Abstract
Legionella pneumophila, the causative agent of Legionnaires’ pneumonia, resides in a distinct vacuole structure called Legionella-containing vacuole (LCV). The LCV resists fusion with the lysosome and permits efficient bacterial replication in host macrophages, which requires a Dot/Icm type IVB secretion system. Dot/Icm-translocated effector SdhA is critical for L. pneumophila intracellular growth and functions to prevent host cell death. Here, we show that the absence of SdhA resulted in elevated caspase-1 activation and IL-1β secretion as well as macrophage pyroptosis during Legionella infection. These inflammasome activation phenotypes were independent of the established flagellin-NAIP5-NLRC4 axis, but relied on the DNA-sensing AIM2 inflammasome. We further demonstrate that Legionella DNA was released into macrophage cytosol, and this effect was significantly exaggerated by the absence of SdhA. SdhA bears a functional Golgi-targeting GRIP domain that is required for preventing AIM2 inflammasome activation. Ectopically expressed SdhA formed a unique ring-shape membrane structure, further indicating a role in membrane trafficking and maintaining LCV membrane integrity. Our data together suggest a possible link, mediated by the function of SdhA, between LCV trafficking/maturation and suppression of host innate immune detection.
Gram-negative Legionella pneumophila, the causative agent of Legionnaires’ disease, infects and replicates within alveolar macrophages. L. pneumophila resides in a distinct vacuole structure called Legionella-containing vacuole (LCV). Dynamic and extensive engagement with host membrane transport system diverts LCV trafficking from normal eukaryotic endocytic pathway and, thereby, results in resistance to fusion with the lysosome (1), a process that requires a Dot/Icm type IVB secretion system (2, 3). The primitive function of the Dot/Icm system is to transfer DNA by bacterial conjugation (4), but its predominant role in infection is to translocate protein substrates across the LCV membrane into host cells. A prevalent function for known Dot/Icm effectors is to subvert eukaryotic vesicular trafficking (1, 5, 6). A large number of >250 Dot/Icm effectors are experimentally confirmed (7). Few Dot/Icm effectors are genetically required for LCV avoidance of lysosomal fusion, suggesting a functional redundancy. However, the Dot/Icm system is critical for establishment of the LCV. The LCV serves as an intracellular niche, where L. pneumophila can efficiently replicate, as has been observed in human U937 monocytes (8).
The inflammasome pathway, generally mediated by cytosolic NOD-like receptors (NLRs), is a critical component in macrophage innate immunity (9). Similar to the Toll-like receptor pathway that senses pathogen-associated molecular patterns (PAMPs), inflammasome also responds to microbial products for counteracting infection. Inflammasomes are large cytoplasmic complexes that induce activation of downstream inflammatory caspases, mainly caspase-1. Caspase-1 activation leads to maturation and secretion of IL-1β and IL-18 as well as macrophage pyroptosis. NAIP5, a mouse NLR protein, functions as a receptor for bacterial flagellin and forms an inflammasome complex with NLRC4 and the ASC adaptor (10, 11). Upon infection, L. pneumophila flagellin triggers potent NAIP5-dependent caspase-1 activation and pyroptosis in C57/BL6-derived macrophages that are nonpermissive for L. pneumophila growth. Both interference with host vesicular trafficking and modulation of the innate immune response are crucial for Dot/Icm-dependent L. pneumophila intracellular survival/replication, but it is not known whether the two pathogenic aspects of L. pneumophila are functionally connected.
Different from the NAIP family of inflammasome receptors (11), the non-NLR protein AIM2 recognizes cytosolic double-strand DNA (dsDNA), such as that from intracellular bacteria, and forms an active inflammasome complex with ASC (12–15). Here, we discover that SdhA, the sole Dot/Icm effector whose mutation leads to severe L. pneumophila growth defects in macrophages (16), negatively regulates AIM2 inflammasome activation during infection. L. pneumophila ΔsdhA-stimulated AIM2 activation correlates with much-elevated bacterial DNA release from the ΔsdhA mutant into macrophage cytosol. Further characterization of SdhA function suggests that SdhA is intimately linked to the membrane trafficking system, including the presence of a functionally important Golgi-targeting GRIP domain. Thus, the function of SdhA represents a possible link between LCV trafficking/maturation and suppression of inflammasome-mediated innate immune detection.
