR family exhibits divergence in expression, sequence, and gene number among species, the basic biology and downstream signaling is generally thought to be conserved. The most widely studied pattern recognition receptor is TLR4, which recognizes bacterial lipopolysaccharide (LPS) from Gram-negative bacteria, as well as several host-derived danger signals. TLR4 is essential for host resistance to a range of bacterial infections, but dysregulated responses through this receptor lead to systemic inflammation (e.g., septic shock). Some species differences in the transcriptional response to inflammatory stimuli such as LPS are already appreciated. For example, the inducible nitric oxide synthase (iNOS) gene, the product of which generates the antimicrobial agent nitric oxide, is robustly induced by IFN-γ and LPS in macrophages from mice, but not humans (6) or several other mammalian species (7). Conversely, the gene encoding the anti-microbial peptide cathelicidin is TLR-inducible via a vitamin D-dependent pathway in human but not mouse macrophages (8). Moreover, mice are relatively resistant to LPS-mediated toxicity (9), and many therapeutic agents that reduce inflammation and mortality in mouse septic shock models show no benefit clinically (10) or even increase mortality (e.g., nitric oxide antagonists) (11).
Multiple mechanisms can contribute to phenotypic differences between species. These include the evolution of nonorthologous genes, functional evolution of orthologous genes (e.g., changes in amino acid sequence affecting function), and evolution in transcriptional regulation of orthologous genes. Extensive gene expression divergence between even closely related species clearly exists (12–18), but our understanding of how gene regulation evolves, and the impact of this on phenotypic diversity is limited. A number of mechanisms could contribute to expression divergence between species, including divergence in cis (e.g., promoter/enhancer sequence change) or in trans (e.g., altered transcription factor expression, activation or motif specificity, chromatin remodeling, or cellular environment). As with protein coding sequence, gene regulatory sequences evolve as a consequence of mutation and the subsequent action of genetic drift and natural selection. Adaptive evolution is particularly prevalent in immune response genes as a result of sustained selective pressure exerted by rapid pathogen evolution (19–21).
In the present study, we developed a custom platform that permits interspecies gene expression comparison and, in parallel, performed genome-wide transcription start site (TSS) identification by deeply sequencing mRNA 5′ ends on matched samples from human and mouse. This study represents the most comprehensive and detailed study to date of a gene expression time course, measured in parallel and anchored to experimentally defined promoters, in two mammalian species. In so doing, we systematically identified conserved and divergent TLR4-regulated gene expression, with demonstrated biological consequences, and defined promoter properties that are associated with evolutionary divergence in innate immune genes.
Results
LPS-Regulated mRNA Regulation in Human and Mouse Macrophages.
Interspecies comparisons that use whole-genome arrays exaggerate transcriptional divergence because of low transcript coverage and poor internal replication. We therefore undertook a rigorous, two-step approach. We first interrogated whole-genome microarrays (Illumina; Affymetrix) to identify transcripts significantly regulated over a time course of LPS stimulation in mouse or human macrophages. LPS-regulated genes identified in this screen were then interrogated using custom-designed mouse and human arrays. These afforded a high level of internal replication, comprehensive coverage of transcript variants, and superior probe design to commercially available whole-genome arrays (Table S1).
We chose 2 h LPS stimulation as representative of direct response genes (validated for the early response gene, TNF), 6 h as representative of the peak of inflammatory gene responses, and 24 h to sample the subsequent “resolution” phase during which feedback control is exerted (22, 23). To minimize differences caused by cellular environment, we compared primary macrophage populations derived under very similar conditions ex vivo: human monocyte-derived macrophages (HMDMs) and mouse bone marrow-derived macrophages (BMMs). Both are differentiated ex vivo with colony-stimulating factor (CSF)-1 for 7 d. We also included a second mouse macrophage population, thioglycollate-elicited peritoneal macrophages (TEPMs), that are derived from mouse monocytes in vivo and are commonly used as a cellular model of inflammatory macrophages. Our choice of human and mouse macrophages reflects the most widely used primary macrophage populations in studies of innate immunity.
For one-to-one orthologues, we adopted a stringent approach to capturing interspecies divergence by selecting a “representative profile” for each gene (summarized in Fig. S1; see SI Glossary of Technical Terms for definitions of all technical terms). This is defined as the most similar expression profile between HMDM and either mouse macrophage population, for any transcript (or set of transcripts, wherein individual probes detect multiple transcripts) emanating from that gene. Probe, transcript, and representative profile data and statistical analyses are available from the Macrophage Comparative Expression Gateway (MacGate, http://www.macgate.qfab.org), and summary data are available in Dataset S1A. Although our study focused on conservation and divergence in LPS regulation between human and mouse genes displaying one-to-one orthology relationships, we also analyzed coregulation between genes with expanded orthology relationships (e.g., one-to-many, many-to-many) and found regulatory divergence between and within species was common, reflecting the trend for paralogous genes to evolve specialized functions (Fig. S2).
Conservation in LPS-Regulated Gene Expression in Human and Mouse Macrophages.
The relative importance of selection vs. random genetic drift in shaping gene expression patterns is controversial (24). Under a simple model of neutral drift, we would expect extensive gene expression divergence between species, approaching saturation (24). In fact, gene regulation over the time course was strikingly similar between the human (i.e., HMDM) and both mouse (i.e., BMM, TEPM) macrophage populations (Fig. 1A). Permutation testing of summed normalized expression difference (SDiff, the sum of absolute differences between orthologous expression profile values over all time points; adapted from ref. 25) revealed that this similarity was significantly greater than would be expected by chance (P < 0.05) for 30.30% (759 of 2,505) regulated gene pairs. These results are consistent with the prevailing action of purifying selection constraining regulatory divergence. The overall similarities in the LPS-regulated mRNA expression profiles of the human and mouse macrophages support the view that the experimental systems are broadly comparable and that the transcription milieu triggered by LPS is also broadly conserved.
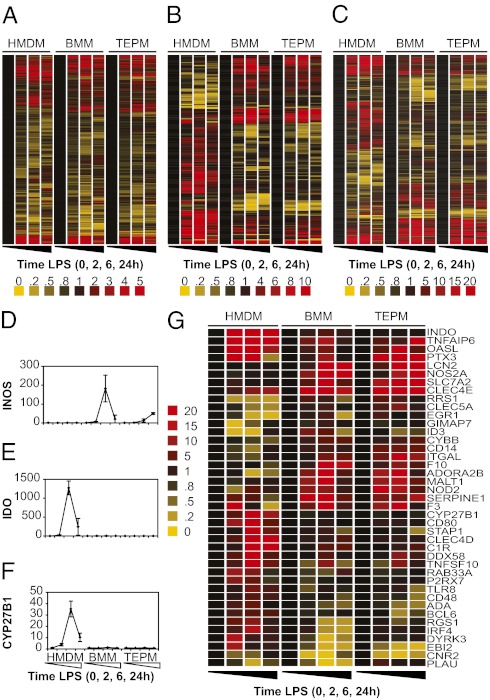
Fig. 1.
