Abstract
Carbohydrate moieties are important components of natural products, which are often imperative for the solubility and biological activity of the compounds. The aromatic polyketide alnumycin A contains an extraordinary sugar-like 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane moiety attached via a carbon-carbon bond to the aglycone. Here we have extensively investigated the biosynthesis of the dioxane unit through 13C labeling studies, gene inactivation experiments and enzymatic synthesis. We show that AlnA and AlnB, members of the pseudouridine glycosidase and haloacid dehalogenase enzyme families, respectively, catalyze C-ribosylation conceivably through Michael-type addition of d-ribose-5-phosphate and dephosphorylation. The ribose moiety may be attached both in furanose (alnumycin C) and pyranose (alnumycin D) forms. The C1′-C2′ bond of alnumycin C is subsequently cleaved and the ribose unit is rearranged into an unprecedented dioxolane (cis-bicyclo[3.3.0]-2′,4′,6′-trioxaoctan-3′β-ol) structure present in alnumycin B. The reaction is catalyzed by Aln6, which belongs to a previously uncharacterized enzyme family. The conversion was accompanied with consumption of O2 and formation of H2O2, which allowed us to propose that the reaction may proceed via hydroxylation of C1′ followed by retro-aldol cleavage and acetal formation. Interestingly, no cofactors could be detected and the reaction was also conducted in the presence of metal chelating agents. The last step is the conversion of alnumycin B into the final end-product alnumycin A catalyzed by Aln4, an NADPH-dependent aldo-keto reductase. This characterization of the dioxane biosynthetic pathway sets the basis for the utilization of C-C bound ribose, dioxolane and dioxane moieties in the generation of improved biologically active compounds.
Keywords: ribose-5-phosphate, natural product, biosynthesis, Streptomyces
Carbohydrates are abundant constituents of natural products, which are often essential for the biological activity and solubility of the compounds (1). Structure/function studies have identified that many medically important compounds in clinical use, such as the antibiotic erythromycin (2) and the anthracycline anticancer agents doxorubicin (3) and aclacinomycin (4), owe their activity to the deoxysugar moieties. Therefore it is not surprising that the formation and attachment of these units has been the focus of intensive research, which has led to the observation that the carbohydrate moieties are modified as 4-keto-6-deoxy-d-hexose nucleoside diphosphates prior to attachment by glycosyl transferases (GTs) (5, 6). The most common means of attachment is via an O-glycosidic bond, which unfortunately is susceptible to hydrolysis in the acidic environment of the stomach or by the action of glycosidases in the small intestine (7). On the other hand, C-glycosylation provides an enzymatically resistant and chemically stable bond (8), but it is, by comparison, rare and only found in a handful of natural products (9, 10), such as the angucycline antibiotic urdamycin (11).
Alnumycin A (1, Fig. 1) is an exceptional member of the aromatic polyketides in the sense that instead of typical deoxysugars, it contains an unusual 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane moiety that mimics the structure of deoxysugars and is furthermore attached to the isochromanequinone aglycone via a C8-C1′ bond (12, 13). To date the biosynthetic pathway utilized for synthesis of the dioxane unit has been unknown, although we have previously cloned and successfully expressed the alnumycin gene cluster heterologously in Streptomyces albus (Fig. 2 A and B) to enable efficient manipulation of the pathway (14). Initial knockout studies also identified the involvement of two genes, alnA and alnB, in the biosynthesis and transfer of the dioxane moiety as both single-gene mutants produced prealnumycin (2, Fig. 1) as their main metabolite (14).
Fig. 1.
Structures of the metabolites investigated in this study and the pathway for dioxane biosynthesis. Structures of alnumycin A (1), prealnumycin (2), K1115 A (3), alnumycin B1 (4a) and B2 (4b), alnumycin C1 (5a), C2 (5b) and D, (6), and alnumycin S (7). Proposed shunt products of the pathway are shown in gray and the various hydroxyl groups of the attached ribose are color coded for clarity.
Fig. 2.