Results
L. pneumophila ΔsdhA Infection Triggers Pyroptosis and Caspase-1 Activation in Human Macrophage Cells.
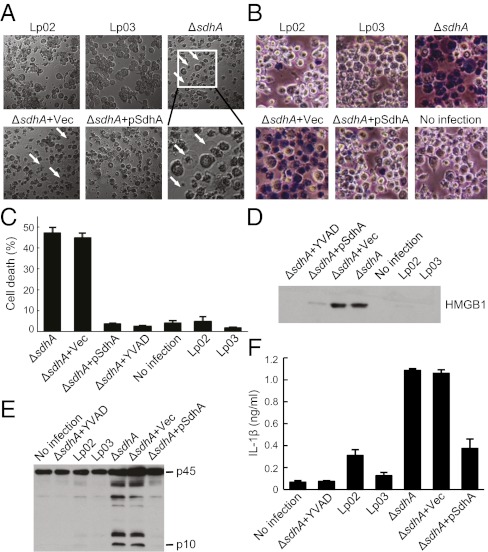
Dot/Icm-translocated SdhA has been shown to function to prevent macrophage death (16). To investigate the mechanism underlying SdhA function, PMA-differentiated U937 human macrophage cells were infected with L. pneumophila strain proficient (Lp02) or deficient in Dot/Icm-mediated secretion (ΔdotA, Lp03) or the isogenic ΔsdhA strain. By 5 h of infection, U937 cells infected with ΔsdhA exhibited evident osmotic pressure-induced cell swelling and nuclear condensation (Fig. 1A), two typical morphological features of macrophage pyroptosis. In contrast, cells infected with Lp02 and Lp03 strains underwent little such morphological changes. Trypan blue staining further showed extensive nuclear staining signals in cells infected with L. pneumophila ΔsdhA but not in Lp02 and Lp03 infected cells (Fig. 1B). ΔsdhA-triggered loss of cell membrane integrity was confirmed by massive releases of lactose dehydrogenase (LDH) and high-mobility group protein B1 (HMGB1) (Fig. 1 C and D). Quantification of LDH release further indicated that nearly 50% of ΔsdhA-infected cells underwent pyroptosis within 5 h of infection, whereas the percentages of cell death observed with Lp02 and Lp03 infections were comparable to that in uninfected cells (Fig. 1B).
Fig. 1.
Deletion of sdhA triggers pyroptosis and caspase-1 activation in U937 cells. (A and B) Morphological examination of effects of sdhA deletion during L. pneumophila infection of PMA-differentiated U937 cells (MOI = 10). Lp02 is used as the wild-type strain; Lp03 is a dotA mutant. pSdhA is a complementation plasmid expressing SdhA. Differential interference contrast images (5 h after infection) are shown with cells stained with Trypan blue in B. Arrows indicate pyroptotic cells in A. (C and D) Lactate dehydrogenase (LDH) (C) and HMGB1 (D) release assays of effects of sdhA deletion. Shown in C are percentages of cell death as mean values ± SD (error bars) from four independent experiments. Anti-HMGB1 immunoblot of culture media is shown in D. YVAD, a caspase-1 inhibitor. (E and F) Caspase-1 activation and IL-1β release assays of sdhA deletion. Shown in E is anti–caspase-1 immunoblot of culture supernatants. p45, procaspase-1; p10, the processed mature form of caspase-1. IL-1β ELISA data shown in F are as mean values ± SD (error bars) from three independent experiments.
Consistent with the morphological feature of inflammasome activation, robust caspase-1 activation was detected in the culture supernatant of ΔsdhA-infected U937 macrophages, but not in that of uninfected or Lp02/Lp03-infected macrophages (Fig. 1E). ΔsdhA-induced caspase-1 activation and macrophage pyroptosis were completely diminished by YVAD, a caspase-1 specific inhibitor (Fig. 1 C and E). Furthermore, compared with Lp02 and Lp03 infection, infection of U937 cells with L. pneumophila ΔsdhA resulted in secretion of a much larger amount of mature IL-1β, which was also sensitive to YVAD pretreatment (Fig. 1F).
We then tested whether loss of SdhA is responsible for the above observed increased inflammasome activation. An SdhA-expressing plasmid was found to be able to completely reverse ΔsdhA-induced morphological changes (Fig. 1 A and B), LDH and HMGB1 release (Fig. 1 C and D), caspase-1 activation (Fig. 1E), and IL-1β production (Fig. 1F), whereas a control empty vector showed so such rescue effects. These analyses clearly establish that SdhA prevents human macrophage cell death by blocking inflammasome activation.