Global overview of LPS regulation between human and mouse macrophage populations. Clustered representative profiles for (A) all one-to-one orthologues significantly regulated by LPS in any macrophage population, (B) all profile-divergent orthologues, and (C) all time point-divergent orthologues. Known examples of expression divergence between the species were confirmed, including (D) INOS (P = 1.41 × 10−7), (E) IDO (P = 1.46 × 10−24), and (F) CYP27B1 (P = 2.31 × 10−7; microarray data average fold induction ± SEM, n = 3–4, significance assessed by ANOVA). (G) Hierarchical clustering of profile-divergent orthologues involved in immune defense or antimicrobial pathways. Genes encoding cytokine, chemokine, and growth factor ligands and receptors, as well as matrix metalloproteinases, were subtracted from this list as they are considered separately in Fig. 3A. Gene fold induction is indicated by the color-code key.
Identification of Interspecies Divergence Between Primary Human and Mouse Macrophages.
Given the overall conservation in LPS responses, regulatory divergence should be viewed against a background of overall similarity. By using the stringent approach described earlier, representative profiles for 186 (7.4%) LPS-regulated genes, whose profile in HMDM was significantly different (P < 0.05, two-way ANOVA with variables of sample and time) to both mouse populations (i.e., BMM and TEPM), were classified as differentially regulated across the LPS time course (“profile divergent”; Fig. 1B and Fig. S1). To capture more subtle differences in gene regulation between the species, we also performed t tests to compare regulation at each time point (“time-point divergent”; Fig. 1C). A total of 23.9% (598 of 2,505) of regulated genes exhibited differential profile or time-point regulation between human and both mouse macrophage populations [“divergently regulated” (DR); Fig. S1 and Dataset S1B]. As expected, our analysis captured TLR4 target genes already known to be differentially regulated between human and mouse macrophages including INOS/NOS2 (Fig. 1D), IDO (Fig. 1E), and CYP27B1 (Fig. 1F). A number of genes known to be involved in immune defense and antimicrobial pathways showed profile divergence between species (Fig. 1G and Dataset S1C).
Macrophages exhibit a degree of plasticity with respect to phenotype and function. It was therefore conceivable that some of the differences attributed to species divergence actually reflected heterogeneity between different macrophage populations. Our choice of specific mouse macrophage populations was designed to minimize such effects. CSF-1–replete BMMs are a proliferating cell population, and LPS treatment causes growth inhibition. Consequently, LPS regulates gene expression programs associated with the cell cycle, as well as inflammatory and antimicrobial responses in BMMs. TEPMs are post-proliferative, so LPS does not cause growth inhibition in these cells. We therefore predicted overlapping but distinct LPS-regulated gene expression programs in these cells. Indeed, BMMs and TEPMs did exhibit striking differences in LPS-regulated gene expression (Fig. S3), as we previously documented for individual LPS target genes (26). Importantly, our analysis required that the HMDM profile was distinct from those of both mouse macrophage populations. This selection criterion enabled us to identify a wide spectrum of LPS target genes in the mouse, and to ensure that these genes were differentially regulated in the primary human macrophage population examined.
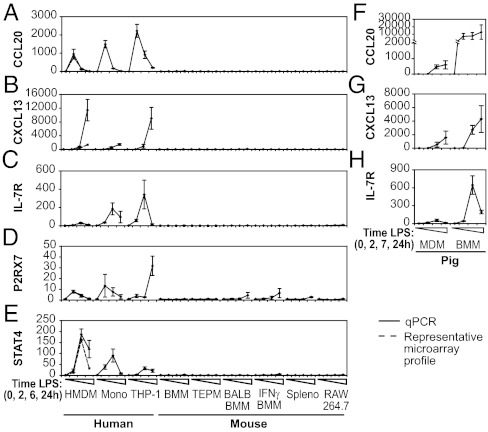
To confirm that the aforementioned strategy had successfully identified true species differences, we next tested expression of individual human-specific TLR4 target genes by quantitative PCR across an extensive panel of human and mouse primary cells and cell lines that were not used in our initial expression profiling analysis. The human-specific LPS target genes CCL20, CXCL13, IL-7R, P2RX7, and STAT4 were all robustly up-regulated in the human monocyte/macrophage populations examined compared with the mouse macrophage populations (Fig. 2 A–E). In each of these cases, the lack of induction by LPS in mouse is supported by Cap-Analysis Gene Expression (CAGE) data from the Functional Annotation of the Mouse (FANTOM) 3 project (http://fantom3.gsc.riken.jp/) (27), as well as previous data with the use of the Affymetrix array platform. Importantly, not all DR genes showed an all-or-nothing response. Indeed, differences in the timing or the magnitude of the response were frequently observed (Fig. 1C and Fig. S1). In the case of the P2RX7 gene, for example, rapid up-regulation was apparent in all human myeloid populations (2 h after stimulation). In mouse cells, this early response was not apparent, but up-regulation did occur for some cell populations at a later time point (24 h after stimulation; Fig. 2D). To further ensure that apparent species differences were not merely a result of differences in the cellular systems used, we also sampled pig macrophages. In this species, it is possible to generate both BMMs and monocyte-derived macrophages (MDMs) in the presence of human CSF-1. LPS robustly induced CCL20, CXCL13, and IL-7R in MDMs and BMMs from pigs (Fig. 2 F–H). IDO and STAT4 were also LPS-inducible in pig macrophages, whereas iNOS was not (28). Thus, regardless of their origin (MDM or BMM), pig macrophages resembled HMDMs rather than mouse BMMs or TEPMs in their TLR4-regulated responses, thus confirming, at least for these genes, that cell-type differences are not sufficient to explain the divergent regulation we observe.
Fig. 2.
Validation of human-specific TLR4 target genes and confirmed coregulation in porcine macrophages. LPS-regulated mRNA expression of selected human-specific TLR4 target genes was quantified by quantitative PCR in cells from human (HMDMs, primary CD14+ monocytes, PMA-differentiated THP-1 macrophage-like cells) or mouse (BMMs, TEPMs, BMMs derived from BALB/c mice, C57BL/6 BMMs primed overnight with IFN-γ, C57BL/6 splenocytes, macrophage-like cell line RAW264.7). Human-specific LPS-regulated gene expression is displayed for CCL20 (A), CXCL13 (B), IL-7R (C), P2RX7 (D), and STAT4 (E). Quantitative PCR data represent average fold induction ± range (BALB/c BMMs, n = 2) or ± SEM (n ≥ 3, all other profiles) of measurements with independent samples. Microarray data are shown only for HMDMs, BMMs, and TEPMs (dotted line, average fold induction ± SEM, n = 3–4). Expression of CCL20 (F), CXCL13 (G), and IL-7R (H) were further investigated by profiling LPS-dependent gene expression in MDMs and BMMs derived from pigs; data represent average fold induction ± SEM of measurements with three or four independently prepared samples.
Functional Impact of Regulatory Divergence in TLR4 Responses.
