Abstract
The translationally controlled tumor protein (TCTP) is essential for survival by mechanisms that as yet are incompletely defined. Here we describe an important role of TCTP in response to DNA damage. Upon exposure of normal human cells to low-dose γ rays, the TCTP protein level was greatly increased, with a significant enrichment in nuclei. TCTP up-regulation occurred in a manner dependent on ataxia-telangiectasia mutated (ATM) kinase and the DNA-dependent protein kinase and was associated with protective effects against DNA damage. In chromatin of irradiated cells, coimmunoprecipitation experiments showed that TCTP forms a complex with ATM and γH2A.X, in agreement with its distinct localization with the foci of the DNA damage-marker proteins γH2A.X, 53BP1, and P-ATM. In cells lacking TCTP, repair of chromosomal damage induced by γ rays was compromised significantly. TCTP also was shown to interact with p53 and the DNA-binding subunits, Ku70 and Ku80, of DNA-dependent protein kinase. TCTP knockdown led to decreased levels of Ku70 and Ku80 in nuclei of irradiated cells and attenuated their DNA-binding activity. It also attenuated the radiation-induced G1 delay but prolonged the G2 delay. TCTP therefore may play a critical role in maintaining genomic integrity in response to DNA-damaging agents.
Keywords: low dose ionizing radiation, adaptive responses, DNA repair, cell cycle checkpoints, genomic stability
Ionizing radiation is used extensively to investigate the molecular events involved in the sensing and repair of DNA damage. Our major focus is to understand the cellular responses to low doses of radiation that mimic human exposure during diagnostic radiography or occupational activities. We previously observed important adaptive responses when normal human cells were exposed to low doses of 137Cs γ rays. Specifically, these irradiated cells exhibited significantly less chromosomal damage than occurred spontaneously in the corresponding nonirradiated cells (1). To gain insight into this protective mechanism elicited by low doses of γ rays, we used a proteomic approach to identify differentially expressed proteins. Among many regulated proteins, the translationally controlled tumor protein (TCTP) was found to be most sensitive, having its level increased by twofold in low-dose–irradiated cells.
TCTP is highly conserved and abundantly expressed in eukaryotes. It is unique in that it has no sequence similarity to other known proteins. Homozygous mutant (TCTP−/−) mice are embryonically lethal (2). TCTP is known to participate in numerous cellular processes including protein synthesis and cell growth, thereby explaining the essential nature of the protein in cell survival. Interestingly, down-regulation of TCTP leads to tumor reversion (reviewed in refs. 3 and 4). In general, TCTP is thought to promote cell viability through an antiapoptotic role (5). However, a role for TCTP in other prosurvival mechanisms is unclear.
Here we tested our hypothesis that TCTP supports the survival and genomic integrity of irradiated cells through a role in the DNA damage response (DDR). Our data show that TCTP interacts with ataxia-telangiectasia mutated (ATM) and p53 proteins, two major early mediators of DNA damage sensing. We also find that in irradiated cells the level of TCTP is regulated in an ATM- and DNA-dependent protein kinase (DNA-PK)–dependent manner. After DNA damage, TCTP accumulates in the nucleus where it colocalizes at sites of DNA damage with proteins involved in DNA damage sensing and repair. In G1-phase cells, it promotes binding to damaged DNA of the Ku proteins that are components of the nonhomologous end-joining (NHEJ) mode of DNA double-strand break (DSB) repair. In TCTP knockdown cells, DNA damage accumulates, and repair of radiation-induced chromosomal damage is compromised. TCTP also was found to regulate the radiation-induced G1 and G2 checkpoints, therefore likely maintaining genomic stability. These data identify a role for TCTP in DNA damage-sensing and repair pathways.
Results
TCTP Induced by Low-Dose Radiation Protects Against DNA Damage.
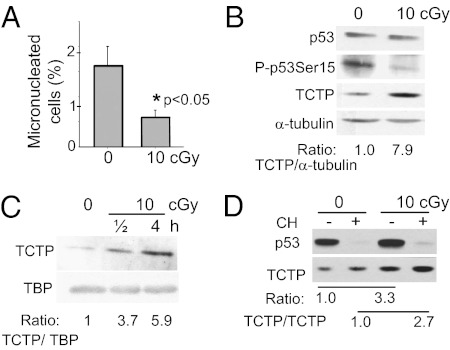
Micronuclei (MN) are indicators of chromosomal damage and arise mainly from DSBs. When confluent AG1522 normal human fibroblasts were subjected to protracted exposure to low-dose 137Cs γ rays (10 cGy delivered over 50 h), MN formation decreased (P < 0.05) even below the spontaneous level (Fig. 1A). Likewise, phosphorylation of p53 on serine 15, another marker of DNA damage, also was reduced (Fig. 1B). These results suggest that low-dose radiation triggers mechanisms that protect DNA from basal oxidative damage. To understand the mediating events, we used the quantitative amine-specific isobaric tags for relative and absolute quantitation (iTRAQ) shotgun proteomics approach (6). Proteins were isolated from cells immediately after irradiation and analyzed. Levels of several proteins were found to be altered relative to sham-irradiated samples (Table S1). Among the proteins with increased levels, we focused on TCTP and confirmed, by Western blotting, that it was highly up-regulated in cells exposed to 10-cGy radiation delivered at a low dose rate (0.2 cGy/h) (Fig. 1B). TCTP up-regulation also was induced after acute (6 cGy/min) low-dose γ irradiation and resulted in approximately sixfold enrichment in nuclei by 4 h after exposure (Fig. 1C). Increased TCTP levels were detected as early as 30 min and as late as 48 h after acute 10 cGy irradiation (Fig. S1A). The induction of TCTP by low-dose γ rays appeared to be a general phenomenon that occurred in multiple cell strains grown in 2D or 3D architecture, in cells at the different phases of the cell cycle, and in tissues of irradiated mice (Fig. S1 A–E). TCTP also was induced following exposure to energetic protons(Fig. S1F).
Fig. 1.
Low-dose 137Cs γ rays up-regulate TCTP and induce protective effects. (A) MN formation in AG1522 cells exposed to no or 10-cGy radiation delivered at a low dose rate (0.2 cGy/h). Control and irradiated confluent cells were subcultured, and MN formation was evaluated in binucleated cells following 72 h incubation. (B) Western blot analyses in confluent AG1522 cells exposed to no or 10-cGy radiation (0.2 cGy/h) and harvested for analysis within 5 min after irradiation. (C) Western blot analyses of TCTP in nuclei of AG1522 confluent cell cultures exposed to no or to 10-cGy radiation at an acute dose rate (6 cGy/min). TBP was used as the loading control. (D) Western blot analysis of TCTP in AG1522 confluent cell cultures exposed to no or to 10-cGy radiation at an acute dose rate (6 cGy/min) in the presence (+) or absence (−) of cycloheximide (CH). Cells were incubated in cycloheximide 30 min before irradiation and were harvested for analysis 4 h later.