Flagellin Is Dispensable for ΔsdhA-Induced Inflammasome Activation.
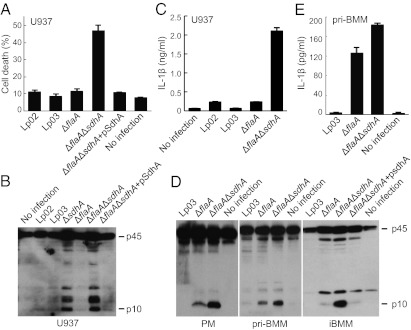
Cytosolic flagellin triggers strong caspase-1 activation during L. pneumophila infection (17–19). To test whether SdhA prevents or negatively regulates flagellin-induced inflammasome activation, flagellin-deficient (ΔflaA) and SdhA/flagellin (ΔflaAΔsdhA) double deletion strains were constructed and assayed for inflammasome activation. In human U937 macrophages, ΔflaAΔsdhA induced significantly higher levels of LDH release (Fig. 2A), caspase-1 activation (Fig. 2B), and IL-1β maturation (Fig. 2C) than the background ΔflaA strain, which could be fully rescued by plasmid-encoded SdhA. Flagellin-independent inflammasome activation observed with ΔsdhA infection agrees with our recent discovery that human macrophages lack the flagellin-sensing NAIP5 inflammasome receptor (11). Consistently, deletion of flagellin from wild-type L. pneumophila also did not alter caspase-1 activation in U937 cells (compare ΔflaA with Lp02 in Fig. 2 A–C).
Fig. 2.
ΔsdhA-induced inflammasome activation is flagellin-independent. U937 cells were used for infection in A–C and mouse macrophages were assayed in D and E. LDH release in A shows percentages of cell death as mean values ± SD (error bars) from four independent experiments. Shown in B and D are anti–caspase-1 immunoblots of culture supernatants. IL-1β ELISA data shown in C and E are mean values ± SD (error bars) from three independent experiments. iBMM, immortalized bone marrow macrophages (C57/BL6 background); PM, peritoneal macrophages (129S background); pri-BMM, primary bone marrow macrophages (129S background). Culture supernatants collected from 2 to 6 h after infection were subjected to analyses in D and E.
Different from human cells, mouse macrophages express the NAIP5 receptor and are responsive to Legionella flagellin stimulation of caspase-1 activation. Several types of mouse macrophages, including primary peritoneal macrophage (PM) and primary/immortalized bone marrow-derived macrophage (BMM), were then assayed for the inhibitory effects of SdhA on caspase-1 activation. Consistent with that observed in U937 macrophages, ablation of sdhA from flagellin-deficient L. pneumophila could still lead to elevated caspase-1 activation and, to a lesser extent, IL-1β production (Fig. 2 D and E). These results suggest that the function of SdhA in inhibiting caspase-1 activation is flagellin-independent and SdhA likely blocks inflammasome activation triggered by other Legionella-derived PAMPs.
ΔsdhA-Induced Caspase-1 Activation and Pyroptosis Require AIM2 and ASC.
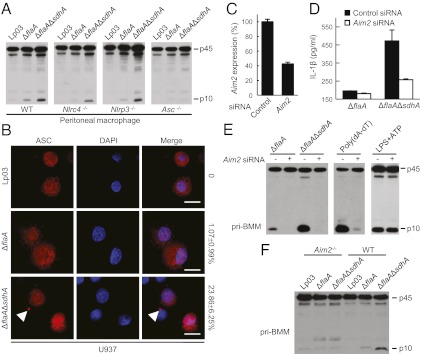
To determine ΔsdhA-induced inflammasome activation, primary peritoneal macrophages deficient in various known inflammasome components including NLRP3, NLRC4, and ASC were subjected to infection with ΔsdhA strain. As shown in Fig. 3A, Nlrp3−/− and Nlrc4−/− macrophages behaved similarly as wild-type macrophages and showed increased caspase-1 activation in response to sdhA deletion. In contrast, no caspase-1 activation was detected in Asc−/− macrophages infected with either ΔflaA or ΔsdhAΔflaA strains (Fig. 3A). ASC often aggregates to form a single speck-like large structure called pyroptosome that functions to mediate caspase-1 autoprocessing (20). We also observed ASC pyroptosome formation in ≈25% of U937 cells infected with ΔsdhA, but not in cells infected with Lp02, Lp03, or the ΔflaA strain (Fig. 3B).
Fig. 3.