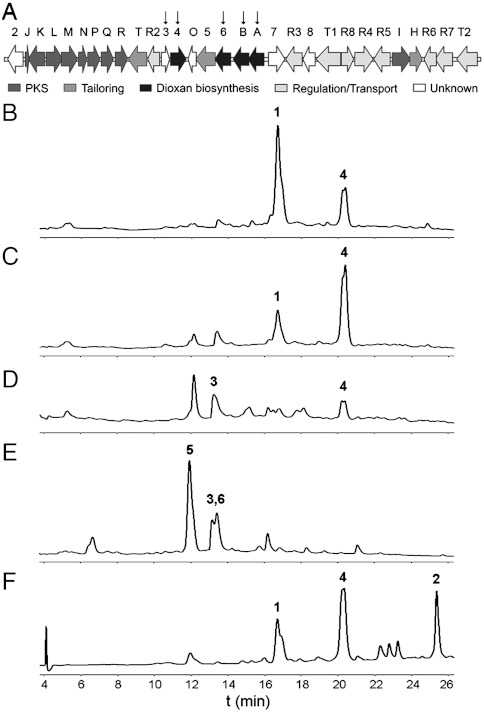
Schematic of the gene cluster and secondary metabolite production profiles. (A) The alnumycin biosynthetic gene cluster with the genes investigated in this study marked with an arrow; HPLC chromatograms (detection at 470 nm) of the secondary metabolite production profiles of (B) the heterologous expression system S. albus/pAlnuori, (C) S. albus/pAlnuoriΔaln3, (D) S. albus/pAlnuoriΔaln4, (E) S. albus/pAlnuoriΔaln6, and (F) enzymatic one-pot synthesis of alnumycin A (1) from d-ribose-5-phosphate and prealnumycin (2).
To elucidate the biosynthetic pathway responsible for attachment and synthesis of the dioxane unit, in this study we have (i) conducted incorporation experiments with 13C-labeled precursors, (ii) inactivated individual alnumycin biosynthetic genes and isolated several unique alnumycins from the knockout strains and finally, and (iii) used purified biosynthetic enzymes for reconstruction of the complete pathway in vitro.
Results and Discussion
Identification of the Biosynthetic Origin of the Dioxane Moiety.
First, universally labeled [U-13C] d-ribose was fed to cultures of the native alnumycin producer Streptomyces sp. CM020 and the heterologous host S. albus/pAlnuori. NMR analysis of 1 surprisingly indicated the incorporation of all five 13C atoms into the dioxane moiety (SI Appendix, Table S1). The result was verified with [1-13C] and [5-13C] labeled d-ribose, which enhanced the notion that the whole dioxane unit originated from d-ribose-5-phosphate or a closely related metabolite from the pentose phosphate pathway. Specifically, the origin of C-1′ in 1 was the C-1 carbon of d-ribose, whereas the [5-13C] label was enriched both at C-3′ and C-6′ suggesting alternative incorporation patterns of the four-carbon unit (C3′-C6′).
Gene Inactivation Experiments and Isolation of Unique Alnumycins.
To identify all of the genes required for the biosynthesis of the dioxane unit, we inactivated several open reading frames of unknown function residing in cosmid pAlnuori by λ red recombineering (15) in Escherichia coli K12. The modified cosmids were conjugated into S. albus from E. coli ET12567/pUZ8002 and the strains were screened for production of compounds with the distinct alnumycin UV-visible spectrum, but a different retention time to 1 and 2 in HPLC. Three mutants with desired production profiles were found indicating that the gene products might be involved in late-stage modification steps after formation of the polyketide aglycone.
For one strain, inactivation of a small open reading frame aln3 (Fig. 2A) resulted in the accumulation of an unknown secondary metabolite with the expected properties, along with minor quantities of 1 (Fig. 2C). The metabolite was also detected in lesser amounts from a previously reported aln4 mutant (Fig. 2D) that produced K1115 A (3, Fig. 1) as its main metabolite, but crucially the biosynthesis of 1 was completely abolished in the strain. At the time, Aln4 was annotated to be involved in formation of the alnumycin chromophore due to the incorrect cyclization of the third ring in 3 (14). The unidentified metabolite was duly isolated and purified from a S. albus/pAlnuoriΔaln3 cultivation, and structure elucidation of the compound (SI Appendix) confirmed the presence of a unique dioxolane (cis-bicyclo[3.3.0]-2′,4′,6′-trioxaoctan-3′β-ol) moiety in place of the dioxane unit. Furthermore, the structural analysis showed that, in fact, two isomers differing at C-1′ were present, denoted as alnumycin B1 (4a, major isomer, 57%, Fig. 1) and alnumycin B2 (4b, 43%, Fig. 1).
In cultures of the aln6 mutant strain S. albus/pAlnuoriΔaln6, two previously undetected metabolites were observed (Fig. 2E). Purification and structure determination of the compounds (SI Appendix) revealed that the main metabolite was yet again present as a C-1′ epimeric pair, as alnumycin C1 (5a, major isomer 76%, Fig. 1) and alnumycin C2 (5b, 24%, Fig. 1). The structural data verified that the compounds contained a ribose unit in furanose form attached to the alnumycin chromophore via a C8-C1′ bond. Spectroscopic analysis of the minor metabolite, alnumycin D (6, Fig. 1), produced by the strain revealed that the compound contained a ribose unit attached in a pyranose form and was found to be a stereochemically pure β anomer (SI Appendix).