Incubation of control and γ-irradiated AG1522 cells for up to 30 h with cycloheximide, an inhibitor of cytosolic protein synthesis, had no significant effect on TCTP levels (Fig. S1G), indicating a long half-life of the protein in these cells. In contrast, p53 was undetectable in the same cells following 4.5 h-incubation (Fig. 1D). Acute irradiation of 10 cGy of γ rays up-regulated TCTP by threefold at 4 h after irradiation; incubation in cycloheximide for 0.5 h before irradiation and 4 h postirradiation increased TCTP by about the same amount. These results therefore imply stabilization of TCTP in low-dose–irradiated cells.
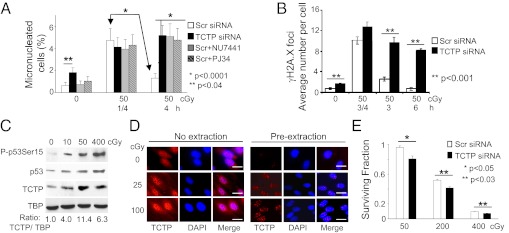
To determine the protective role of TCTP against DNA damage, we knocked down the protein in AG1522 cells using RNAi technology (Fig. S2A). We then assessed the accumulation of DSBs by MN (Fig. 2A) and the formation of γH2A.X foci as a function of time after γ irradiation (Fig. 2B). Confluent cells were subjected to no or 50-cGy acute-dose irradiation; they were subcultured 0.25 or 4 h later and were grown for additional 72 h to evaluate MN formation. In irradiated cells transfected with scrambled (Scr) siRNA, the sevenfold increase in MN frequency observed in cells held in quiescence for 0.25 h decreased by 3.5-fold (P < 0.0001) when the incubation period was prolonged to 4 h (Fig. 2A). These expected results are consistent with repair of DNA damage during the longer incubation period. In contrast, irradiated cells that were treated with siRNA targeting TCTP (siTCTP) showed no decrease in MN formation when held in the confluent state for 4 h before subculture (P < 0.0001) (Fig. 2A), perhaps because of the failure of DNA repair in the absence of TCTP. This notion is supported further by the observation that, even in nonirradiated cells, the knockdown of TCTP also resulted in an enhanced basal MN level (P < 0.04). These data suggest that knockdown of TCTP interferes with repair of DNA damage. This concept was substantiated when similar results were obtained with Scr siRNA-treated cells that were irradiated in the presence of the DNA repair inhibitors PJ34 or NU7441, which inhibit poly(ADP-ribose) polymerase and DNA-PK, respectively (7–9). Incubation of the drug-treated and irradiated cells for 4 h did not result in reduced MN formation (P < 0.0001) (Fig. 2A). Likewise, when Scr siRNA-transfected cells were exposed to acute-dose 50-cGy radiation and were held in confluence for 3 or 6 h after irradiation, the number of γH2A.X foci (a marker of DSBs) per cell decreased to 2.6 ± 0.4 and 0.7 ± 0.3, respectively, reflecting ongoing DNA repair (10). By contrast, the foci numbers in TCTP knockdown cells were higher (9.6 ± 1.0, P < 0.001, and 8.6 ± 0.4, P < 0.001, respectively) (Fig. 2B and Fig. S2B), suggestive of persistent DNA damage.
Fig. 2.
Involvement of TCTP in protection against DNA damage in γ-irradiated AG1522 cells. (A) MN formation in confluent cells that were subcultured 0.25 or 4 h after acute 50-cGy exposure. The cells were transfected with Scr or TCTP siRNA. Scr siRNA-transfected cells also were treated with PJ34 or NU7441, which inhibit DNA repair. (B) Analyses of γH2A.X foci in cells transfected with Scr or TCTP siRNA at different time points after irradiation with 50 cGy. For each time point 75–100 cells were counted. Data represent mean ± SD of three independent experiments. (C) Immunoblot analyses show that exposure to low- or high-dose γ rays up-regulates TCTP in the nuclei of confluent cells 30 min after exposure. (D) In situ immunodetection of TCTP. AG1522 cells were exposed to acute γ rays (25 or 100 cGy) 1 h before immunostaining for TCTP and DAPI counterstaining. Where indicated, the cells were preextracted with a buffer containing 0.5% Triton X-100 for 5 min before fixation. (Scale bars, 10 μm.) (E) Clonogenic survival of γ-irradiated cells transfected with Scr or TCTP siRNA.
Consistent with a function of TCTP in DDR (Fig. 2 A and B and Fig. S2B), Western blot analyses revealed four- to 11-fold increases in TCTP level in nuclei of AG1522 cells by 0.5 h after low- (10 cGy), moderate- (50 cGy), or high-dose (400 cGy) acute γ ray exposure (Fig. 2C), and these observations were confirmed further by in situ immunodetection (Fig. 2D, Left). When nucleoplasmic proteins were removed by detergent treatment, nuclear TCTP foci were clearly visible, and their number increased as a function of radiation dose (Fig. 2D, Right). Further, the induction of TCTP occurred in both the soluble nuclear fraction and the chromatin-enriched fraction of irradiated cells (Fig. S2C). Together, these results (Figs. 1 and 2) strongly support a role for TCTP in repair and/or sensing of DNA damage induced by low to moderate doses of radiation. The protective effects of TCTP against the lethal effects of high therapeutic doses of γ rays also were apparent. Knockdown of TCTP enhanced reproductive failure in AG1522 cells exposed to 200- or 400-cGy radiation (Fig. 2E). Although the effect was modest, it was statistically significant (P < 0.03). These protective effects at both low and high radiation doses are consistent with prosurvival functions of TCTP (3, 4).
Upstream Regulatory Events.