ΔsdhA-induced inflammasome activation requires Aim2 and Asc. (A) ΔsdhA-induced caspase-1 activation in Nlrc4−/−, Nlrp3−/−, and Asc−/− macrophages. Peritoneal macrophages prepared from C57/BL6 (WT) or indicated knockout mice were infected with indicated L. pneumophila strains. (B) ASC foci formation in U937 cells infected with sdhA+ or sdhA− strains (MOI = 10). Statistics of percentages of cells showing the ASC pyroptosome (arrowhead) are listed on the right to the merged confocal images. (Scale bars: 7.5 μm.) (C–E) Effects of Aim2 knockdown on ΔsdhA-induced inflammasome activation. BMMs from 129S mice were transfected with Aim2 targeting or a control siRNA before indicated infections. qRT-PCR measurements of knockdown efficiency (C) are shown as mean values ± SEM (error bars) from three independent experiments. The supernatants were subjected to IL-1β ELISA (D) or anti–caspase-1 immunoblotting (E). Mean values ± SEM (error bars) from two independent experiments are shown in D. (F) ΔsdhA-induced caspase-1 activation in Aim2−/− macrophages. Peritoneal macrophages prepared from C57/BL6 (WT) or Aim2−/− mice were infected with indicated L. pneumophila strains.
Among known inflammasome complexes, ASC is strictly required for NLRP3 and AIM2 mediated caspase-1 activation due to pyrin–pyrin interaction between NLRP3/AIM2 and ASC. The insensitivity of ΔsdhA-induced caspase-1 activation to Nlrp3 deficiency directed our focus to AIM2. In fact, small interference RNA (siRNA) knockdown of AIM2 expression in 129S mice-derived BMMs (Fig. 3C) could largely diminish ΔsdhA-induced caspase-1 activation and IL-1β production (Fig. 3 D and E). Knockdown of AIM2 abolished poly(dA-dT)-induced caspase-1 activation, but not that by lipopolysaccharide (LPS) plus ATP (Fig. 3E), verifying the functional specificity of the siRNA oligoes used. The requirement of AIM2 for ΔsdhA-induced caspase-1 activation was further confirmed in primary BMMs derived from Aim2 knockout C57/BL6 mice (Fig. 3F). Caspase-1 activation in C57/BL6-derived primary BMMs, compared with that in 129S-derived BMMs, appeared to be less responsive to ΔsdhA infection likely due to the strain difference. Taken together, our data suggest that elevated caspase-1 activation induced by L. pneumophila ΔsdhA infection is a result of AIM2 inflammasome activation.
Bacterial DNA Is Released into Macrophage Cytosol, which Is Enhanced by the Absence of sdhA.
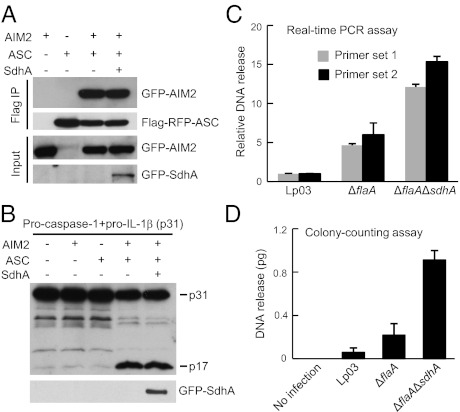
We then examined whether SdhA could directly target the AIM2–ASC–caspase-1 signaling axis. Coimmunoprecipitation of AIM2 and ASC in the ectopic expression system was not affected by SdhA (Fig. 4A). The AIM2/ASC complex produced in 293T cells could induce IL-1β maturation in the presence of caspase-1, but this effect was also not subjected to inhibition by SdhA (Fig. 4B). Because SdhA is unlikely to be a direct inhibitor of the AIM2 inflammasome complex, we then hypothesized that loss of SdhA might lead to an increased bacterial DNA release into macrophage cytosol. To this end, an ampicillin-resistant Escherichia coli/Legionella shuttle plasmid (pJB908) was transformed into SdhA-positive or -negative strains. After 4 h of infection of U937 cells or mouse BMMs, macrophages were lysed in a hypotonic buffer and cytosolic DNA containing the released plasmid was extracted. Before DNA extraction, macrophage cytosol was supplemented with a kanamycin-resistant plasmid (pEGFP-N1) that served as an internal control for subsequent experimental procedures. The extracted DNA was subjected to both quantitative real-time PCR (qRT-PCR) analysis and a colony-counting assay after E. coli transformation. The ratio of pJB908 quantification to pEGFP-N1 quantification calculated from both assays clearly showed that Legionella DNA was released into macrophage cytosol and deletion of sdhA resulted in a significant enhancement of such effects (Fig. 4 C and D). The DNA release pattern for the different L. pneumophila strains assayed correlated with their profiles of inducing caspase-1 activation and IL-1β release (Fig. 2 C and D). Thus, SdhA blocks AIM2 inflammasome-mediated innate immune response by preventing bacterial DNA release into the cytosol of infected macrophages. The pJB908 reporter plasmid used here lacks the oriT origin and was defective for Dot/Icm-mediated conjugal transfer (4), regardless of the presence and absence of sdhA (Table S1). This result suggests that increased DNA release induced by ΔsdhA is independent of Dot/Icm-mediated plasmid conjugation.