The C-Ribosylation of Prealnumycin.
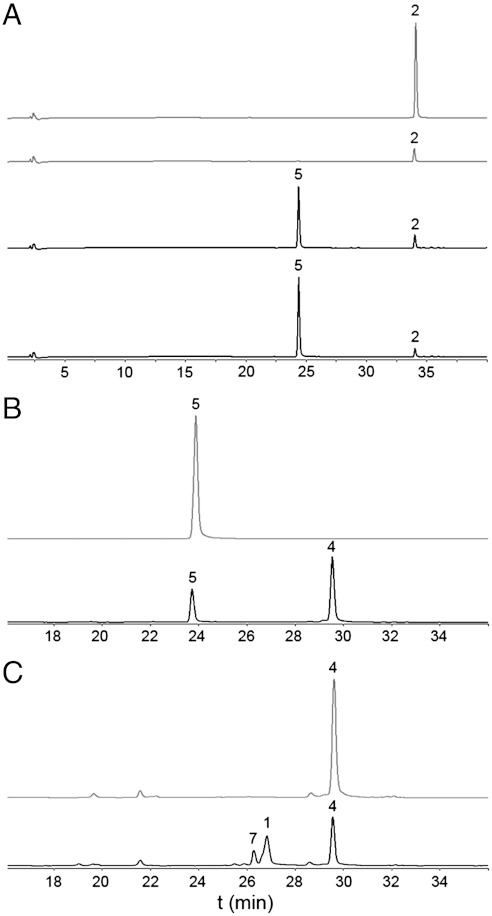
Previous gene inactivation experiments suggested that AlnA and AlnB were responsible for formation of the C8-C1′ bond due to the observation that both single-gene knockout mutants produced 2 as their main metabolite. Sequence analysis revealed that AlnA (305 amino acids, 31.5 kDa) belongs to the recently discovered enzyme family of pseudouridine C-glycosidases such as YeiN (41% identity) and TM1464 (28% identity) from E. coli and Thermotoga maritima, respectively. Recently these enzymes have been shown to cleave pseudouridine-5′-phosphate to uracil and d-ribose-5-phosphate (16). On the other hand, AlnB (226 amino acids, 22.8 kDa) is predicted to be a phosphatase from the well-characterized haloacid dehalogenase (HAD) superfamily (17, 18). When the two enzymes were incubated in a coupled reaction together with 2 and d-ribose-5-phosphate, 5 could be isolated from the reaction mixture. The presence of both enzymes was requisite for the synthesis of 5 and lowering the oxygen concentration was found to be critical for efficient synthesis (Fig. 3A). Curiously, formation of 6 was detected serendipitously only under reducing conditions in the presence of NAD(P)H.
Fig. 3.
Stepwise in vitro synthesis of the dioxane unit in alnumycin A. The enzymatic reactions are drawn in black, and control reactions with boiled enzymes are in gray in the individual chromatograms shown at 470 nm. The reactions are from top to bottom. (A) Control with boiled AlnA, control with boiled AlnB, enzymatic synthesis of alnumycin C (5) from 1 mM d-ribulose-5-phosphate and 0.3 mM prealnumycin (2) by 30 μM AlnA and 10 μM AlnB (3.5 h incubation), and enzymatic synthesis of 5 with d-ribose-5-phosphate in place of d-ribulose-5-phosphate. (B) Control reaction with boiled Aln6, enzymatic synthesis of alnumycin B (4) from 0.3 mM 5 by 3 μM Aln6 (aerobic 1 h incubation). (C) Control reaction with boiled Aln4, enzymatic synthesis of alnumycin A (1), and alnumycin S (7) from 0.3 mM 4 and 4 mM NADPH by 15 μM Aln4 (90 min incubation).