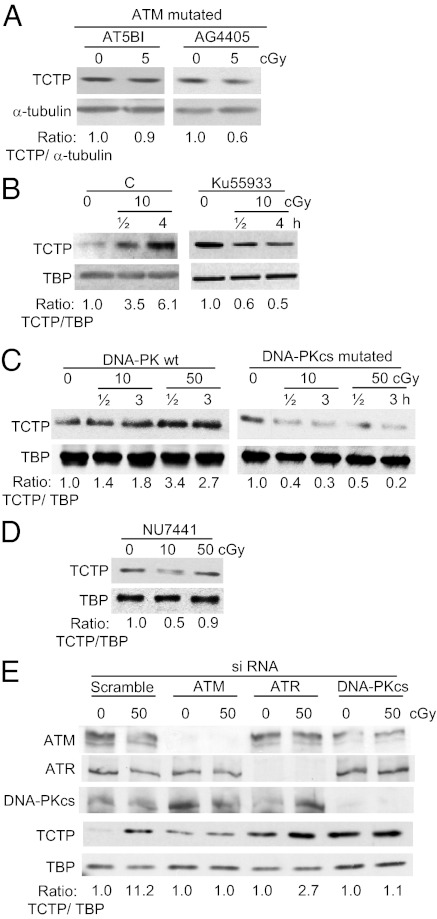
The role of ATM in mediating the cellular responses to DNA damage is well established (11). To investigate whether ATM mediates up-regulation of TCTP by low-dose γ rays, we exposed confluent radiosensitive ATM mutant cells (AT5B1 and AG4405) to acute-dose 5-cGy radiation. Unlike WT cells (Figs. 1D and Fig. S1 A–D), a slight decrease in TCTP level was detected in the mutant cells at 1, 4, or 24 h after irradiation; representative data at 4 h are shown in Fig. 3A. The mutant cells may suffer from various modifications that might alter TCTP levels nonspecifically. To rule out this possibility, we treated moderately radiosensitive WT AG1522 confluent cells with Ku55933 (10 μM), a selective inhibitor of ATM kinase, 0.5 h before exposure to acute 10-cGy radiation. Cells were harvested at 0.5 and 4 h after exposure, and nuclei were extracted. The TCTP level was found to be decreased (Fig. 3B), suggesting a prominent role of ATM in TCTP expression. By contrast, vehicle-treated and irradiated WT cells exhibited increased levels of TCTP. Importantly, knockdown of ATM by siRNA abrogated the radiation-induced increase in TCTP level (Fig. 3E). These results show that ATM has a role in regulating and/or maintaining the increased level of TCTP in cells exposed to low-dose γ radiation.
Fig. 3.
Effects of ATM, DNA-PK, or ATR signaling in the regulation of TCTP levels in γ-irradiated confluent human cells. (A) Immunoblotting of ATM-mutated cells shows lack of TCTP induction by low-dose γ rays. (B) Immunoblots of nuclear extracts of AG1522 fibroblasts exposed to no or 10-cGy radiation in the absence (“C”) or presence of Ku55933 (10 μM), a selective inhibitor of ATM. (C) Immunoblot analysis in nuclear extracts from cells that are WT or mutated for DNA-PKcs. (D) Immunoblots of nuclear extracts from AG1522 cells irradiated in the presence of NU7441, a selective inhibitor of DNA-PK. (E) Immunoblots of nuclear extracts from nonirradiated and irradiated AG1522 cells transfected with Scr siRNA or siRNAs targeting ATM, ATR, or DNA-PKcs.
ATM belongs to the PI3KK family of proteins. To investigate whether other members of this family are involved in regulating the TCTP level in irradiated cells, DNA-PK WT and mutated cells were exposed to 10- or 50-cGy radiation. At 0.5 and 3 h postirradiation, the TCTP level was increased in nuclei of WT but not mutated cells (Fig. 3C). The role of DNA-PK in regulation of TCTP expression in irradiated cells was supported further in experiments with AG1522 cells incubated with NU7441 (Fig. 3D) or treated with siRNA against the catalytic subunit of DNA-PK, DNA-PKcs, (Fig. 3E). In contrast, knockdown of ATR seemed to have less effect on TCTP up-regulation in irradiated cells (Fig. 3E). Together, the data reveal TCTP as a target of ATM/DNA-PK signaling that is pivotal in the cellular response to DNA damage and general oxidative or reductive stresses. It is noteworthy, that inhibition/down-regulation of ATM, DNA-PKcs, or ATR in nonirradiated AG1522 cells resulted in increased levels of TCTP (Fig. 3 B and E), suggesting these proteins have differential effect(s) on TCTP expression in control and irradiated cells.
TCTP Exists in Complex with Components of DNA Damage Sensing and Repair.
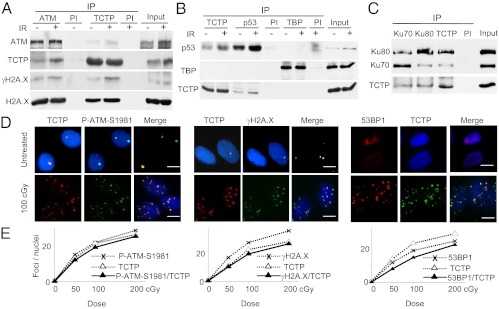
TCTP also was up-regulated in nuclei of low-dose γ-irradiated osteosarcoma U2OS cells with abundant ATM (Fig. S2D). We therefore determined if TCTP in these cells interacts with ATM and/or other proteins involved in DNA damage sensing and repair. A combination of immunoprecipitation (IP), quadrupole TOF (Q-TOF) MS, and immune-blotting showed that ATM exists in complex with TCTP in nuclei of both control and irradiated cells. Notably, coimmunoprecipitation in chromatin-enriched fractions revealed a reproducible interaction between TCTP and ATM in irradiated cells, and these proteins were found in a complex with γH2A.X (Fig. 4A). Further, TCTP and p53, another critical protein involved in DDR (12), also coimmunoprecipitated reciprocally in nuclear extracts of U2OS cells (Fig. 4B). The association of TCTP with p53 was enhanced at ∼0.5 h after exposure to 50 cGy radiation, suggesting that radiation-induced lesions may have a role in this interaction at early stage (Fig. 4B).
Fig. 4.
TCTP interacts with components of DNA damage sensing and repair. (A) Benzonase-treated chromatin-enriched fractions that were isolated from confluent U2OS cells 30 min after no (−) or acute [50 cGy, (+)] irradiation (IR) were immunoprecipitated with anti-ATM or anti-TCTP antibodies or normal mouse or rabbit serum (PI). Immunoblots then were reacted with antibodies against ATM, TCTP, γH2A.X, or H2A. X. (B) Benzonase-treated nuclear extracts isolated 30 min after exposure of U2OS confluent cells to no or acute (50 cGy) irradiation were immunoprecipitated with anti-TCTP, anti-p53, or control anti-TBP antibodies. Mouse or rabbit preimmune serum (PI) was used as a control. Immunoblotting was performed using antibodies against p53, TCTP, or TBP. (C) Immunoblotting of TCTP, Ku70, and Ku80 in Benzonase-treated nuclear extracts of control nonirradiated U2OS confluent cells after IP with either normal serum (PI) or antibodies against TCTP, Ku70, or Ku80. (D) Untreated or γ-irradiated (acute, 100 cGy) AG1522 asynchronous cells were preextracted, fixed 1 h later, and immunostained in situ with anti-TCTP, anti–P-ATM (S1981), anti-γH2A.X, or anti-53BP1 antibodies. (Scale bars, 10 μm.) (E) Quantitative assessment of colocalization of TCTP foci with those of P-ATM (S1981) (Left), γH2A.X (Center), and 53BP1 (Right) in AG1522 asynchronous cells at 1 h after exposure to 50, 100, or 200 cGy.