Fig. 4.
SdhA inhibits Legionella DNA release into macrophage cytosol. (A and B) SdhA does not directly target the AIM2–ASC–caspase-1 pathway. 293T cells transfected with indicated plasmid combination were subjected to anti-Flag immunoprecipitation and/or immunoblotting. p17, the processed mature form of IL-1β. (C and D) Bacterial DNA release into macrophage cytosol during infection and effects of sdhA deletion. PMA-differentiated U937 cells were infected with indicated L. pneumophila strains harboring a reporter plasmid. DNA fractions extracted from infected macrophage cytosol were subjected to qRT-PCR analysis (C) or colony-counting assay (D) to measure relative plasmid DNA release (detailed in SI Materials and Methods). Mean values ± SEM (error bars) from three independent experiments are shown.
L. pneumophila infection also potently activates IRF3-dependent type I IFN response, which plays a role in restricting Legionella intracellular growth (21, 22). A genetic screen also identifies SdhA as a suppressor of L. pneumophila-stimulated type I IFN response (23). Our model of SdhA function is consistent with these observations because cytosolic bacterial DNA is known to be a potent stimulator of type I IFN response. It has been debated whether Legionella DNA or RNA is responsible for activation of the type I IFN response (23, 24). Our data indicate that at least bacterial DNA is released into host cytosol and exposed to host innate immune receptors.
ΔsdhA-Induced AIM2 Inflammasome Activation Is Independent of IFN and TNFα Signaling.
Francisella tularensis also releases its DNA into host cytosol and triggers both type I IFN response and AIM2-dependent caspase-1 activation (25). Full inflammasome activation requires Francisella DNA-stimulated type I IFN signaling (26, 27). We then examined whether such requirement also applies to L. pneumophila ΔsdhA-induced inflammasome activation. To this end, Ifnar−/−, Ifngr−/−, Irf3−/−, Irf7−/−, or Irf3−/−Irf7−/− peritoneal macrophages that are deficient in either type I or type II IFN response were infected with sdhA+ or sdhA− strains. Similarly to that observed in wild-type macrophages, deletion of sdhA still resulted in elevated caspase-1 activation in all these IFN signaling-deficient macrophages (Fig. S1A). Moreover, pretreatment of mouse BMMs or U937 cells with IFN-β did not vary the extent of ΔsdhA-induced caspase-1 activation (Fig. S1B). Thus, L. pneumophila differs from F. tularensis and induces AIM2 activation independently of the type I IFN response.
Bacterial infection often triggers production of the proinflammatory cytokine TNFα that widely regulates macrophage innate immunity. We also observed that ΔsdhA-induced caspase-1 activation remained unchanged in Tnfrsf1a−/−Tnfrsf1b−/− macrophages (Fig. S1C). Moreover, TNFα pretreatment of U937 cells had little effects on caspase-1 activation in response to ΔsdhA infection (Fig. S1C). These data suggest that TNFα signaling is not involved in L. pneumophila ΔsdhA-induced AIM2 inflammasome activation.
SdhA Is Targeted to the Endomembrane Structure and Membrane Targeting Is Required for SdhA Function in Preventing Inflammasome Activation.