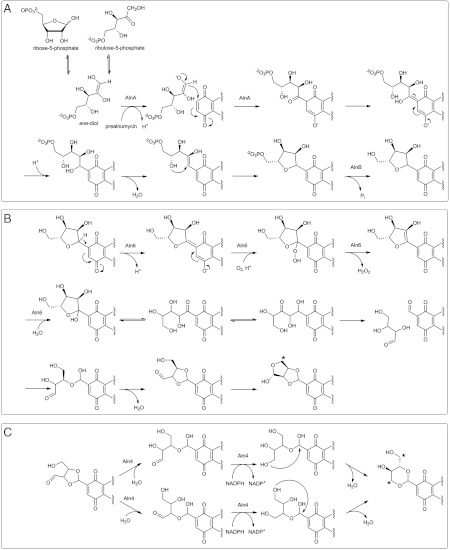
The experiment indicated that AlnA was responsible for formation of the C8-C1′ bond and attachment of d-ribose-5-phosphate in a reaction similar to that of YeiN, although in the opposite direction, whereas AlnB was a phosphatase in accordance to the function predicted from the sequence analysis (18) (Fig. 4A). This observation led us to classify AlnA as a natural C-glycosynthase, in analogy to the unrelated classical O- and S-glycosidases that have been engineered to perform as glycosynthases (19). Surprisingly, the reaction could also be conducted efficiently using d-ribulose-5-phosphate as an alternative substrate (Fig. 3A), which indicated that it may be the ene-diol intermediate of d-ribose-5-phosphate and d-ribulose-5-phosphate, which is the reactive species in the AlnA reaction. More specifically, it is plausible that the C8-C1′ bond is formed through a Michael reaction where the ene-diolate form of d-ribose-5-phosphate acts as a nucleophile and attacks the C-8 of 2 (Fig. 4A). Next, the polyaromatic chromophore system of 2 could be restored, followed by C1′-C2′ dehydration that could allow cyclization of the ribose side chain through an electrophilic addition of an alcohol to an alkene (Fig. 4A). The cyclization might even occur nonenzymatically, which would best explain formation of both 6 and the two stereoisomers 5a and 5b after dephosphorylation by AlnB. In any case, the experiments indisputably confirmed that the AlnA/AlnB system presents a unique means for attachment of carbohydrates into natural products in addition to the unrelated and much studied glycosyl transferases. Furthermore, inspection of available genome sequences reveals that genes homologous to alnA and alnB are found clustered in many cases, which suggests that similar C-ribosylation may occur on various other pathways that are yet to be discovered.
Fig. 4.
Proposed mechanisms for reactions catalyzed by AlnA, AlnB, Aln6, and Aln4. (A) Attachment of ribose onto 2 by AlnA and AlnB. (B) Formation of dioxolane from the ribose moiety in 5 by Aln6. (C) Conversion of the dioxolane in 4 into a dioxane unit by Aln4. The position of the incorporated label from [5-13C] d-ribose feeding experiments is marked with an asterisk.
Rearrangement of Ribose into the Unique Dioxolane Unit of Alnumycin Bs.
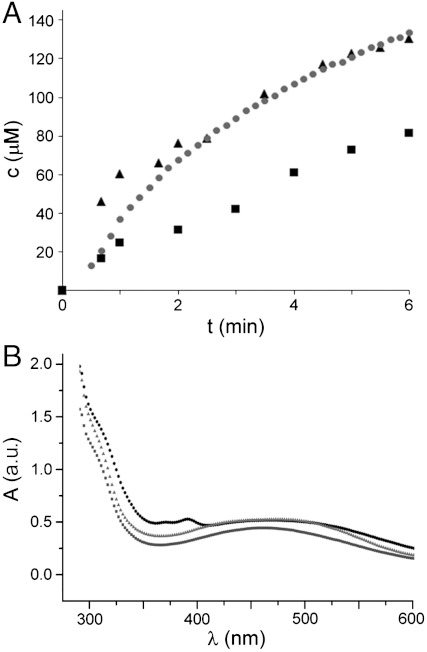
According to molecular genetic studies, Aln6 (358 amino acids, 39.1 kDa), which is homologous to enzymes found in the related benzoisochromanequinone (BIQ) pathways such as ActVA-ORF3 (29% identity) (20) and Gra-ORF28/Gra-ORF30 (26/28% identity) (21) from the actinorhodin and granaticin pathways, respectively, would catalyze the next biosynthetic step (Fig. 1). This supposition was confirmed in vitro as 5 was readily converted into 4 by Aln6 (Fig. 3B), though 6 was not accepted as a substrate and appeared to be a shunt product of the pathway. The rearrangement of the ribose unit was accompanied with consumption of O2 and formation of H2O2, which led us to identify Aln6 as an oxidase (Fig. 5A). The C1′-C2′ cleavage site was also verified by feeding [5-13C] and [1-13C] d-ribose in vivo, which showed clear enrichment at positions 5′ and 1′ in 4, respectively (SI Appendix, Table S2).
Fig. 5.
