Abstract
Mismatch repair (MMR) is a major DNA repair pathway in cells from all branches of life that removes replication errors in a strand-specific manner, such that mismatched nucleotides are preferentially removed from the newly replicated strand of DNA. Here we demonstrate a role for MMR in helping create new phenotypes in nondividing cells. We show that mispairs in yeast that escape MMR during replication can later be subject to MMR activity in a replication strand-independent manner in nondividing cells, resulting in either fully wild-type or mutant DNA sequence. In one case, this activity is responsible for what appears to be adaptive mutation. This replication strand-independent MMR activity could contribute to the formation of tumors arising in nondividing cells and could also contribute to mutagenesis observed during somatic hypermutation of Ig genes.
Keywords: DNA mismatch repair, oligonucleotide transformation, 8-oxoG
DNA mismatch repair (MMR) recognizes mismatches created in the process of replication and uses some type of strand discrimination signal to selectively remove mismatched nucleotides present on the newly replicated strand of DNA (1–3). In most eukaryotic cells, there are two mismatch recognition complexes with different, but overlapping, specificities: (i) MutSα, a heterodimer of Msh2 and Msh6, recognizing base/base mismatches and small insertion/deletion loops; and (ii) MutSβ, a heterodimer of Msh2 and Msh3, recognizing both small and large loops (1–3). After recognition of a mispair by MutSα or MutSβ, completion of MMR requires association with proteins related to MutL, usually MutLα, which in yeast is a heterodimer of Mlh1 and Pms1 (1–3). DNA on the newly synthesized strand is then excised and the template strand rereplicated.
The method of strand discrimination in eukaryotic MMR is still not solved, although major advances have recently been made. Several components of MMR are known to have association with proliferating cell nuclear antigen (PCNA), a sliding clamp that tracks with the replication fork (1–3). As would be predicted by that model, it has recently been shown that MMR is temporally coupled to replication (4) and that one pathway of MutSα-dependent MMR is through recruitment by a PCNA–Msh6 interaction (5). Whatever signals are used for strand discrimination are presumably lost as replication proceeds.
To study various aspects of MMR, we have used single-stranded oligonucleotides (oligos) to introduce specific mispairs into Saccharomyces cerevisiae chromosomal DNA. We have shown previously that oligos can be introduced into cells by electroporation and can correct frame-shift mutations in LYS2 and that this process is inhibited by MMR (6). Our results are most consistent with a mechanism in which the oligo anneals to either the leading or lagging strand of replication at the replication fork, with subsequent extension. Mispairs created by the oligos are recognized by MMR, but those mispairs that escape MMR recognition create mutations in the next round of replication (6). For the experiments reported here, we used a mutation in an essential codon of the yeast TRP5 gene, the reversion of which occurs solely by restoration of the original codon (7). In this case, we have found that mispairs in the TRP5 gene, created either by oligos or incorporation of 8-oxodGTP, that escape MMR correction during replication are subject to random MMR activity in cells that have stopped growing. Such MMR activity, acting without proper strand discrimination, could be an important source of mutations, both unwanted as in cancer, and beneficial as in somatic hypermutation.
Results
Nontranscribed Strand Oligos Require a Second Round of Replication for Transformation of MMR-Deficient Cells.