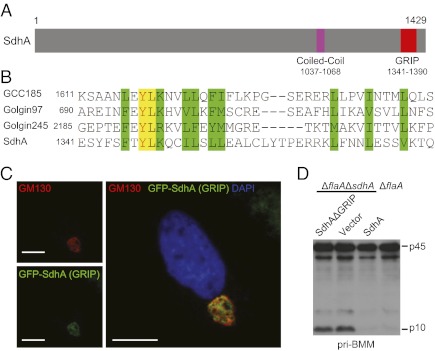
SdhA contains 1,429 amino acids and sequence homology analysis revealed a C-terminal GRIP domain (residues 1341–1390) (Fig. 5 A and B). The GRIP domain of ≈50 residues is present in a group of large coiled-coil membrane proteins, usually at their carboxyl termini (28, 29). The GRIP domain, such as that from many golgins, is necessary and sufficient for Golgi targeting (30). Consistently, the GRIP domain of SdhA was found to be exclusively localized to the Golgi apparatus in mammalian cells as indicated by immunostaining of a Golgi marker GM130 (Fig. 5C). Notably, a GRIP domain deletion mutant of SdhA (SdhAΔGRIP) failed to reverse ΔsdhA-induced caspase-1 hyperactivation (Fig. 5D) despite the mutant was expressed at a higher level than endogenous SdhA (Fig. S2). (A SidC translocation signal was added to SdhAΔGRIP for Dot/Icm-dependent translocation.)
Fig. 5.
SdhA harbors a Golgi-targeting GRIP domain that is required for inhibiting inflammasome. (A) The domain structure of SdhA. (B) Sequence alignment of residues 1341–1381 in SdhA with GRIP domains from several mammalian golgins. Similar residues are highlighted in green, and identical ones are in yellow. (C) SdhA GRIP domain localization in HeLa cells. The Golgi was visualized by GM130 staining. Representative confocal immunofluorescence images are shown. (Scale bars: 7.5 μm.) (D) Complementation of ΔflaAΔsdhA strain by plasmid-encoded SdhA or the SdhAΔGRIP mutant. BMMs from 129S mice were infected with L. pneumophila deletion or rescue strains as indicated. Caspase-1 immunoblot of culture supernatant is shown.
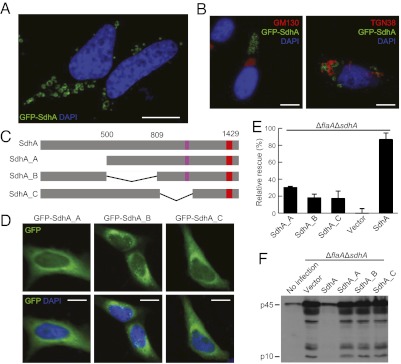
Interestingly, we also observed that GFP-SdhA formed distinct ring-shape membrane structures with a diameter ≈0.5–2 μm in HeLa cells (Fig. 6A). These membrane structures did not adopt cis- (GM130) and trans-Golgi (TGN38) markers (Fig. 6B) and were not sensitive to Golgi-disassembling drugs such as brefeldin A, colchicine, and nocodazole (Fig. S3). The SdhA ring-shape structures were also negative for a panel of subcellular organelle markers including GFP-PTS1 for peroxisome (Fig. S4A), VDAC1, cytochrome c, and Mitotracker for mitochondria (Fig. S4 B–D), Lysotracker and LAMP1 for lysosome (Fig. S5), calreticulin for endoplasmic reticulum (Fig. S6A), Rab5 and EEA1 for early endosome (Fig. S6 B and C), and Rab7 for late endosome (Fig. S6D). Disruption of lysosome biogenesis by bafilomycin A1, a vacuolar-type H+-ATPase inhibitor, had no effects on SdhA formation of the ring-shape structure (Fig. S5A). This membrane structure formed by N-terminal Flag-tagged SdhA-GFP could be visualized by anti-Flag immunofluorescence staining even when digitonin was used for cell permeabilization (Fig. S7A). This result suggests that the amino terminus of SdhA is likely exposed to the cytosol because digitonin was known to be incapable of permeabilizing the endomembrane (Fig. S7B). These extensive analyses, although not revealing for the identity of SdhA-marked membrane structures, provide another support to the potential function of SdhA in membrane trafficking.
Fig. 6.
SdhA forms a distinct ring-shape membrane structure and mutants deficient in ring formation fail to complement ΔsdhA strain. (A and B) GFP-SdhA forms a ring-shape membrane structure that is negative for Golgi markers. GM130 and TGN38 antibodies stain the Golgi structure. (Scale bars: 7.5 μm.) (C) A schematic drawing of SdhA truncation mutant assayed in (D–F). SdhA_A, Δ1–499aa; SdhA_B, Δ501–809aa; SdhA_C, Δ847–1104aa. (D) Localization of GFP-tagged SdhA truncation mutants in HeLa cells. (Scale bars: 7.5 μm.) (E and F) Complementation assays of SdhA truncation mutants. PMA-differentiated U937 cells were infected with ΔflaAΔsdhA strain transformed with a plasmid expressing SdhA or the indicated truncation mutants. Cell death measured by LDH release was assayed in E, and rescue effects relative to vector-complemented strain are shown as mean values ± SD (error bars) from three independent experiments.