Monitoring the Aln6 reaction. (A) Simultaneous formation of 4 (black triangles) and hydrogen peroxide (black squares) as estimated by an end-point assay vs. consumption of molecular oxygen (gray dots) in a continuous measurement. All reactions contained 170 μM 5 and 1.4 μM Aln6. (B) Observed changes in the UV-visible spectra during 80% conversion of 230 μM 5 to 4 by 1.6 μM Aln6. Reaction mixture is shown at the initial state (dark gray squares), after 2 min (black circles), and after 10 min incubation (light gray triangles).
Direct addition of O2 to organic molecules is a spin-forbidden reaction, which is usually overcome by utilization of transition metals or organic cofactors to break the spin barrier (22). Therefore, it was unexpected that no cofactors (heme, flavin, etc.) were detected during protein purification and the Aln6 reaction could be performed under high concentrations of metal chelating compounds (EDTA/EGTA). In spite of the complications in oxygen activation, an increasing number of monooxygenases, which are devoid of any coenzymes, have been described in recent years. A common theme emerging from studies of these diverse enzymes is that dioxygen is activated by the organic substrate rather than the protein (22). In the case of Aln6, we speculate that in the initial step abstraction of a proton from 5 generates a highly conjugated anion intermediate, which might react with oxygen at position C1′ (Fig. 4B). After formation of a hydroperoxy intermediate, the reaction might proceed in a similar manner to the well-characterized urate oxidase (23, 24) through elimination of H2O2 and hydration of the C1′-C2′ alkene to form a C1′ hydroxylated intermediate. Distinctive changes can be detected in the UV-visible spectral properties during the reaction (Fig. 5B), which supports our hypothesis that the conjugated alnumycin chromophore is utilized in the reaction.
The unusual carbon-carbon bond cleavage is likely to be as complex as the initial step requiring molecular oxygen. We propose that after opening of the lactol ring of the C1′ hydroxylated intermediate, enzyme catalyzed tautomerization could plausibly result in formation of a β-hydroxy carbonyl compound, which would be suitable for retro-aldol cleavage. Finally, acetal formation would yield the end product of the reaction (Fig. 4B).
The exceedingly complex oxidative rearrangement process of 5 to 4 undoubtedly requires further extensive biochemical and structural characterization, but our data unequivocally identifies Aln6 as an alnumycin oxidase/riboisomerase. Based on sequence similarities Aln6 belongs to a previously uncharacterized enzyme family, as the functions of the homologous enzymes from the related BIQ pathways have been highly elusive.
Formation of the Dioxane Moiety and Enzymatic Synthesis of Alnumycin S.
When 4 was incubated together with Aln4 in the presence of NADPH, 1 could be isolated from the reaction mixture (Fig. 1, Fig. 3C). This finding was surprising because based on previous molecular genetic evidence, the aldo-keto reductase Aln4 (354 amino acids, 39.2 kDa) was implicated to be involved in cyclization of the third ring of the polyketide aglycone (14). Nonetheless, the aldoreduction of position 3′ of 4 by Aln4 was in agreement with the proposed function of the homologous enzyme AFB1-AR (26% identity) involved in the degradation of aflatoxin (25), although the reaction appeared to be more complicated due to the additional rearrangement of the dioxolane unit of 4 to the dioxane moiety seen in 1. The Aln4 in vitro reaction products included, in addition to 1, another unidentified compound with the characteristic alnumycin chromophore (Fig. 3C). The enzymatic synthesis was therefore conducted on a preparative scale and the 1H NMR (SI Appendix) data confirmed the presence of a unique cis-3′,3″-di(hydroxymethyl)-2′,2″-dioxolane unit in alnumycin S (7, Fig. 1). Compound 7, however, appeared to be a shunt product of the pathway because it could not be further converted to 1 by Aln4. The formation of 7 from 4 can be easily explained by aldoreduction of the open-chain form of the γ-butyrolactol moiety. Instead, conversion into 1 requires opening of both lactol and dioxolane rings. The incorporation of the [5-13C] d-ribose label at C-3′ and C-6′ indicates that the dioxolane rings may be opened in two different ways (Fig. 4C). Subsequent aldoreduction and acetal formation at the newly formed hydroxyl group would then result in the formation of the end product of the pathway (Fig. 4C).
The in vitro confirmation that Aln4 alone was able to catalyze the last biosynthetic step refuted the initial anticipation that also Aln3 (155 amino acids, 16.5 kDa), which belongs to a very recently described family of F420-dependent reductases involved in aflatoxin degradation (26), would be required. The genes aln3 and aln4 putatively reside in the same operon in the alnumycin gene cluster (Fig. 2A) and therefore a downstream effect might explain the phenotypes of the mutants. In any case, the fact that the aln3 knockout strain was still capable of producing 1 demonstrates that Aln3 only has an accessory role, if any, in the biosynthesis.