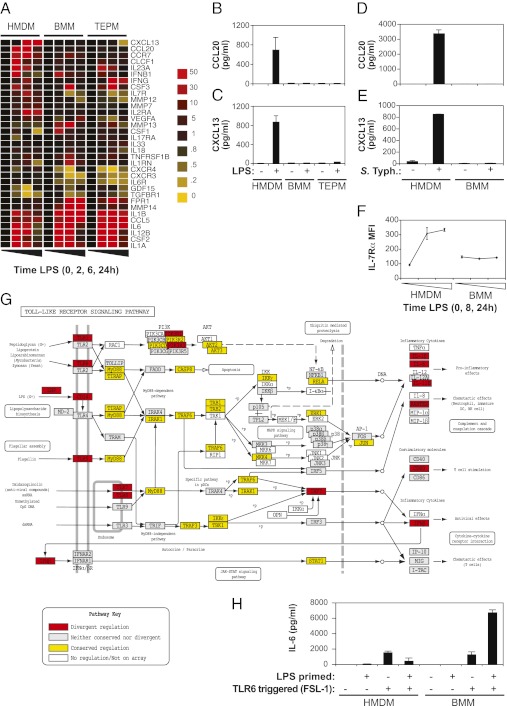
We next assessed the functional consequences of species differences in TLR4 responses. A number of Gene Ontology terms were significantly overrepresented in DR orthologues (Table S2), many of which correspond to secreted or plasma membrane proteins (156 genes; Dataset S1D), including a number of matrix metalloproteinases and cytokine, chemokine, and growth factor ligands and receptors. Fig. 3A shows a heat map for profile divergent orthologues within this subset of secreted or plasma membrane proteins. Such DR orthologues include CCL20, CXCL13, IL-7R, and P2RX7 (Fig. 2 A–D). We confirmed human-specific secretion of CCL20 and CXCL13 in response to LPS (Fig. 3 B and C). Similarly, infection with the Gram-negative pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) triggered CCL20 and CXCL13 production in human but not mouse macrophages (Fig. 3 D and E). We also confirmed induction of cell surface expression of the cytokine receptor IL-7Rα (CD127) in HMDMs but not BMMs (Fig. 3F). In confirmation of the ontology enrichment analysis, annotation of the core TLR signaling pathway itself according to regulatory divergence highlights that signaling components were generally conserved in their regulation, whereas regulatory divergence was concentrated in cellular inputs and outputs (Fig. 3G). To examine potential regulatory divergence in TLR signaling and TLRs themselves, we interrogated a broad, hand-curated set of TLR-associated genes on the focused arrays. Twenty of these were DR, including TLRs (e.g., TLR6, TLR8) and several negative feedback regulators (e.g., IRAK3, JDP2, SOCS1, and ATF3; Fig. S4 A–D and Dataset S1E). The prevalence of regulatory divergence in immune receptors suggests that LPS may differentially affect sensitivity to a second immune challenge. Interspecies differences in ligand sensitization are likely to be important in the context of infection or inflammation in which multiple receptors are engaged. To verify this model, we selected the pathogen recognition receptor TLR6 for functional analysis. TLR6 mRNA was up-regulated by LPS in mouse but not human macrophages (Fig. 3G and Fig. S4A; also see MacGate, http://www.macgate.qfab.org). As predicted, LPS priming boosted IL-6 production in response to the TLR2/TLR6 agonist FSL-1 in mouse macrophages, whereas this effect was not apparent in human macrophages (Fig. 3H).
Fig. 3.
Functional consequences of regulatory divergence in macrophage inputs and outputs. (A) Hierarchical clustering of DR orthologues that encode matrix metalloproteinases or cytokines, chemokines, and growth factors and their receptors. Gene fold induction is indicated by the color-code key. Human-specific production of the DR secreted proteins, CCL20 (B and D) and CXCL13 (C and E) in response to 24 h LPS treatment (B and C) or 24 h S. Typhimurium infection (D and E) was validated by ELISA. Data represent mean ± SD of technical replicates (n = 4, LPS-stimulated HMDMs or BMMs) or mean ± range of technical replicates (n = 2, LPS-stimulated TEPMs and Salmonella-infected HMDMs and BMMs). Profiles are representative of two to four experiments with independent cellular sources. (F) Cell-surface IL-7Rα/CD127 expression on HMDMs and BMMs over a time course of LPS stimulation was measured by flow cytometry [average mean fluorescence intensity (MFI) ± SD of n = 4 technical replicates, prepared, stimulated, and harvested in parallel]. Data are representative of two to five experiments with independent cellular sources. (G) The reference Kyoto Encyclopedia of Genes and Genomes pathway for TLR signaling was color-coded according to regulatory conservation (yellow) or divergence (profile or time point divergence; red) between species. LPS-regulated orthologues displaying neither striking conservation nor divergence are shaded in gray, and genes that were not significantly LPS-regulated or not represented on the focused microarray are white. This striking separation of DR genes according to pathway input/output vs. pathway core is highly unlikely to occur by chance (P = 4 × 10−7). (H) Macrophages were primed with LPS or medium for 11 h, washed, and restimulated with the TLR2/6 ligand FSL-1 for 8 h. IL-6 production was measured by ELISA. Data are average ± SD of three technical replicates, prepared, stimulated, and harvested in parallel, and are representative of three or four independent experiments.
Species Divergence in Transcription Factor Gene Regulation.
LPS differentially regulated 49 transcriptional regulators between the species (Fig. S4E and Dataset S1F). For example, HEY1 and HESX1 were strikingly induced only in HMDMs (Fig. S4 F and G). As with mRNA regulation (Fig. 2E), STAT4 protein was markedly LPS-inducible in HMDMs and the human monocytic cell line THP-1, but not in BMMs (Fig. S4H). Such data imply that inducibly expressed, DR transcription factors may affect downstream gene expression, thus contributing to regulatory divergence. However, the extent of regulatory divergence between species declined with time when LPS-regulated genes were partitioned according to the time point of their peak regulation (DR genes were 10.5% of total regulated genes peaking at 2 h, 10.3% at 6 h, and 4% at 24 h after LPS stimulation; Fisher exact test, P = 0.9101, P = 1.73 × 10−5, and P = 7.858 × 10−4, respectively, for early/mid, mid/late, and early/late comparisons; χ2 test, P = 1.27 × 10−4). This argues against inducible transcription factors acting on downstream genes as a major driver of regulatory divergence, although it is still possible that there is some contribution from this mechanism. Contraction in interspecies regulatory divergence across time could reflect differing mechanisms for feedback control of LPS-regulated gene expression, which ultimately achieves the same end (Fig. S4 A–C). It also suggests that evolution selects for the final LPS-induced cellular state (e.g., 24 h and beyond) rather than the specific mechanisms to achieve this state (2–6 h). Interestingly, the kinetics of peak LPS regulation appeared to be more rapid in HMDMs compared with mouse macrophages [502 human and 355 mouse profiles showed peak LPS regulation at the earliest (2 h) time point; Fisher exact test, odds ratio of 1.52, P = 4.052 × 10−8 for 2 h vs. not-2 h data partitions].