Truncation analyses were further performed to map the regions in SdhA that are required for formation of the ring-shape structure. Three SdhA truncation mutants with deletions of various amino-terminal regions (SdhA_A, SdhA_B, and SdhA_C) lost the ability to form the unique ring-shape structure and became dispersed in the cytoplasm (Fig. 6 C and D). All three mutants were expressed at a much higher level than endogenous SdhA (Fig. S2) but failed to reverse ΔsdhA-induced pyroptosis and caspas-1 activation in infected U937 cells (Fig. 6 E and F). Similar results were obtained in mouse BMM cells (Fig. S8). Consistent with our proposed role of SdhA in preventing bacterial release into macrophage cytosol, the relative amount of bacterial DNA released from the three mutants-complemented strains was at a similarly higher level as that from the ΔsdhA deletion strain (Fig. S9). These data, together with the functional requirement of the Golgi-targeting GRIP domain, indicate that Dot/Icm-translocated SdhA likely targets the membrane system to prevent bacterial DNA release and activation of AIM2 inflammasome, which is possibly through maintaining LCV membrane integrity.
Discussion
In this study, we show that L. pneumophila triggers inflammasome activation in human monocyte-derived macrophages. Consistent with the absence of NAIP5 inflammasome receptor for bacterial flagellin in human system (11), we identify the DNA-sensing AIM2 inflammasome that is responsible for L. pneumophila-induced caspase-1 activation in human macrophages. Legionella activation of AIM2 also occurs in mouse macrophages, but is relatively hysteretic compared with the more predominant activation of the NAIP5 inflammasome by flagellin. The difference between the human and mouse inflammasome system might explain the long known permissiveness to L. pneumophila intracellular replication observed with U937 cells, but not mouse macrophages that harbor wild-type Naip5. We further provide direct evidences that L. pneumophila DNA leaks out of the bacteria into macrophage cytosol, which can also explain the previously observed induction of the type I IFN response (21–23). Release of bacterial DNA to trigger AIM2 inflammasome activation and type I IFN response has been observed with other bacteria such as F. tularensis and L. monocytogenes (27, 31). Thus, DNA derived from intracellular bacteria is likely a general PAMP for host innate immune system. Meanwhile, host-derived DNA, particularly mitochondrial DNA, may also possibly contribute to L. pneumophila stimulation of the innate immune response. In fact, mitochondria are extensively recruited to the LCV and mitochondria disruption has been noted in ΔsdhA-infected macrophages (16).
SdhA is critically required for L. pneumophila growth in macrophages (16). Our genetic studies demonstrate that SdhA functions to prevent bacterial DNA release into macrophage cytosol. Loss of SdhA, therefore, results in much increased activation of AIM2 inflammasome and the type I IFN response observed (23). Caspase-1–mediated secretion of IL-1β /IL-18 trigger strong inflammatory responses that alert the immune system for pathogen clearance. Caspase-1–induced pyroptosis can also clear intracellular bacteria independently of cytokine secretion (32). Nuclear acid-activated type I IFN response has been established as a crucial immune defense mechanism to restrict intracellular bacteria including L. pneumophila (21, 33, 34). Thus, our discovery of SdhA function in preventing DNA release provides a possible mechanistic explanation for the severe growth defect observed with ΔsdhA mutant in macrophage hosts. Our observation may also explain that ΔsdhA mutant has no intracellular growth defects in amoebae hosts that do not have the nuclear acid-sensing innate immune system.
SdhA contains a functionally important GRIP domain that alone is targeted to the Golgi apparatus. Our extensive analyses establish that SdhA is most likely involved in membrane trafficking. This observation is not unexpected given that a large majority of Dot/Icm-secreted substrates appear to modulate the membrane system for appropriate LCV trafficking (6). A plausible model for SdhA function is that it is involved in a certain aspect of vesicular trafficking and thereby maintains LCV membrane integrity to prevent bacterial DNA from leaking out of the LCV. Supporting our model, a recent study that came out during the revision process of our manuscript provides direct evidences that LCV containing ΔsdhA mutant is unstable and, therefore, accessible to host cytosol (35). Despite that it is not known whether preventing DNA release by SdhA is a bacterial “intentional” virulence mechanism, the mode of SdhA action through maintaining the LCV membrane integrity represents another paradigm in bacterial effector-mediated counteraction of host innate immune defense (36).