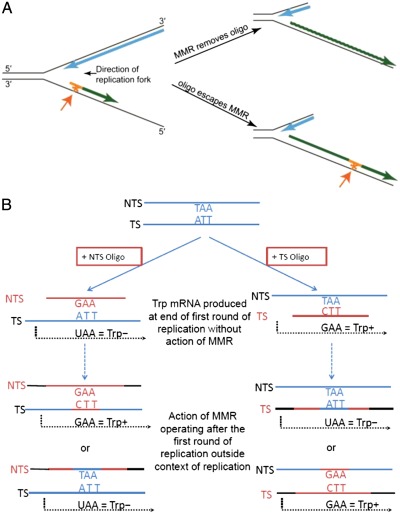
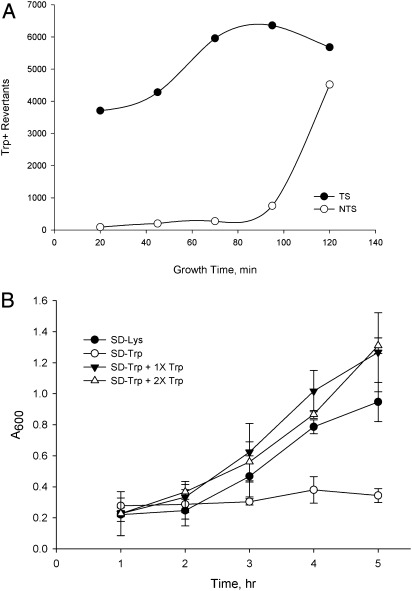
We had previously created a series of point mutations in the TRP5 gene that revert only by restoring the wild-type sequence and, thus, have an extremely low reversion rate and are placed in both orientations near a dependable origin of replication (7). These strains were ideal for studying the effect of MMR on single base mispairs, in contrast to the loop mispairs that had been created in our previous experiments (6). The oligos we used created a base-base mismatch that would restore the wild-type sequence and were the sequence either of the transcribed strand (TS) of TRP5 or of the nontranscribed strand (NTS). We found previously that oligo transformation in MMR-proficient cells is much less efficient than in MMR-deficient cells, because the mismatches created by the oligos are recognized by MMR and removed (6); thus, one observes transformation only by those oligos that have escaped MMR, as illustrated in Fig. 1A. Strains deleted for MSH6 should be devoid of MutSα, and thus should not recognize the mismatch created by these oligos. As indicated in Fig. 1A, a round of replication must precede transformation to allow the oligo to anneal at the replication fork. In addition, as illustrated in Fig. 1B, a cell containing the TS oligo incorporated into the genome can transcribe wild-type TRP5 mRNA even before the cell has completed replication, but cells transformed with an NTS oligo must undergo a second round of replication before a wild-type transcription template is produced. Oligos were introduced into yeast by electroporation; following electroporation, cells were allowed to recover in rich medium before washing and plating on selective media. As we were testing electroporation conditions, we found that TS and NTS oligos responded to the length of recovery time in very different ways in msh6 strains (Fig. 2A): TS oligos required only a short recovery time to produce large numbers of revertants, whereas NTS oligos required a long recovery time. This surprising result could most easily be explained if the cells stopped growth as soon as they were plated on selective medium. The selection for Trp+ is extremely tight; in liquid medium cells do not go through even one cell-doubling in the absence of tryptophan, unlike the same cells when placed in medium lacking lysine (Fig. 2B). The doubling time for these strains in rich medium is ∼2 h; the average increase in cell number after electroporation from 0 to 2 h is 1.8-fold. That growth time fits with what is observed in msh6 cells with the NTS oligo, which requires over 90 min of growth in rich medium before substantial transformation is observed. The small percentage of transformants observed with NTS oligos at earlier growth times is presumably because of cells that stochastically happen to have enough tryptophan to undergo a second cell cycle.
Fig. 1.
Model for transformation of trp5 G148T strains with TS or NTS oligos. (A) An oligo (orange) anneals at the replication fork, creating a mismatch (asterisk and orange arrow), and is extended to form an Okazaki fragment. MMR will remove the oligo, but if the oligo escapes MMR it will be incorporated into the genome. (B) An oligo (orange) is introduced into a cell and anneals with chromosomal DNA (blue). TAA is the NTS sequence of the mutant glutamic acid codon that must be mutated to GAA to give Trp+ revertants in the trp5 G148T strains. At the end of the first round of replication, Trp+ mRNA can be transcribed from the TS oligo, but RNA produced from transformation with a NTS oligo will still be Trp−. MMR correction operating outside the context of the replication fork after the first round of replication could result in two alternatives for each oligo, including Trp+ mRNA from an NTS oligo and Trp− mRNA from a TS oligo.
Fig. 2.