We also examined the time course of regulatory divergence between the mouse macrophage populations. As these cells are genetically identical, regulatory divergence must be mediated by differences in the basal or inducible trans environment. In stark contrast to the interspecies comparison, regulatory divergence between the two mouse macrophage populations modestly increased with time (1.27 and 1.65 fold for 2 h and 6 h vs. 24 h after LPS stimulation; Fisher exact test, P = 0.08576, P = 1.42 × 10−5, and P = 0.1171, respectively, for early/mid, mid/late, and early/late comparisons; χ2 test, P = 5.14 × 10−5). The distinct temporal signature in inter- vs. intraspecies gene regulatory divergence further argues against a major contribution from macrophage heterogeneity in the species divergence signature. It also suggests that differences in cis (i.e., DNA sequence changes near the DR gene) and/or the trans environment (e.g., availability of specific transcriptional regulators) are likely to make a substantial contribution to interspecies regulatory divergence.
Paradox of Promoter Conservation and Expression Divergence.
To examine the contribution of promoter sequence evolution to regulatory divergence between species, we precisely defined TSSs using genome-wide deep CAGE performed in parallel with expression profiling. This yielded 5.3 million human macrophage and 2.6 million mouse macrophage tags that define TSSs to single nucleotide resolution (Fig. S5 A and B; data available at http://fantom.gsc.riken.jp/4/download/Supplemental_Materials/Schroder_et_al_2012/). CAGE-defined TSSs were projected onto multiple genome sequence alignments of as many as nine placental mammals with deeply sequenced genomes (Fig. S5C). Each gene was assigned a primary TSS in each species, being the TSS with the most CAGE tags over the LPS time course, after correcting for depth of library coverage. We initially investigated the role of TSS switching in expression divergence (i.e., the primary TSS for a gene in one species is not orthologous to the primary TSS in the other species), but found that the DR gene set was not significantly enriched for genes showing evidence of TSS switching between species (odds ratios of 1.12 and 1.33 for profile and time-point divergent genes, respectively; Fisher test, P > 0.2 in both cases). Subsequent analyses focused on the stringently filtered set of 1,210 TSSs that occur in orthologous positions and represent the primary TSSs in human and mouse.
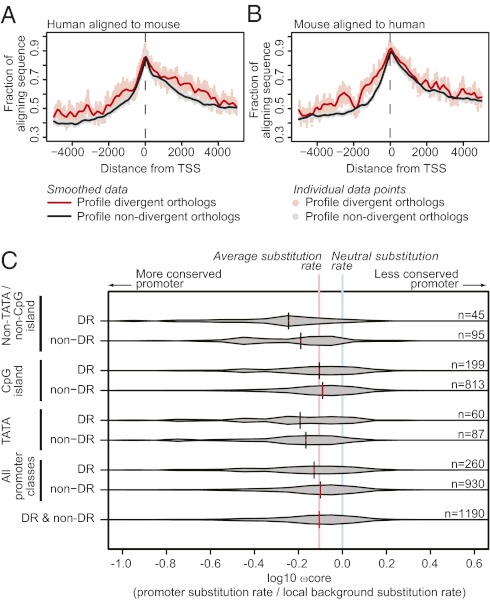
As we hypothesized that cis-regulatory sequence evolution would be a major driver of gene regulatory divergence, we analyzed sequence conservation in promoters of DR vs. non-DR genes. Several approaches confirmed a very surprising finding: promoters driving DR genes were demonstrably more conserved than non-DR promoters. Relative to non-DR genes, DR gene “TSS regions” (−5 kb to +5 kb) exhibited a greater fraction of human:mouse-aligning nucleotides than those of non-DR genes (Fig. 4 A and B). We next analyzed the extent of core promoter sequence divergence between species. DR gene core promoter substitution rates (Rcore, −200 to −1 nt relative to TSS; further information is provided in SI Glossary of Technical Terms) were lower than non-DR substitution rates (Kolmogorov–Smirnov test, D = 0.18, P = 3.4 × 10−4; Table S3), and Rcore was negatively correlated with expression divergence (Spearman correlation, ρ = −0.15, P = 3.6 × 10−6). One possible explanation for this finding was that DR genes are biased toward regions in the genome that are highly conserved. To address this possibility, we quantified evolutionary constraint on a per-promoter basis, correcting for the local neutral rate of evolution. We thus calculated Rcore to the substitution rate in nearby interspersed repetitive elements (Rire) to give ωcore (i.e., Rcore/Rire), a measure analogous to protein coding sequence ω (i.e., Rsynonymous/Rnonsynonymous). The ωcore significantly negatively correlated with regulatory divergence (SDiff; Spearman ρ = −0.13, P = 5 × 10−6; Table S3), and the promoters of DR genes were significantly biased to lower values of ωcore (Table S3 and Fig. 4C). Thus, higher sequence conservation is associated with increasing regulatory divergence, even when the local substitution rate was accounted for. Even at a wider (1 Mb) interval, DR genes were significantly enriched for association with conserved noncoding regions (as defined in ref. 29) relative to non-DR genes (one-tailed Wilcoxon test, P = 0.0038). Together, these data indicate that regulatory sequences of DR genes are under greater evolutionary constraint than those associated with non-DR genes.
Fig. 4.
The promoters of DR genes exhibit a higher fraction of aligning sequence and lower substitution rates than non-DR promoters. (A and B) Spatial profile of human/mouse aligning sequence. The fraction of human TSS region nucleotides aligned with mouse (A) and mouse TSS aligned with human (B) for profile-divergent genes (red) vs. non–profile-divergent subset (black), for which an orthologous macrophage TSS was defined in the other species (mouse or human) by CAGE. (C) Distributions of core promoter corrected substitution rate ratios [log10(ωcore)] for primary TSSs that are orthologous between human and mouse (n = 1,190). log10(ωcore) < 0 indicates predominantly purifying selection; log10(ωcore) > 0 indicates predominantly diversifying selection. Also shown is the distribution median (vertical black/red bar), the fraction of all TSSs represented by the distribution (red to black fraction of vertical bar), and the median of all TSSs (pale red). Distributions are shown for specific promoter types (TATA, CpG island, neither TATA nor CpG island).
Despite the predominant trend for evolutionary constraint within DR gene promoters, the small population of core promoters with substitution rates significantly in excess of the neutral substitution rate (ωcore >1, <5% FDR), indicative of positive selection, were modestly enriched for DR genes (odds ratio = 1.4; Dataset S1G). Thus, for a small number of DR genes, positive selection on cis-regulatory sequences within the proximal promoter is likely to drive gene regulatory divergence. Conversely, a small population of non-DR genes also exhibited strong evidence of positive selection, demonstrating that rapid promoter evolution may not always lead to a measurable impact on gene expression, at least in our restricted sampling of cells and conditions.
Regulatory Divergence Is Associated with Specific Promoter Architecture.