Like γH2A.X foci, P-ATM (S1981) and 53BP1 foci are markers of DSBs. Interestingly, TCTP foci colocalized with all of them in AG1522 (Fig. 4 D and E) and U2OS cells exposed to 100 cGy-acute dose of γ radiation (Fig. S3A), thereby identifying TCTP at sites of DNA breaks. In addition, Q-TOF MS and IP/Western blots identified Ku70 and Ku80, the DNA-binding subunits of DNA-PK that participate in NHEJ of DSBs (8, 13), as factors that form a complex with TCTP in nuclear extracts of U2OS (Fig. 4C and Fig. S2E) and AG1522 (Fig. S2F) cells. Taken together, these results strongly support a function of TCTP in sensing and repair of DNA damage induced by low (10 cGy), moderate (50 cGy), and high doses (100 cGy) of ionizing radiation.
Knockdown of TCTP Interferes with Subcellular Distribution and DNA-Binding Activity of Ku70/80 After Irradiation.
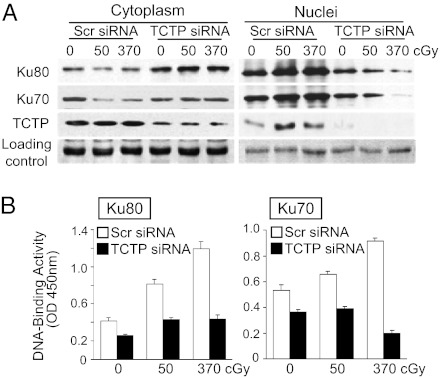
Compared with cells transfected with Scr siRNA, depletion of TCTP by siRNA resulted in opposite abundance patterns of Ku proteins in cytoplasm and nucleus of γ-irradiated cells (Fig. 5A and Fig. S3B). Whereas Ku70 and Ku80 proteins were increased in nuclei of control cells by 0.5 h after irradiation, presumably because of translocation from the cytoplasm to the nucleus in response to DNA damage (14), this increase did not occur in siTCTP-transfected cells. In the latter, Ku70 and Ku80 accumulated in cytoplasm (Fig. 5A, Left and Fig. S3B); in nuclei, the dramatic reduction in TCTP level was associated with radiation dose-dependent decrease in Ku70 and Ku80 (Fig. 5A, Right and Fig. S3B). Relative to Scr siRNA-treated cells, the decreases in Ku70 and Ku80 abundance in nuclei of irradiated siTCTP-transfected cells were associated with significant attenuation (>50%, P < 0.001) in the DNA-binding activity of Ku70 and Ku80 from extracts of irradiated cells (Fig. 5B). These data further support a role of TCTP in the function of main elements of NHEJ mode of DNA repair.
Fig. 5.
TCTP is involved in regulation of subcellular distribution and DNA-binding activity of Ku70 and Ku80 after γ irradiation. (A) Confluent U2OS cells transfected with Scr or TCTP siRNA were exposed to no, 50-, or 370-cGy radiation. Expression of TCTP, Ku70, and Ku80 in nuclei and cytoplasm fractions 30 min after irradiation was examined by Western blotting. Reaction of goat anti-rabbit IgG (sc-2030; Santa Cruz Biotechnology) with a protein of ∼30 kDa was used to verify equal loading of samples (loading control). (B) Ku70 (Right) and Ku80 (Left) DNA-binding activity in irradiated U2OS confluent cells expressing Scr or TCTP siRNA at 30 min after irradiation.
TCTP and Regulation of Radiation-Induced G1 and G2 Delays.
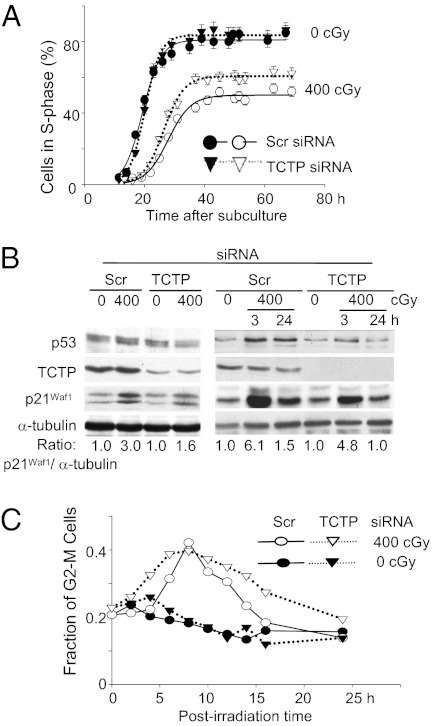
Although TCTP knockdown inhibits cell growth and induces apoptosis in tumor cells (15), down-regulation of TCTP for up to 72 h did not cause apparent structural changes in control AG1522 normal cells (Fig. S2A) or alter their cloning efficiency (28 ± 3% in Scr siRNA vs. 29 ± 1% in siTCTP-transfected cells). To examine whether TCTP affects other cell-growth parameters, we investigated its effect on the G1- to S-phase transition in control and γ-irradiated cells. Scr or TCTP siRNA-treated AG1522 cells synchronized in G0/G1 by density-inhibition of growth were exposed to no or 400-cGy radiation. Within 10 min after irradiation, they were subcultured to lower density and were incubated in the presence of [3H]-thymidine. Movement into the S phase was monitored by autoradiography by measuring the cumulative labeling indices at multiple time points up to 70 h after subculture. The transition time from G1 to S phase in nonirradiated cells was similar whether the cells were transfected with Scr siRNA or siTCTP (Fig. 6A). However, compared with Scr siRNA-treated cells, a 5.5-h reduction in the radiation-induced G1 delay occurred in cells treated with TCTP siRNA (∼9 h in TCTP siRNA-treated vs. 14.5 h in Scr siRNA-treated cells) (Fig. 6A). As well, relative to control, depletion of TCTP attenuated the induction of p21Waf1, a cyclin-dependent kinase inhibitor (16, 17) in irradiated confluent cells that were subcultured and harvested for analyses 24 h later (Fig. 6B, Left). The induction level of p21Waf1 also was reduced at 3 and 24 h after exposure of quiescent siTCTP-transfected cells to 400-cGy radiation (Fig. 6B, Right). The loss of normal G1 checkpoint control disrupts DNA repair and is an early step in carcinogenesis (18); therefore, these results are consistent with a role of TCTP in regulating a process that maintains genomic integrity.