One-Pot Enzymatic Synthesis of Alnumycin A.
In addition to the stepwise enzymatic synthesis described above, we were able to establish a one-pot enzymatic synthesis of 1 by incubating AlnA (8 μM), AlnB (7 μM), Aln6 (3 μM), and Aln4 (21 μM) in the presence of NADPH (4 mM) and the substrates 2 (0.5 mM) and d-ribose-5-phosphate (2 mM) (Fig. 2F). The use of the glucose oxidase system to reduce the oxygen concentration was paramount for successful conversion, and consistent with the production profiles of the heterologous host in vivo (Fig. 2B), substantial quantities of 4 remained in the reaction mixture.
Concluding Remarks
Carbohydrate moieties are arguably one of the most important means to generate molecular diversity in biologically active natural products. Here we have demonstrated in detail how nature has found an alternative means to generate structures that mimic deoxysugars. Importantly, this attachment is accomplished via a carbon-carbon bond, which has been discussed in context with glycosyl transferases as an exciting means to generate hydrolytically resistant natural products. The experiments described here unambiguously demonstrate that we have successfully characterized and reconstituted the entire pathway required for the biosynthesis and attachment of the 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane moiety of 1. The results set the stage for more detailed mechanistic studies of the exceptional biosynthetic enzymes to gain a better understanding of the chemistry underlying the biological synthesis of the sugar-like dioxane unit. The work also enables interesting opportunities for utilization of ribose, dioxolane, and dioxane moieties in generation of unique natural products with improved biological activities. Both avenues are currently being actively pursued in our laboratory.
Materials and Methods
Bacterial Strains and Culture Conditions.
For production of the metabolites, Streptomyces strains were routinely cultivated in 25 mL of NoS-E1soy medium (27) with added Amberlite XAD 7HP (Rohm and Haas) resin (1 g/50 mL) for 5 d at 300 rpm, 301 K. MS plates (28) were used as a solid medium for S. albus (29). Apramycin at 50 μg/mL and thiostrepton at 50 μg/mL and 40 μg/mL were used for selection in solid and liquid media, respectively. E. coli ET12567/pUZ8002 (28) and TOP10 (Invitrogen) were used for intergeneric conjugation and protein production, respectively. E. coli strains were cultivated in Luria–Bertoli or 2 × yeast extract/Tryptone medium supplemented with appropriate antibiotics.
General DNA Techniques and Inactivation of Genes.
Isolation of plasmid and cosmid DNA from Streptomyces species was performed using conventional techniques. The open reading frames aln3 (468 bp) and aln6 (1,080 bp) were deleted from the shuttle cosmid pAlnuori, as described previously (14), in two steps using 70 nt primers (SI Appendix, Table S3) and the λ red recombinase-Flp recombinase (15) system, resulting in pAlnuoriΔaln3 and pAlnuoriΔaln6, respectively.
Analysis of Metabolites.
Samples were analyzed using an SCL-10Avp HPLC system equipped with an SPD-M10Avp diode array detector (Shimadzu) and a LiChroCART 250-4 RP-18 column (5 μm, Merck) eluted with 0.1% aqueous formic acid and a 55–100% acetonitrile gradient. Preparative HPLC was conducted using L-6200A HPLC system (Merck Hitachi). High resolution electron impact-MS data was measured using a ZABSpec mass spectrometer (VG Analytical). HPLC-electrospray ionization (ESI)-MS were acquired using a MicrOTOF-Q mass spectrometer (Bruker) and an Agilent Technologies 1200 Series HPLC system equipped with a diode array detector and a SunFire C18 column (3.5 μm, Waters) eluted with 0.1% aqueous formic acid and a 15–100% acetonitrile gradient. A full description on the production, isolation, and analysis of Streptomyces secondary metabolites is presented in SI Appendix.
Production and Purification of Recombinant Proteins.