DR genes exhibited lower substitution rates 20 to 35 bases upstream of the TSS, and a series of pronounced peaks and troughs in sequence conservation downstream of the TSS (Fig. 4), which are hallmarks of TATA box-containing promoters (30). Indeed, promoters of DR genes were enriched for the presence of a TATA box. 17.2% of human and 14.8% of mouse DR gene promoters contained a TATA box, compared with 9.1% of human and 7.7% of mouse nondivergent gene promoters (Fisher test, P = 1.2 × 10−7 and P = 4.8 × 10−7 for human and mouse comparisons, respectively). To identify other features of DR genes, we also surveyed transcription factor binding sites (TFBSs) within proximal promoters. No position constraint was applied in this analysis, as recent evidence suggests that functional TFBSs are often not conserved across species (31). Although our approach identified TFBSs in DR and concordantly regulated genes that are commonly associated with LPS-regulated transcription (e.g., Rel, Stat, Ets), we did not find any strong association between specific TFBSs and regulatory divergence (Table S4). As described earlier, the one obvious exception was the TATA motif, which was more likely to be present in DR genes (odds ratio, 1.42; P = 3.32 × 10−3; Table S4). However, even when using this approach that applied no position constraint, the TATA box was present in fewer than 50% of DR genes. Thus, not all DR genes contain a TATA box in their promoter regions.
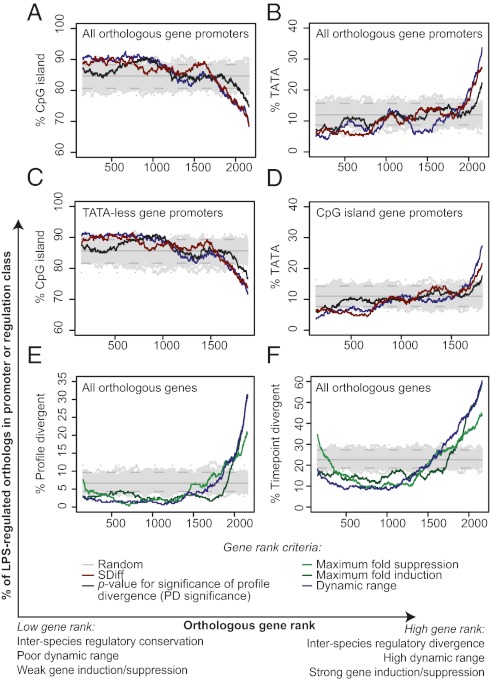
Profile divergent genes (Fisher test, P = 2.67 × 10−7 and P = 9.69 × 10−10 for human and mouse comparisons, respectively) and time point-divergent genes (Fisher test, P = 4.58 × 10−7 and P = 1.39 × 10−7 for human and mouse comparisons, respectively) were also significantly depleted of 5′ CpG islands, compared with nondivergent genes. TATA enrichment and CpG island depletion were not only associated with the degree of regulatory divergence (ranking by SDiff or ANOVA P value), but also the extent to which gene expression changes over the LPS time course (dynamic range, calculated as the difference between maximum fold induction and maximum fold suppression over the time course; Fig. 5 A and B). Conversely, genes exhibiting a relatively small dynamic range and low interspecies divergence were enriched for CpG island promoters and depleted of TATA promoters. TATA enrichment and CpG island depletion are clearly not redundant signals, as conditioning on one does not remove the effect of the other (Fig. 5 C and D). A gene's dynamic range, as well as its maximum LPS-dependent regulation (i.e., induction or suppression from unstimulated), was clearly associated with regulatory divergence, such that highly regulated genes were enriched for regulatory divergence (Fig. 5 E and F). In total, these results indicate that TATA-containing and non-CpG island promoters confer a high degree of inherent transcriptional plasticity (i.e., the capacity of a gene to change its transcription level under different environmental or evolutionary conditions), as TATA-containing and non-CpG island promoters were associated with both a large magnitude of response to LPS and expression divergence between species. TATA boxes tend to be associated with slowly evolving promoter sequences, and CpG islands with rapidly evolving promoter sequences (30), so it is important to note that the greater sequence constraint observed in DR gene promoters (Table S3) held true even within a promoter class (Fig. 4C). Thus, the effects observed are not just a manifestation of differences in promoter architecture between DR and non-DR genes, but rather are a general property of DR macrophage gene promoters. These distinct properties ultimately validate our approach for identifying DR genes.
Fig. 5.
DR genes are enriched for TATA box promoters, are depleted of CpG islands, and tend to exhibit a large dynamic range of expression in response to LPS. (A and B) Orthologous genes were ranked based on variation between species (SDiff or P value for significance of profile divergence; high rank is high significance and low P value) or dynamic range, such that genes with a high variation within or between species achieve a high rank, and analyzed according to percentage of genes with (A) CpG island-type orthologous gene promoters or (B) TATA box-containing orthologous gene promoters. (C and D) analyses were performed as in A and B, but considering only orthologues without a TATA box in the promoter (C) or only those that had CpG island-containing promoters (D). (E and F) Genes were ranked based on dynamic range, maximum fold suppression, or maximum fold induction and analyzed for percentage of DR genes according to (E) profile divergence or (F) time-point divergence. For A–F, 100 randomly permuted rankings of the data are shown (gray curves; nonrandom rankings protruding from the gray curves represent P < 0.01). Also indicated are confidence intervals (95%, dashed gray lines; 99%, dotted gray lines) and average for the whole dataset (solid gray lines). Sliding 300-gene windows are shown.
Discussion
Some differences between mouse and human LPS responses at the whole-organism level are already documented (9). In this study, we demonstrate differential gene regulation in response to LPS in human and mouse macrophages that is strongly biased toward genes encoding cell surface receptors and secreted molecules. Individual examples of such DR genes were further examined in porcine BMMs and MDMs, in which LPS profiles for both macrophage populations reflected human-specific up-regulation, tempting speculation that the short generation time of mice may accelerate regulatory evolution in this species. The prevalence of regulatory divergence in cell surface and secreted proteins predicts that differences in LPS regulation underpin divergent cellular and physiological functional responses to LPS, or may function to sensitize the cell to further stimuli that would be present during in vivo inflammation. We verified this at the cellular level by examining downstream responses to one DR “input,” TLR6 (Fig. 3H). Notably, more than 150 other genes encoding inputs and outputs were also differentially regulated, which suggest further differences in macrophage functional responses to inflammation/infection in humans and mice, and/or the mechanisms used to achieve them.
Although it is known that humans are physiologically more sensitive than mice to LPS (9), the contribution of particular DR genes to this phenomenon remains to be established. Our study used a maximal stimulatory dose of LPS to elicit all LPS-regulated transcriptional responses; thus, our data do not yield a direct comparison of LPS sensitivity between human and mouse macrophages. However, our data are suggestive of mechanisms contributing to differential LPS sensitivity between species. We observed more rapid peak regulation kinetics in human vs. mouse macrophages, suggesting that HMDMs may be somewhat “primed” for immediate/early LPS responses, which in turn suggests increased sensitivity. We also observed more rapid and greater induction of several negative feedback regulators of the TLR pathway in mouse macrophages (IRAK3/IRAK-M and JDP2; Fig. S4 B and C). Enhanced feedback regulation may dampen the primary LPS response in mouse macrophages, thereby contributing to the relatively reduced endotoxin sensitivity observed in mice compared with humans. Elevated expression of some of the “human-specific” TLR4 target genes, for example IL-7R (32) and CXCL13 (33), has been linked to pathological human macrophage inflammatory responses. Similarly, the chemokine CCL20, which was specifically induced in human macrophages by LPS or whole pathogen infection in our study, is clearly linked to a range of human autoimmune diseases by virtue of its ability to attract pathogenic Th17 cells to a site of in vivo inflammation (34). Furthermore, polymorphisms in several DR genes, including coding sequence variants in P2RX7 (35) and a promoter polymorphism in IL7R (36), have been associated with infectious and inflammatory diseases. Such data support the contention that DR genes are critical components of the human innate immune system, and are likely to contribute to pathological inflammatory processes.