Fig. 6.
Effect of TCTP knockdown in the γ-ray–induced G1 and G2 checkpoints in AG1522 fibroblasts. (A) Confluent cultures of Scr or TCTP siRNA-treated cells were exposed to no or 400-cGy radiation, were subcultured to lower density within 10 min after irradiation, and were grown in the presence of [3H]-thymidine. Cumulative labeling indices were measured as a function of time after subculture. (B) Western blot analyses reveal that knockdown of TCTP attenuates p21Waf1 induction in irradiated confluent cells that subsequently were subcultured to lower density or were maintained in the confluent, density-inhibited state. (C) The γ-ray–induced G2 delay in Scr or siTCTP asynchronous cells.
In addition to its effect on the stress-induced G1 checkpoint, flow cytometry analyses have shown that TCTP has a role in modulating the magnitude of the radiation-induced G2 checkpoint in asynchronous AG1522 cells. Similar to the absence of an effect on G1- to S-phase progression in control cells, knockdown of TCTP did not affect progression through G2 appreciably. However, siTCTP-transfected AG1522 cells exposed to 400-cGy radiation entered G2 phase faster than Scr siRNA–transfected cells and were arrested in G2 for a longer time (Fig. 6C). Whereas the earlier entry in G2 may be a consequence of the faster progression through G1 to S phase (Fig. 6A), the longer delay in G2 likely is caused by a greater level of DNA damage in irradiated cells (Fig. 2).
Discussion
TCTP is thought to protect cells against harmful conditions through antiapoptotic and antioxidant roles (19). The results described here identify TCTP as a member of the DDR group of proteins. Specifically, the protein is essential for eliciting effective responses to radiation-induced genotoxic stress. Our data establish a functional relation between TCTP and several key proteins such as ATM, Ku70, Ku80, and p53 that participate in DSB sensing and repair and/or regulation of the radiation-induced cell-cycle checkpoints. They are consistent with recent findings identifying p53 as a TCTP-interacting protein (20). In human cells exposed to low/moderate doses of γ rays, TCTP is up-regulated rapidly in an ATM- and DNA-PK–dependent manner (Fig. 3), followed by posttranslational stabilization (Fig. 1D). TCTP forms foci (Fig. 2D) that colocalize with those of P-ATM (S1981), γH2A.X, and 53BP1 that reflect sites of DNA damage (Fig. 4 D and E). This phenomenon is consistent with the observation that TCTP interaction with ATM and γH2A.X is enhanced in chromatin-enriched fractions following exposure to ionizing radiation (Fig. 4A). However, the exact nature of the interaction (direct or indirect) between TCTP and ATM kinase remains unknown. TCTP contains putative PI3KK phosphorylation sites (e.g., T39, S46, and S53), but in experiments involving IP of TCTP from AG1522 (WT ATM), U2OS (WT ATM), or AG4405 (mutated ATM) cells exposed to 50 cGy of γ rays, followed by in-gel trypsin digestion and analysis by liquid chromatography tandem MS (LC-MS/MS) on the LTC Orbitrap Velos instrument, we did not detect any phosphorylation site within the protein.
Further, TCTP coimmunoprecipitated with the Ku proteins (Fig. 4C) involved in NHEJ of DSBs. TCTP therefore may participate in repair of DSBs. This notion is substantiated further by the observation that TCTP knockdown prevents the repair of radiation-induced DNA damage leading to the accumulation of MN and γH2A.X foci (Fig. 2 A and B and Fig. S2B). The DSB is a serious DNA lesion that leads to cell death (21). Consistent with this concept, the accumulation of DNA damage in siTCTP cells results in enhanced cell killing (Fig. 2E). Importantly, TCTP is required for the DNA-binding activity of Ku70 and Ku80 in response to irradiation (Fig. 5B). Moreover, in TCTP-depleted cells exposed to radiation, Ku70 and Ku80 levels in nuclei are reduced (Fig. 5A and Fig. S3B), probably by disruption of the events implicated in Ku translocation to the nucleus. This reduction may imply a chaperone role of TCTP as described previously (22). In this context, our IP and MS analyses indicated that in irradiated cells TCTP also interacts with the stress-induced molecular chaperones, heat shock 90-kDa, 70-kDa, and 60-kDa proteins.
Damage to DNA in the form of DSBs can lead to carcinogenesis and cell death (23). Two mechanisms participate in the repair of DSBs: homologous recombination repair (HRR), acting mainly in the S and G2 phases of the cell cycle, and NHEJ, acting throughout the cell cycle (13). Our data show that TCTP has important effects on the Ku proteins. This finding is particularly important because Ku proteins are involved in DSB repair by NHEJ, and inactivation of Ku proteins leads to defects in telomere maintenance and chromosomal end fusion (24). Our IP/MS experiments also indicate that TCTP interacts with filamin A, which appears to be required for efficient HRR (25). The role of TCTP in such critical mechanisms that maintain genomic integrity may explain in part why homozygous mutation in TCTP is embryonically lethal (2).
TCTP is coimmunoprecipitated with p53 (Fig. 4B), a protein with an essential function in the radiation-induced G1 checkpoint (12). The knockdown of TCTP attenuates the γ-ray–induced G1 delay in p53 WT AG1522 cells but has no effect on normal progression from G1 to S phase (Fig. 6A). The attenuated stress-induced G1 checkpoint in these cells correlates well with the attenuated increase in the levels of p53 and of p21Waf1 that inhibits the activity of cyclin-dependent kinases involved in proliferation (Fig. 6B). A reduction in the duration of the G1 checkpoint presumably deprives irradiated cells of crucial time to repair DNA damage before DNA replication. These data are consistent with yet another role for TCTP in p53-dependent mechanisms that maintain genome integrity in normal cells under stressful conditions. Further, knockdown of TCTP modulates the γ-ray–induced G2 checkpoint (Fig. 6C). Ten to twenty-five hours after exposure to 400 cGy, greater fractions of cells were delayed in G2 phase, likely as a result of accumulation of DNA damage in these cells (Fig. 2). Recently, overexpression of TCTP was shown to destabilize p53 in lung carcinoma cells (20), and knockdown of TCTP interfered with the p53–MDM2 axis (26). Therefore TCTP effects on p53 are multifaceted and may depend on cell type, cellular microenvironment, inherent sensitivity, and whether cells are challenged with low or high doses of environmental agents.