The primers used for cloning of the genes into the modified pBADHisB (Invitrogen) vector (30) are provided in SI Appendix, Table S3. Coexpression of chaperones encoded by pG-KJE8 (Takara Bio, Inc.) increased the yield of His7-Aln4. Protein production was induced at OD600 = 0.5–0.7 with 0.02% l-arabinose, after which the cultures were grown at 295 K overnight, harvested at 277 K, and lysed using French Press. Triton X-100 was then added to a final concentration of 0.5% (vol/vol). The proteins were purified in a single affinity chromatography step using Talon Cobalt resin (Clontech), 1 mL HIS-select Cartridge (Sigma, AlnA), or 5 mL HisTrap Ni2+- column (Aln4) as part of an ÄKTA FPLC system (GE Healthcare). After elution, the buffer was exchanged into either 50 mM Hepes (pH 7.2), 150 mM NaCl, 10% glycerol, 5 mM MgCl2 (AlnA, AlnB and Aln6), or 20 mM Trizma-HCl (pH 6.8 vs. 6), 50 mM NaCl, 50 mM KCl, 10% glycerol (Aln4), and frozen in 50% glycerol. Protein concentrations were estimated by Bradford assay. Typical purity of each enzyme preparation is shown in SI Appendix, Fig. S1.
Enzyme Activity Assays.
Reactions were set up in 10 mM Trizma-HCl (pH 6.8) (pH 6 for Aln6), 25 mM NaCl, 25 mM KCl, ≥10% glycerol, and substrates in 2–5% DMSO. For all enzymes except Aln6, the oxygen concentration was reduced using the glucose oxidase (100 nM)-catalase (1.5 μM) system in 60 mM d-glucose. Incubation for 1–3.5 h at 288 K (296 K for Aln6) was followed by extraction with 3 × 0.5–1 volume of CHCl3. The CHCl3 extracts were air-dried and dissolved in acetonitrile for HPLC analysis, which was conducted using either the LiChroCART 250-4 column eluted with 20 mM aqueous ammonium acetate and a 55–100% acetonitrile gradient (Fig. 2), or the SunFire C18 column eluted with 0.1% aqueous formic acid and a 15–100% acetonitrile gradient (Fig. 3). Oxygen consumption by Aln6 was monitored at 296 K with an MI-730 oxygen electrode (Microelectrodes, Inc.). For end-point assays with Aln6, individual 100 µL reactions were quenched by chloroform extraction followed by quantitative HPLC analysis of each organic phase, and a peroxidase assay of each water phase to estimate the hydrogen peroxide concentration. The latter was monitored spectrophotometrically at 734 nm using 0.23 mg/mL peroxidase, 0.5 mM 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) and known concentrations of hydrogen peroxide as a standard. Formation of a stoichiometric amount of H2O2 and 4 could not be observed because of increased instability of H2O2 caused by Aln6. The preparative scale enzymatic production of 7 is described in SI Appendix. The high resolution mass spectra for both 7 and the enzymatically synthesized 1 and 4–6 were obtained by HPLC-ESI-MS as described above.
NMR Experiments.
A full description of the NMR methodology is given in SI Appendix.
Supplementary Material
Acknowledgments.
We thank Prof. Gunter Schneider for helpful discussions, Nadine Dreiack for assistance with some of the chromatographic separations, and Dr. Olli Martiskainen for acquiring high resolution mass spectra. This study was supported by the National Graduate School in Informational and Structural Biology (ISB); the Academy of Finland Grants 121688, 136060, and 127844; and the Turku University Foundation. We acknowledge CSC–IT Center for Science Ltd. for computing resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201530109/-/DCSupplemental.
References
- 1.Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 2.Schlunzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase center in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 3.Cipollone A, et al. Novel anthracycline oligosaccharides: Influence of chemical modifications of the carbohydrate moiety on biological activity. Bioorg Med Chem. 2002;10:1459–1470. doi: 10.1016/s0968-0896(01)00411-4. [DOI] [PubMed] [Google Scholar]
- 4.Yang DZ, Wang AHJ. Structure by NMR of antitumor drugs aclacinomycin A and aclacinomycin B complexed to D(CGTACG) Biochemistry. 1994;33:6595–6604. doi: 10.1021/bi00187a029. [DOI] [PubMed] [Google Scholar]
- 5.Luzhetskyy A, Mendez C, Salas JA, Bechthold A. Glycosyltransferases, important tools for drug design. Curr Top Med Chem. 2008;8:680–709. doi: 10.2174/156802608784221514. [DOI] [PubMed] [Google Scholar]
- 6.Thibodeaux CJ, Melancon CE, Liu HW. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 7.Kren V, Martínková L. Glycosides in medicine: The role of glycosidic residue in biological activity. Curr Med Chem. 2001;8:1303–1328. doi: 10.2174/0929867013372193. [DOI] [PubMed] [Google Scholar]