Although some differences between species in antimicrobial gene regulation have previously been documented, it is clear from our study that multiple genes within such pathways were coregulated in a species-specific fashion. For example, mouse-specific LPS induction of Inos (Fig. 1D) paralleled that of mRNAs encoding proteins regulating iNOS function (the arginine transporter Slc7a2 and the degradative enzyme arginase; also see MacGate, http://www.macgate.qfab.org). Conversely, IDO and KYNU, which encodes an enzyme downstream of IDO in the tryptophan metabolic pathway, were coinduced in a human-specific manner (Fig. 1E; also see MacGate, http://www.macgate.qfab.org). Importantly, our approach also identified previously unrecognized regulatory divergence in other antimicrobial pathways, such as Lcn2 (mouse-specific) that encodes a secreted protein interfering with iron acquisition by bacterial siderophores (37), and two genes specifically induced in human macrophages (P2X7 receptor, P2RX7, and CXCL13, Fig. 2 B and E) that are required for effective clearance of Mycobacterium tuberculosis (38, 39). Given the evolutionary arms race between host and pathogen, we postulate that many other DR genes are likely to have key roles in direct antimicrobial responses of macrophages.
Despite significant divergence in gene expression profiles, LPS-regulated gene expression in human and mouse macrophages was far more similar than would be expected if expression diverges randomly by neutral drift alone. This is consistent with prior expectation and previous studies on other systems suggesting that gene regulation is often conserved by purifying selection (13, 18, 40, 41). Furthermore, expression divergence declined over time, suggesting that, even in the face of extensive species differences at early time points, the two biological systems ultimately reached a level of commonality, at least at the cellular level. This in turn suggests that evolution selects for the final LPS-reprogrammed cellular state (at 24 h and beyond) rather than the means to achieve this state. Thus, robustness in innate immune pathways may ultimately enable a level of functional conservation in at least some biological responses.
Recent expression quantitative trait locus studies have highlighted the importance of cis-regulatory changes, and particularly promoter region polymorphisms, in determining gene expression level within the human population (42, 43). The strongest expression quantitative trait locus effects are preferentially clustered around gene TSSs. We thus hypothesized that gene expression divergence would be mirrored to some extent by sequence divergence in cis-regulatory sequences, particularly in core promoters. Indeed, numerous promoter polymorphisms have been identified that alter the expression of inflammation-associated TLR4 target genes (e.g., IL1, IL6, TNF, IL10) (44). This was indeed the case for a small number of DR genes, which exhibited significantly faster than neutral substitution rates in their promoters. These genes appear to possess very rapidly evolving promoters, suggestive of sustained or repeated rounds of diversifying selection. Notable among them is DDX58, encoding the protein RIG-I, an RNA helicase that recognizes viral RNAs and is a key regulator of antiviral innate immunity (45). However, we consistently found the opposite of the expected trend in the data set as a whole; interspecies regulatory divergence was associated with constraint in promoter sequences. This was seen in the per-promoter substitution rates (including the ω estimates that correct for local background effects), the fraction of nucleotides aligning between species, and the association of highly conserved proximal noncoding regions with DR genes.
DR genes are significantly enriched for TATA-containing promoters. Such promoters are known to possess a higher density of regulatory motifs relative to other promoters (46). This likely explains the observation that such promoters evolve rapidly in their transcriptional regulation (14), despite their propensity for enhanced conservation at the nucleotide level. The higher levels of promoter constraint we observed for DR relative to non-DR genes also applied across promoter “categories”; DR genes with TATA promoters have more constrained promoter regions than non-DR genes with TATA promoters, and DR genes have more constrained cis-regulatory sequences for the CpG island promoter and non-CpG island promoter data partitions (Fig. 4C). Although enhanced promoter sequence conservation seems to be a generalized feature of DR genes, it is important to note that not all DR genes are characterized by the presence of a TATA box and/or lack of CpG islands. Indeed, when applying a position constraint on the motif, only 17.2% of human and 14.8% of mouse DR genes contained a TATA box. STAT4 and CCL20, for example, were both induced in a human-specific fashion (Fig. 2 A and E), yet only the latter is a TATA-containing promoter in human, pig, and mouse. In some cases, differences in the actual TATA box may directly contribute to expression divergence. The IL-7Rα promoter, for example, is a TATA-containing promoter in human and pig, but not mouse, which is consistent with its human/pig-specific induction by LPS.
Dynamic range was also strongly correlated to the extent of regulatory divergence. The most regulated genes in each species were among the least likely to be similarly regulated in the other. A large dynamic range in mRNA expression in response to a TLR agonist indicates a gene is highly sensitive to changes in the trans-regulatory environment. It follows, then, that such genes might also be sensitive to the many alterations to the trans environment concomitant with species divergence, consistent with previous studies in yeast (14, 17, 46). Our data support such a model for higher eukaryotes, and we extend this model by showing the connection between greater transcriptional plasticity (i.e., highly dynamic, evolvable transcriptional responses) and higher levels of promoter constraint. Consider a promoter that responds to multiple trans-regulatory inputs. The integration of more inputs suggests more cis-regulatory sites, both in distant enhancers and in the core promoter, where these signals are ultimately assimilated for gene expression. A greater number of cis-regulatory sites leads to a higher fraction of constrained sites and promoter/cis-regulatory sequence conservation. Such a gene is also likely to be hypersensitive to evolutionary perturbation of the trans-acting milieu or specific cis-regulatory sites.
LPS induces transcriptional changes in hundreds of genes and their products, indicative of complex gene regulatory networks (22, 23, 47). Widespread divergence in TLR-regulated transcription factor expression between mouse and human (Fig. S4 E–H) predicts species-specific transcriptional networks leading to regulatory divergence. There are some correlative suggestions of this from our analysis. For example, HEY1, which was specifically induced by LPS in human macrophages (Fig. S4F), was recently identified as a negative feedback regulator of IL6 in LPS-activated human macrophages (48). In our study, peak IL6 induction in HMDMs was significantly lower and faster than in either BMMs or TEPMs (MacGate, http://www.macgate.qfab.org), implicating this transcription factor in feedback regulation in humans but not mice. Similarly, STAT4 was LPS-inducible in human but not mouse macrophages (Fig. 2E and Fig. S4H), as was the known STAT4 target gene, IRF4 (49) (MacGate, http://www.macgate.qfab.org). Nonetheless, regulatory divergence significantly contracted as the LPS time course proceeded. This finding argues against a major input from inducible trans factors to regulatory divergence, although we cannot discount the possibility that they make some contribution. Instead, we suggest that different sets of LPS-inducible transcription factors in human vs. mouse enable conserved regulation of late response genes and/or that DR inducible feedback regulators ultimately limit functional divergence over time. It is also possible that many of the TLR4-inducible transcription factors are not involved in secondary LPS responses, but rather are induced to permit responses to other stimuli in the in vivo setting. Importantly, the finding that divergence is most pronounced in primary LPS responses suggests changes in cis and/or the basal trans environment are the major drivers of regulatory divergence. We have provided evidence for the former, but it is likely that the latter also contributes.