Low doses of toxic agents, including chemicals and ionizing radiation, often induce protective mechanisms that enhance the ability of the organism to cope with stress from normal metabolism or exposures to exogenous agents (27). Here, we show that doses of γ rays as low as 1–10 cGy (Fig. S1 A–E), particularly when delivered at a low dose rate (0.2 cGy/h), up-regulate TCTP expression and reduce the frequency of MN formation to below the spontaneous rate in human fibroblasts (Fig. 1A). MN arise predominantly from DSBs, and in the case of spontaneous formation, such as when TCTP was knocked down in AG1522 cells (Fig. 2A), they likely are induced by closely spaced oxidative base lesions caused by reactive oxygen species generated during normal cellular metabolism (23). These MN formation results and those showing that knockdown of TCTP attenuates the repair of radiation-induced DNA breaks (DSBs, closely spaced single-strand breaks) that lead to MN formation and γH2A.X foci (Fig. 2) suggest a role for TCTP in the expression of radiation-induced adaptive responses (27, 28).
TCTP up-regulation by ionizing radiation is not restricted to tissue-culture phenomena but also occurs in tissues of mice after low-dose irradiation to the whole body (Fig. S1E). The fact that TCTP induction occurs in cells at all phases of the cell cycle and is of long (at least 48 h) duration (Fig. S1A) is consistent with its role as a surveillance sentinel that guards against prolonged radiation-induced oxidative damage (29). In addition, TCTP also is responsive to high-dose irradiation (Figs. 2, 5, and 6). Understanding the role of TCTP in the sensing and repair of radiation-induced DNA damage as described here (schematic in Fig. 7) will be informative for understanding the system responses to low-dose radiation exposures, and this understanding would help in estimating the health risks of such exposures. It also may lead to greater insight in the molecular events induced by therapeutic doses of radiation.
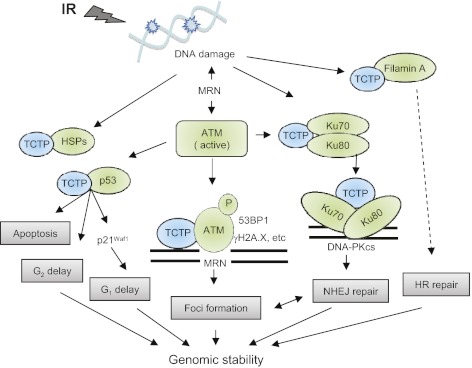
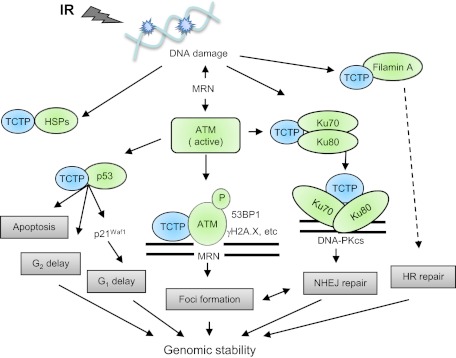
Fig. 7.
The role of TCTP in DNA damage sensing and repair. TCTP is up-regulated by ionizing radiation, interacts with elements of DNA damage sensing and repair, and modulates radiation-induced cell-cycle checkpoints. HR, homologous recombination; HSPs, heat shock proteins; IR, ionizing radiation; MRN, MRE11–RAD50–NBS1; NHEJ, nonhomologous end joining; P, phosphorylation.
Materials and Methods
Cells.
The human skin fibroblast strains AG1522 and GM5758 are apparently normal; AT5BI and AG4405 were derived from patients with ataxia telangiectasia (Coriell Institute for Medical Research, Camden, NJ). U2OS cells are osteosarcoma cells (ATCC). M059K (WT) and M059J (DNA-PK–mutated cells) are human glioblastoma cells (ATCC). Unless indicated, confluent cells were used in experiments and were grown as described (1). For growth in 3D architecture, cells were adapted to grow on a Cytomatrix carbon scaffold (Cell Science Therapeutics) (1). Experiments were repeated two to six times, and comparisons between treatment groups and respective controls were performed using the Pearson's χ2-test. A P value of ≤ 0.05 between groups was considered significant.
Animals.
The 5- to 6-wk-old C3H/HeJ mice were obtained from Jackson Laboratory. When irradiated, they were 7 to 9 wk old.
Irradiation.
Cell cultures were exposed to γ rays at 37 °C in a humidified atmosphere of 5% (vol/vol) CO2 in air in a Mark I 137Cs irradiator (J. L. Shepherd) at a low dose rate (0.2 cGy/h or 6 cGy/min) or an acute dose rate (330 cGy/min). Exposure to 1 GeV protons was carried at the National Aeronautics Space Agency Space Radiation Laboratory (Upton, NY) at 5 cGy/min.
Inhibitors.
PJ34 (Alexis Biochemicals) was used at 30 μM and was added to cells 3 h before irradiation. Ku 55933 (KuDOS Pharmaceuticals) and NU7441 (Tocris) were added at 10 μM 30 min before irradiation. Cycloheximide (Calbiochem) was added at 2 μg/mL 30 min before irradiation. Cells were incubated with the various inhibitors until harvest. Controls were incubated with the dissolving vehicles.
Immunoblotting and Antibodies.
Immunoblotting was performed as described (1). The primary antibodies were TCTP [ab37506 (Abcam) and sc-30124 (Santa Cruz Biotechnology)]; ATM [sc-23291 (Santa Cruz Biotechnology), A1106 (Sigma), and GTX 70103 (GeneTex)]; P-ATM (S1981) (05-740; Upstate Biotechnology); ATR (A300-138A; Bethyl Laboratories); DNA-PKcs (sc-5282; Santa Cruz Biotechnology); Ku70 (sc-1486 and sc-9033; Santa Cruz Biotechnology); Ku80 [sc-9034 (Santa Cruz Biotechnology) and GTX 22173 (GeneTex)], p53 [OP03 and OP43 (VWR) and 9282 (Cell Signaling)]; P-p53 (S15) (9284; Cell Signaling); p21Waf1 [OP64, Ab-1 (VWR), 05 345MI (Fisher), and 53BP1 (A300-272A; Bethyl Laboratories)]; γH2A.X (05-636MI; Fisher); H2A.X (50-230-9763; Fisher); and ORC2 (559255; BD Biosciences). Secondary antibodies for Western blotting were from BioRad. To verify equal loading of samples, membranes were stained with Ponceau S Red (Sigma) and reacted with anti-tubulin (CP06; Calbiochem), anti-TATA box-binding protein (TBP) (ab818; Abcam), or goat anti-rabbit IgG (sc-2030; Santa Cruz Biotechnology) that recognizes a protein of ∼30 kDa (loading control).
Immunofluorescence.