- 8.Weatherman RV, Mortell KH, Chervenak M, Kiessling LL, Toone EJ. Specificity of C-glycoside complexation by mannose/glucose specific lectins. Biochemistry. 1996;35:3619–3624. doi: 10.1021/bi951916z. [DOI] [PubMed] [Google Scholar]
- 9.Bililign T, Griffith BR, Thorson JS. Structure, activity, synthesis and biosynthesis of aryl-C-glycosides. Nat Prod Rep. 2005;22:742–760. doi: 10.1039/b407364a. [DOI] [PubMed] [Google Scholar]
- 10.Hultin PG. Bioactive C-glycosides from bacterial secondary metabolism. Curr Top Med Chem. 2005;5:1299–1331. doi: 10.2174/156802605774643015. [DOI] [PubMed] [Google Scholar]
- 11.Mittler M, Bechthold A, Schulz GE. Structure and action of the C-C bond-forming glycosyltransferase UrdGT2 involved in the biosynthesis of the antibiotic urdamycin. J Mol Biol. 2007;372:67–76. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Bieber B, Nuske J, Ritzau M, Grafe U. Alnumycin a new naphthoquinone antibiotic produced by an endophytic Streptomyces sp. J Antibiot. 1998;51:381–382. doi: 10.7164/antibiotics.51.381. [DOI] [PubMed] [Google Scholar]
- 13.Naruse N, Goto M, Watanabe Y, Terasawa T, Dobashi K. K1115 A, a new anthraquinone that inhibits the binding of activator protein-1 (AP-1) to its recognition sites. II. Taxonomy, fermentation, isolation, physico-chemical properties and structure determination. J Antibiot. 1998;51:545–552. doi: 10.7164/antibiotics.51.545. [DOI] [PubMed] [Google Scholar]
- 14.Oja T, et al. Characterization of the alnumycin gene cluster reveals unusual gene products for pyran ring formation and dioxan biosynthesis. Chem Biol. 2008;15:1046–1057. doi: 10.1016/j.chembiol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preumont A, Snoussi K, Stroobant V, Collet JF, Van Schaftingen E. Molecular identification of pseudouridine-metabolizing enzymes. J Biol Chem. 2008;283:25238–25246. doi: 10.1074/jbc.M804122200. [DOI] [PubMed] [Google Scholar]
- 17.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 19.Hancock SM, Vaughan MD, Withers SG. Engineering of glycosidases and glycosyltransferases. Curr Opin Chem Biol. 2006;10:509–519. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Caballero JL, Martinez E, Malpartida F, Hopwood DA. Organization and functions of the actVA region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor. Mol Gen Genet. 1991;230:401–412. doi: 10.1007/BF00280297. [DOI] [PubMed] [Google Scholar]
- 21.Ichinose K, et al. The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tu22: Sequence analysis and expression in a heterologous host. Chem Biol. 1998;5:647–659. doi: 10.1016/s1074-5521(98)90292-7. [DOI] [PubMed] [Google Scholar]
- 22.Fetzner S, Steiner R. Cofactor-independent oxidases and oxygenases. Appl Microbiol Biotechnol. 2010;86:791–804. doi: 10.1007/s00253-010-2455-0. [DOI] [PubMed] [Google Scholar]
- 23.Gabison L, et al. Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: Mechanistic implications. BMC Struct Biol. 2008;8:32–39. doi: 10.1186/1472-6807-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tipton PA. Urate oxidase: Single-turnover stopped-flow techniques for detecting two discrete enzyme-bound intermediates. Methods Enzymol. 2002;354:310–319. doi: 10.1016/s0076-6879(02)54025-8. [DOI] [PubMed] [Google Scholar]
- 25.Hayes JD, Judah DJ, Neal GE. Resistance to aflatoxin B1 is associated with the expression of a novel aldo-keto reductase which has catalytic activity towards a cytotoxic aldehyde-containing metabolite of the toxin. Cancer Res. 1993;53:3887–3894. [PubMed] [Google Scholar]
- 26.Taylor MC, et al. Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol Microbiol. 2010;78:561–575. doi: 10.1111/j.1365-2958.2010.07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siitonen V, et al. Identification of late-stage glycosylation steps in the biosynthetic pathway of the anthracycline nogalamycin. ChemBioChem. 2012;13:120–128. doi: 10.1002/cbic.201100637. [DOI] [PubMed] [Google Scholar]
- 28.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 29.Chater KF, Wilde LC. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol. 1980;116:323–334. doi: 10.1099/00221287-116-2-323. [DOI] [PubMed] [Google Scholar]
- 30.Kallio P, Sultana A, Niemi J, Mäntsälä P, Schneider G. Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: Implications for catalytic mechanism and product stereoselectivity. J Mol Biol. 2006;357:210–220. doi: 10.1016/j.jmb.2005.12.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.