In summary, we have performed the most stringent human:mouse comparison of innate immune gene regulation reported to date to the best of our knowledge. Many LPS-responsive genes in mouse and human macrophages are conserved in their regulation, but we also identified and validated considerable divergence in TLR4 responses between these species. We suggest this regulatory divergence is likely to be one contributing factor to phenotypic differences between species in innate immunity. Importantly, our findings show that transcriptional plasticity in response to a stimulus within species and variability between species are intrinsically linked. The capacity for expression variation in both cases is associated with specific promoter properties (high promoter conservation, TATA box enrichment, and CpG island depletion). Paradoxically, differences in transcriptional regulation between species may be taken as evidence that the absolute level of the transcript, and its protein product, is crucial for innate immunity. By extension, functional analyses of TLR4 target genes uniquely regulated in human macrophages are likely to provide important insights into human infectious and inflammatory diseases. Furthermore, by revealing control points not already used in humans, the alternative evolutionary path trodden by the mouse may also provide opportunities for therapeutic intervention in such diseases.
Materials and Methods
A summary of the methods used is presented in the subsequent sections; further details are provided in SI Materials and Methods.
Ethics Statement.
Before undertaking the studies described, approvals for all experiments using primary human and mouse cells were obtained from the relevant University of Queensland Ethics Committee.
Preparation of Primary Macrophages.
HMDMs were differentiated in vitro from CD14+ monocytes isolated from peripheral blood mononuclear cells using MACS technology (Miltenyi Biotech), and BMMs were derived from bone marrow progenitors. HMDMs and BMMs were differentiated from progenitors in complete media containing recombinant CSF-1 for 7 d. Mouse TEPMs were harvested by PBS solution lavage 5 d after i.p. injection of thioglycollate broth. All mice were C57BL/6 unless otherwise stated.
Design, Hybridization, and Signal Processing of Custom Microarrays.
By using in-house and public access microarrays, we identified LPS-regulated genes in HMDMs, BMMs, or TEPMs. We then designed human and mouse custom 15,000 probe microarrays (Agilent Technologies) targeting genes regulated by LPS in any of these macrophage populations, for which orthology could be predicted with high confidence. RNA from three independent cell preparations (BMMs, TEPMs) or four independent biological replicates (HMDMs, in which each replicate RNA was a pool of RNA from two independent blood donors; n = 8 donors in total) was analyzed by microarray.
Custom Microarray Data Analysis.
Processed signal intensities were quantile normalized in R using the Affymetrix package in Bioconductor, and median-centered. Because of the high level of internal replication for the quantification of transcript expression as a result of multiple probes targeting the same transcript(s), probe-level data were collapsed into transcript-level data if profiles from multiple probes were highly correlated. For one-to-one orthologues, all transcript-level profiles significantly regulated by LPS were compared with all transcript-level profiles for the orthologous gene. A two-way ANOVA with two fixed effects (time and species) was fitted to normalized, log10 data averaged over the replicates, and representative profiles were chosen by taking the least divergent profile pair (highest Benjamini–Hochberg corrected P value in either the HMDM/BMM or HMDM/TEPM comparison). If this P value was below a threshold of 0.05, the orthologous genes were classified as exhibiting significant profile divergence between the species. A second statistical analysis was performed on the representative profiles for human and mouse to identify more subtle differences in expression at single time points (more than threefold difference in expression and Student two-tailed t test P value <0.05). All P values for time point or profile divergence calculated were corrected for multiple testing using the Benjamini–Hochberg method. Expression profile similarity was further assessed by using SDiff (25), calculated as the sum of absolute differences between orthologous expression profile values over all time points. Concordantly regulated genes were defined as those with SDiff scores lower than 0.8 and lower than 0.6 (corresponding to P < 0.05 for randomly permuted data) for HMDM/BMM and HMDM/TEPM profile comparisons, respectively. All microarray data (probe-level, transcript profile-level, and representative human/mouse profiles) are freely available through MacGate (http://www.macgate.qfab.org), and are also accessible via the National Center for Biotechnology Information Gene Expression Omnibus repository (GSE19492 SuperSeries containing human and mouse data for Agilent custom and Illumina whole-genome microarrays). Custom microarrays used in this study are freely available for order from Agilent Technologies (probe files available via GSE19492).
TSS Identification and Evolutionary Analyses.
CAGE libraries from HMDM (0, 2, 6, 24 h LPS) and BMM (0, 6 h LPS) total RNA were prepared as described and subjected to deep sequencing to identify TSSs (50). BMM deep CAGE data were merged with comparable data from FANTOM3 for LPS-stimulated BMMs (Fig. S5) (27). CAGE tags were mapped to genome assemblies (51) (Hg18, Mm9) and mapped to probe sets via Ensembl and Refseq transcripts. CAGE data are freely accessible through FANTOM (http://fantom.gsc.riken.jp/4/download/Supplemental_Materials/Schroder_et_al_2012/). By this method, at least one TSS was defined for 89% (2,238 of 2,505) of human and 92% (2,297 of 2,505) of mouse genes on the targeted microarray (Fig. S5). The EPO-9 whole genome alignment dataset from Ensembl was used for coordinate transformations between species to extract orthologous sequence alignments from the reference genomes for available eutherian mammals and for the calculation of substitution rates.
TATA Box and CpG Island Mapping.
CAGE-defined TSSs were defined as “TATA-TSS” if they had a match to the Jaspar TATA box matrix (TFMscan, options: −l, −p, 6), with the 5′ end of the match located 25 to 35 nt upstream of the TSS reference position (the interval that shows TATA box enrichment over the whole dataset). Genes were defined as “TATA-genes” if they had one or more TATA-TSS. Mouse (mm9) and human (hg18) CpG islands were downloaded from the University of California, Santa Cruz, genome table browser, and CpG island genes were defined as those with a CpG island overlapping their core promoter (−300 to +100 with respect to the TSS reference position).