The cells were fixed in 4% (wt/vol) formaldehyde for 10 min at room temperature, permeabilized in 0.2% Triton X-100 in PBS for 10 min, blocked with 4% (wt/vol) BSA, and incubated with the primary antibodies. Signals were visualized by use of secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 594, or DAPI (Invitrogen). For preextraction, cells were subjected to detergent extraction with Triton X-100 in PBS (0.5% for 5–10 min) to remove the majority of non–chromatin-bound proteins before fixation and immunostaining.
Foci Analyses.
Digital images were acquired from random fields (Zeiss Axiovert 200M) and analyzed with AxioVision LE 4.6 software. The Apotome function of the microscope was used for foci colocalization. Freely available FociCounter software (30) was used to count foci, and colocalization was scored manually in at least 30 randomly chosen cells.
IP.
Monolayer cells were washed with PBS and lysed on ice for 10 min in buffer A [10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05% Nonidet P-40 with protease and phosphatase inhibitor mixtures] (Sigma). After centrifugation (800 × g for 10 min at 4 °C), the nuclei pellet was resuspended in high-salt buffer B [5 mM Hepes (pH 7.9), 300 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol (vol/vol), and 0.5% Nonidet P-40] and left on ice for 30 min. Supernatants were collected by centrifugation at 14,000 × g for 20 min at 4 °C and were treated with 100 U/mL Benzonase (EMD Millipore) for 1 h at room temperature or with 50 μg/mL ethidium bromide on ice for 30 min to release chromatin-bound proteins. Following incubation with appropriate antibodies overnight at 4 °C, a mixture of protein A and protein G beads (Pierce) was added, and the mixture was incubated for >3 h at 4 °C. After three washings with Hepes-buffered saline buffer [50 mM Hepes (pH 7.5), 1 mM EDTA, and 0.2% Nonidet P-40], proteins were solubilized in SDS/PAGE loading buffer and analyzed by Western blotting. Inputs represent 1/20 of the extract used for IP.
Subcellular Fractionation.
For nuclei and chromatin-bound protein isolation, 107 cells were lysed in 1 mL of buffer A [10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 340 mM sucrose, 10% (vol/vol) glycerol, 0.1% Triton X-100, 1 mM DTT, 0.1 mM PMSF, protease inhibitor mixture] for 10 min on ice. Cytoplasmic proteins were separated from nuclei by low-speed centrifugation (500 × g for 5 min at 4 °C). Isolated nuclei were washed once with buffer A and then lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and protease inhibitors) on ice for 15 min. Soluble nuclear proteins were separated from insoluble chromatin by centrifugation at 1,500 × g for 5 min at 4 °C. Isolated chromatin was washed with buffer B and centrifuged at 10,000 × g for 1 min. The final chromatin pellet was resuspended in buffer A without glycerol. Then 100 U/mL Benzonase (EMD Millipore) was added and incubated for 1 h at room temperature to release chromatin-bound proteins.
Ku70 and Ku80 Activity.
Ku70 and Ku80 DNA-binding activity was analyzed by a quantitative Ku70/80 DNA repair kit (Active Motif). Briefly, cells were washed and resuspended in hypotonic buffer. The nuclear extract was isolated for Ku activity analysis. Then 5 μg of nuclear protein was incubated with immobilized, blunt-ended linear DNA in wells of a 96-well plate for 1 h at room temperature. Bound Ku70 or Ku80 was detected using respective antibodies followed by detection with HRP-linked secondary antibody. Each sample was assayed with Ku competitor oligonucleotide to ensure specificity, and a positive control consisting of a nuclear lysate of Raji cells, which are provided with the kit, was included in every run. The color product was detected at 450 nm with use of a Perkin-Elmer Victor plate reader, and absorbance values were corrected for assay blank (activity in the absence of sample) and nonspecific signal (activity in the presence of competitor). Each experiment was repeated three times, and data represent the mean ± SD of three separate determinations.
siRNA Silencing.
siRNAs capable of targeting mRNAs encoding TCTP (also known as tpt1) were from Ambion. Scr siRNA Duplex (Ambion) was included as control. The siRNAs to knock down ATM, ATR, and DNA-PKcs were from Thermo Scientific. Treatments were performed 72 h posttransfection when the cells were in G0/G1 phase of the cell cycle.
MN Formation.
DNA damage and its repair were assessed by measuring the frequency of MN formation by the cytokinesis-block technique as previously described (1). After treatments, confluent AG1522 fibroblasts were subcultured, and ∼3 × 104 cells were seeded in chamber flaskettes (Nunc) and allowed to progress in the cell cycle in the presence of 2 μg/mL cytochalasin B (Sigma). After 72 h incubation, an ample time to account for radiation-induced cell-cycle delays and for replication of AG1522 cells that have a doubling time of 26 h, the cells were rinsed in PBS, fixed in cold methanol, stained with Hoechst 33342 (1 μg/mL PBS), and viewed with a fluorescence microscope. At least 1,000 cells per treatment were examined, and only MN in binucleated cells were considered for analysis. At the concentration used, cytochalasin B was nontoxic. Binomial statistics were applied in analysis of the data.
Cell Survival.
Confluent cultures were exposed to γ rays and assayed for clonogenic survival within 5–10 min after exposure.
G1 to S Phase Traverse.
Quiescent cell populations were subcultured to low density in growth medium containing [3H]-thymidine (1 μCi/mL, specific activity 20 Ci/mmol) (PerkinElmer LAS, Inc), seeded in 30-mm dishes, and incubated at 37 °C. At regular intervals, duplicate dishes were rinsed with PBS, fixed with ethanol, and subjected to autoradiography. To determine labeling indices, a minimum of 1,000 cells per dish was scored. The use of this continuous-labeling technique allows precise determination of G1 delays.
Delay in G2 Phase.
Actively growing cells were γ-irradiated, and samples were harvested by trypsinization at different times after exposure. They were rinsed in PBS, fixed in 4% (wt/vol) formaldehyde for 10 min at room temperature, and then permeabilized in 0.2% Triton X-100 in PBS/1% BSA for 10 min. Following RNase treatment (0.5 mg/mL in PBS/1% BSA) at 37 °C for 1 h, the cells were stained with propidium iodide and submitted to flow cytometry (FACSCalibur; Becton Dickinson).
iTRAQ Labeling, MS, and Protein Quantification.
Soluble proteins were sequentially reduced, alkylated, digested, and labeled with four iTRAQ reagents according to the manufacturer's instructions (Applied Biosystems). The labeled peptides were combined and separated by strong cation-exchange chromatography followed by reversed-phase LC and analysis on a 4700 Proteomics Analyzer tandem mass spectrometer (ABI) (6). GPS Explorer software (v. 3.5; ABI) was used to process the MS/MS spectra and to submit peak lists to the MASCOT (v. 1.9) search engine for peptide identification. Only peptides identified with confidence interval values ≥95% were used for protein identification and quantification.