Materials and Methods
L. pneumophila strains were cultured on buffered charcoal yeast extract agar plates supplemented with 0.1 mg/mL thymidine. L. pneumophila ΔflaA, ΔsdhA, and ΔflaAΔsdhA strains were generated by standard homologous recombination using the suicide plasmid pSR47s. pJB908-based complementation plasmids were introduced into L. pneumophila by electroporation (2.5 Kv, 200 Ω, 25 μF, and 5 ms). The complementation stains were cultured on buffered charcoal yeast extract agar without thymidine. For macrophage infection, fresh single bacterial colonies were streaked onto culture plates 2 d before infection. Bacteria were scraped off, diluted in sterile water, and added to cells at a multiplicity of infection (MOI) of 10. Infection was facilitated by a centrifugation of 300 × g for 10 min. All infection assays were performed in the media without serum and antibiotics. Caspase-1 activation and cell death assays were performed as described (11, 37). The rest of information about plasmid, antibodies, mice, cell culture and siRNA transfection, DNA release, and immunofluorescence assays is presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. V. Dixit for providing Nlrp3−/−, Nlrc4−/−, Aim2−/−, and Asc−/− mice; Dr. T. Taniguchi for Irf3−/− and Irf7−/− mice; Dr. D. Portnoy for Ifnar−/− mice; Dr. K. Fitzgerald for AIM2 constructs; Dr. J. Vogel for pJB2819 plasmid; Dr. L. Yu for TGN38 antibody; and the imaging facility and animal facility at the National Institute of Biological Sciences for technical assistance. This work was supported by National Basic Research Program of China 973 Programs 2010CB835400 and 2012CB518700 and by an International Early Career Scientist grant from the Howard Hughes Medical Institute (to F.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117490109/-/DCSupplemental.
References
- 1.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 4.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 5.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 6.Ge J, Shao F. Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cell Microbiol. 2011;13:1870–1880. doi: 10.1111/j.1462-5822.2011.01710.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz MA. Characterization of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J Exp Med. 1987;166:1310–1328. doi: 10.1084/jem.166.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 10.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 12.Bürckstümmer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 13.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 16.Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci USA. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 18.Molofsky AB, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes-Alnemri T, et al. The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opitz B, et al. Legionella pneumophila induces IFNbeta in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J Biol Chem. 2006;281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- 22.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 2009;5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones JW, Broz P, Monack DM. Innate immune recognition of francisella tularensis: Activation of type-I interferons and the inflammasome. Front Microbiol. 2011;2:16. doi: 10.3389/fmicb.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kjer-Nielsen L, Teasdale RD, van Vliet C, Gleeson PA. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol. 1999;9:385–388. doi: 10.1016/s0960-9822(99)80168-7. [DOI] [PubMed] [Google Scholar]
- 29.Munro S, Nichols BJ. The GRIP domain - a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol. 1999;9:377–380. doi: 10.1016/s0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- 30.Lu L, Tai G, Wu M, Song H, Hong W. Multilayer interactions determine the Golgi localization of GRIP golgins. Traffic. 2006;7:1399–1407. doi: 10.1111/j.1600-0854.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- 31.Sauer JD, et al. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fortier A, Doiron K, Saleh M, Grinstein S, Gros P. Restriction of Legionella pneumophila replication in macrophages requires concerted action of the transcriptional regulators Irf1 and Irf8 and nod-like receptors Naip5 and Nlrc4. Infect Immun. 2009;77:4794–4805. doi: 10.1128/IAI.01546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plumlee CR, et al. Interferons direct an effective innate response to Legionella pneumophila infection. J Biol Chem. 2009;284:30058–30066. doi: 10.1074/jbc.M109.018283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creasey EA, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci USA. 2012. 109:3481–3486. [DOI] [PMC free article] [PubMed]
- 36.Cui J, Shao F. Biochemistry and cell signaling taught by bacterial effectors. Trends Biochem Sci. 2011;36:532–540. doi: 10.1016/j.tibs.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Gong YN, et al. Chemical probing reveals insights into the signaling mechanism of inflammasome activation. Cell Res. 2010;20:1289–1305. doi: 10.1038/cr.2010.135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.