Supplementary Material
Acknowledgments
We thank the Institute for Molecular Bioscience Special Research Centre Microarray Facility for performing the microarray hybridizations, as well as David Wood and Dr. Cas Simons (Queensland Facility for Advanced Bioinformatics, University of Queensland) for building the MacGate Web site. This work was supported by C. J. Martin Fellowship 490993 from the Australian National Health and Medical Research Council (to K.S.); Australian Research Council under the ARC Centres of Excellence program (ARC Centre of Excellence in Bioinformatics, K.-A.L.C.); Australian Research Council Future Fellowship FT100100657 (to M.J.S.); honorary Australian National Health and Medical Research Council Senior Research Fellowship APP1003470 (to M.J.S.); Australian National Health and Medical Research Council Grant 631531, a research grant to the RIKEN Omics Science Center from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT); and an Innovative Cell Biology by Innovative Technology Project grant from MEXT (to Y.H.), which supported Cap Analysis of Gene Expression data collection and mapping.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE19492). The mRNA deep sequencing data reported in this paper is available for download from http://fantom.gsc.riken.jp/4/download/Supplemental_Materials/Schroder_et_al_2012/. The analyzed interspecies comparative data reported in this paper is available for perusal and interrogation at http://www.macgate.qfab.org.
See Author Summary on page 5925 (volume 109, number 16).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110156109/-/DCSupplemental.
References
- 1.Waterston RH, et al. Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Mestas J, Hughes CC. Of mice and not men: Differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 3.Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayday AC, Peakman M. The habitual, diverse and surmountable obstacles to human immunology research. Nat Immunol. 2008;9:575–580. doi: 10.1038/ni0608-575. [DOI] [PubMed] [Google Scholar]
- 5.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 6.Schneemann M, et al. Nitric oxide synthase is not a constituent of the antimicrobial armature of human mononuclear phagocytes. J Infect Dis. 1993;167:1358–1363. doi: 10.1093/infdis/167.6.1358. [DOI] [PubMed] [Google Scholar]
- 7.Jesch NK, et al. Expression of inducible nitric oxide synthase and formation of nitric oxide by alveolar macrophages: An interspecies comparison. Environ Health Perspect. 1997;105(suppl 5):1297–1300. doi: 10.1289/ehp.97105s51297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu PT, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 9.Copeland S, Warren HS, Lowry SF, Calvano SE, Remick D. Inflammation and the Host Response to Injury Investigators Acute inflammatory response to endotoxin in mice and humans. Clin Diagn Lab Immunol. 2005;12:60–67. doi: 10.1128/CDLI.12.1.60-67.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poli-de-Figueiredo LF, Garrido AG, Nakagawa N, Sannomiya P. Experimental models of sepsis and their clinical relevance. Shock. 2008;30(suppl 1):53–59. doi: 10.1097/SHK.0b013e318181a343. [DOI] [PubMed] [Google Scholar]
- 11.López A, et al. Multiple-center, randomized, placebo-controlled, double-blind study of the nitric oxide synthase inhibitor 546C88: Effect on survival in patients with septic shock. Crit Care Med. 2004;32:21–30. doi: 10.1097/01.CCM.0000105581.01815.C6. [DOI] [PubMed] [Google Scholar]
- 12.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 13.McCarroll SA, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 14.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 15.Khaitovich P, et al. A neutral model of transcriptome evolution. PLoS Biol. 2004;2:E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao BY, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol. 2006;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- 17.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 18.Barreiro LB, Marioni JC, Blekhman R, Stephens M, Gilad Y. Functional comparison of innate immune signaling pathways in primates. PLoS Genet. 2010;6:e1001249. doi: 10.1371/journal.pgen.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 20.Kosiol C, et al. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obbard DJ, Welch JJ, Kim KW, Jiggins FM. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 2009;5:e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson R, et al. Transcriptional network dynamics in macrophage activation. Genomics. 2006;88:133–142. doi: 10.1016/j.ygeno.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 24.Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proc Natl Acad Sci USA. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- 26.Lattin JE, et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carninci P, et al. FANTOM Consortium RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 28.Kapetanovic R, et al. Pig bone marrow-derived macrophages resemble human macrophages in their response to bacterial lipopolysaccharide. J Immunol. 2012 doi: 10.4049/jimmunol.1102649. [DOI] [PubMed] [Google Scholar]
- 29.Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 30.Taylor MS, et al. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006;2:e30. doi: 10.1371/journal.pgen.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickens SR, et al. Characterization of interleukin-7 and interleukin-7 receptor in the pathogenesis of rheumatoid arthritis. Arthritis Rheum. 2011;63:2884–2893. doi: 10.1002/art.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlsen HS, Baekkevold ES, Morton HC, Haraldsen G, Brandtzaeg P. Monocyte-like and mature macrophages produce CXCL13 (B cell-attracting chemokine 1) in inflammatory lesions with lymphoid neogenesis. Blood. 2004;104:3021–3027. doi: 10.1182/blood-2004-02-0701. [DOI] [PubMed] [Google Scholar]
- 34.Comerford I, et al. An immune paradox: How can the same chemokine axis regulate both immune tolerance and activation?: CCR6/CCL20: A chemokine axis balancing immunological tolerance and inflammation in autoimmune disease. Bioessays. 2010;32:1067–1076. doi: 10.1002/bies.201000063. [DOI] [PubMed] [Google Scholar]
- 35.Fernando SL, et al. A polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis. Am J Respir Crit Care Med. 2007;175:360–366. doi: 10.1164/rccm.200607-970OC. [DOI] [PubMed] [Google Scholar]
- 36.Gregory SG, et al. Multiple Sclerosis Genetics Group Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 37.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 38.Britton WJ, Fernando SL, Saunders BM, Sluyter R, Wiley JS. The genetic control of susceptibility to Mycobacterium tuberculosis. Novartis Found Symp. 2007;281:79–89. doi: 10.1002/9780470062128.ch8. [DOI] [PubMed] [Google Scholar]
- 39.Khader SA, et al. In a murine tuberculosis model, the absence of homeostatic chemokines delays granuloma formation and protective immunity. J Immunol. 2009;183:8004–8014. doi: 10.4049/jimmunol.0901937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, et al. Cross-species transcriptional profiles establish a functional portrait of embryonic stem cells. Genomics. 2007;89:22–35. doi: 10.1016/j.ygeno.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaix R, Somel M, Kreil DP, Khaitovich P, Lunter GA. Evolution of primate gene expression: drift and corrective sweeps? Genetics. 2008;180:1379–1389. doi: 10.1534/genetics.108.089623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 2009;20:43–59. doi: 10.1016/j.cytogfr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 46.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amit I, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu X, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehtonen A, et al. Differential expression of IFN regulatory factor 4 gene in human monocyte-derived dendritic cells and macrophages. J Immunol. 2005;175:6570–6579. doi: 10.4049/jimmunol.175.10.6570. [DOI] [PubMed] [Google Scholar]
- 50.Maeda N, et al. Development of a DNA barcode tagging method for monitoring dynamic changes in gene expression by using an ultra high-throughput sequencer. Biotechniques. 2008;45:95–97. doi: 10.2144/000112814. [DOI] [PubMed] [Google Scholar]
- 51.Faulkner GJ, et al. A rescue strategy for multimapping short sequence tags refines surveys of transcriptional activity by CAGE. Genomics. 2008;91:281–288. doi: 10.1016/j.ygeno.2007.11.003. [DOI] [PubMed] [Google Scholar]