Phosphorylation Sites in TCTP.
AG1522, U2OS, or AG4405 cells were exposed to 50 cGy of γ rays and incubated for 0.5 h. IP with anti-TCTP was carried in nuclear extracts. Samples were fractionated in SDS/PAGE, and the TCTP band was excised for in-gel trypsin digestion. The resulting peptides were analyzed by LC-MS/MS on an Orbitrap Velos instrument (Thermo Fisher Scientific). The peptides were separated by a reversed-phase column (75 μm × 150 mm, 3 μm, 100 Å, C18; Dionex) on an Ultimate 3000 LC system (Dionex) and were introduced directly into a Proxeon nano-electrospray ionization source on the Orbitrap Velos instrument for MS/ MS analysis. The MS spectra were acquired in the positive ion mode with a spray voltage of 2 kV and a capillary temperature of 275 °C. Collision-induced dissociation was used for peptides fragmentation. The MS/MS spectra were searched against the UniProt human database using both Mascot and Sequest search engines. The protein-sequence coverages for TCTP were 87% for all three samples.
Supplementary Material
Acknowledgments
We thank Dr. R. W. Howell for input in low-dose-rate radiation studies, the staff at the National Aeronautics Space Administration Space Radiation Laboratory for proton irradiation, and Drs. U. Herbig and A. Asaithamby for discussions. E.I.A. was supported by Grant DE-FG02-07ER64344 from the US Department of Energy, Low Dose Radiation Research Program; Grant NNJ06HD91G from the National Aeronautics and Space Administration; and Grants AI067773-04 (a National Institute of Allergy and Infections Disease subaward) and CA049062 from the National Institutes of Health. H.L. was supported by National Institutes of Health Grant NS046593. D.P. was supported by Grant AG030504 from the National Institute on Aging and by Grant 09GRNT2260364 from the American Heart Association. J.Z. received support from the Program for Changjiang Scholars and Innovative Research Team, University of Ministry of Education, China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 5921 (volume 109, number 16).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106300109/-/DCSupplemental.
References
- 1.de Toledo SM, et al. Adaptive responses to low-dose/low-dose-rate γ rays in normal human fibroblasts: The role of growth architecture and oxidative metabolism. Radiat Res. 2006;166:849–857. doi: 10.1667/RR0640.1. [DOI] [PubMed] [Google Scholar]
- 2.Chen SH, et al. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol Biol Cell. 2007;18:2525–2532. doi: 10.1091/mbc.E07-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP) Int J Biochem Cell Biol. 2004;36:379–385. doi: 10.1016/s1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 4.Telerman A, Amson R. The molecular programme of tumour reversion: The steps beyond malignant transformation. Nat Rev Cancer. 2009;9:206–216. doi: 10.1038/nrc2589. [DOI] [PubMed] [Google Scholar]
- 5.Susini L, et al. TCTP protects from apoptotic cell death by antagonizing bax function. Cell Death Differ. 2008;15:1211–1220. doi: 10.1038/cdd.2008.18. [DOI] [PubMed] [Google Scholar]
- 6.Liu T, et al. Identification of differentially expressed proteins in experimental autoimmune encephalomyelitis (EAE) by proteomic analysis of the spinal cord. J Proteome Res. 2007;6:2565–2575. doi: 10.1021/pr070012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 8.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Critchlow SE, Jackson SP. DNA end-joining: From yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 10.Grudzenski S, Raths A, Conrad S, Rübe CE, Löbrich M. Inducible response required for repair of low-dose radiation damage in human fibroblasts. Proc Natl Acad Sci USA. 2010;107:14205–14210. doi: 10.1073/pnas.1002213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 12.Sengupta S, Harris CC. p53: Traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 13.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Koike M. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J Radiat Res (Tokyo) 2002;43:223–236. doi: 10.1269/jrr.43.223. [DOI] [PubMed] [Google Scholar]
- 15.Gnanasekar M, Thirugnanam S, Zheng G, Chen A, Ramaswamy K. Gene silencing of translationally controlled tumor protein (TCTP) by siRNA inhibits cell growth and induces apoptosis of human prostate cancer cells. Int J Oncol. 2009;34:1241–1246. [PubMed] [Google Scholar]
- 16.Abbas T, Dutta A. p21 in cancer: Intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syljuåsen RG, Krolewski B, Little JB. Loss of normal G1 checkpoint control is an early step in carcinogenesis, independent of p53 status. Cancer Res. 1999;59:1008–1014. [PubMed] [Google Scholar]
- 19.Gnanasekar M, Ramaswamy K. Translationally controlled tumor protein of Brugia malayi functions as an antioxidant protein. Parasitol Res. 2007;101:1533–1540. doi: 10.1007/s00436-007-0671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rho SB, et al. Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett. 2011;585:29–35. doi: 10.1016/j.febslet.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Baumstark-Khan C. X-ray-induced DNA double-strand breaks as lethal lesions in diploid human fibroblasts compared to Chinese hamster ovary cells. Int J Radiat Biol. 1993;63:305–311. doi: 10.1080/09553009314550411. [DOI] [PubMed] [Google Scholar]
- 22.Gnanasekar M, Dakshinamoorthy G, Ramaswamy K. Translationally controlled tumor protein is a novel heat shock protein with chaperone-like activity. Biochem Biophys Res Commun. 2009;386:333–337. doi: 10.1016/j.bbrc.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 24.Williams ES, et al. Telomere dysfunction and DNA-PKcs deficiency: Characterization and consequence. Cancer Res. 2009;69:2100–2107. doi: 10.1158/0008-5472.CAN-08-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue J, et al. The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 2009;69:7978–7985. doi: 10.1158/0008-5472.CAN-09-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amson R, et al. Reciprocal repression between P53 and TCTP. Nat Med. 2012;18:91–99. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- 27.Averbeck D. Does scientific evidence support a change from the LNT model for low-dose radiation risk extrapolation? Health Phys. 2009;97:493–504. doi: 10.1097/HP.0b013e3181b08a20. [DOI] [PubMed] [Google Scholar]
- 28.Azzam EI, de Toledo SM, Raaphorst GP, Mitchel RE. Low-dose ionizing radiation decreases the frequency of neoplastic transformation to a level below the spontaneous rate in C3H 10T1/2 cells. Radiat Res. 1996;146:369–373. [PubMed] [Google Scholar]
- 29.Spitz DR, Azzam EI, Li JJ, Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: A unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23:311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 30.Jucha A, et al. FociCounter: A freely available PC programme for quantitative and qualitative analysis of gamma-H2AX foci. Mutat Res. 2010;696:16–20. doi: 10.1016/j.mrgentox.2009.12.004. [DOI] [PubMed] [Google Scholar]