NTS but not TS oligos require a second round of replication for transformation in MutSα-deficient (msh6) strains. (A) trp5 G148T msh6 (Lys−, Trp−) was transformed with a TS or NTS oligo and the number of Trp+ revertants arising on selective plates determined after different growth times in rich medium. (B) One-hundred microliters of a culture of trp5 G148T growing in YPAD was placed in 5 mL of synthetic dextrose (SD) medium lacking lysine, tryptophan, or medium supplemented with the indicated fraction of the normal concentration of tryptophan (20 μg/mL). At the indicated time points, A600 was measured, with each point being the average of three measurements.
NTS Oligos Do Not Require a Second Round of Replication for Transformation of MMR-Proficient Cells.
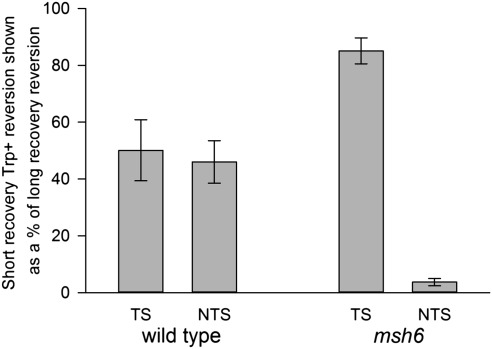
To more extensively study the effect of recovery time on oligo transformation, we chose to examine two time points of recovery growth: 45 min and 2 h. A time of 45 min allows for full recovery of cells from the effects of electroporation but is too short for a second cell cycle to occur. A time of 2 h is sufficient for a second cell cycle but not for a doubling of many transformants, as can be seen in Fig. 2A. Even using the same amount of cells taken at the same growth phase and the same volume of electroporation solution and amount of oligos, we find considerable variation in electroporation efficiency in different experiments (as much as fivefold in the number of Trp+ revertants). By comparing the number of revertants produced in the same electroporation reaction sampled at 45 min and 2 h, we can eliminate the transformation variables and more accurately observe the effect of growth time on transformation. Fig. 3 and Table S1 show the result of multiple experiments in msh6 strains containing the trp5 G148T mutation. TS oligos show little effect of recovery time on transformation (over 80% of the revertants obtained after a long recovery are obtained after a short recovery time), whereas NTS oligos show a greater than 20-fold increase when allowed 2 h of growth in rich medium compared with 45 min (fewer than 4% of the revertants obtained after a long recovery are obtained after a short recovery time).
Fig. 3.
MMR circumvents the requirement of a second round of replication for NTS oligo transformation. Trp+ revertants resulting from transformation with either NTS or TS oligos were determined after 45-min (short) or 2-h (long) recovery time in rich medium. Trp+ revertants obtained at the short recovery time are shown as a percentage of revertants obtained at the long recovery time. Error bars indicate the SD from three or more experiments. NTS oligos create a G-A mismatch; TS oligos create a C-T mismatch.
The results were dramatically different in MMR-proficient strains. NTS oligos showed at most a twofold dependency on recovery time, and TS oligos exhibited approximately the same dependency on recovery time as NTS oligos (Fig. 3) (wild type: 46% for NTS and 51% for TS). The difference between the NTS oligos in wild-type and msh6 strains (and also between the TS oligos, see below) were statistically compared using Bonferroni t tests adjusted for multiple comparisons and were significant at P < 0.01. Similar results were obtained with strains in which the TRP5 gene was in inverted orientation (Table S2).
The results in MMR-proficient strains can most easily be explained by the model illustrated in Fig. 1B. TS oligo transformants can generate Trp+ mRNA after replication is completed in the first cell cycle after transformation. NTS oligos at this same stage still produce mutant Trp mRNA and in the absence of MMR must undergo a second round of replication before a Trp+ mRNA can be produced. However, if the uncorrected mismatches produced by NTS oligo transformation were subject to MMR correction that was replication-strand–independent, Trp+ mRNA could be produced even without a second round of replication. That result is what is observed in wild-type strains, because about 50% of the NTS revertants observed after a long recovery are seen after a short recovery time (Fig. 3). However, Fig. 3 also shows that whereas over 80% of TS revertants are observed after a short recovery time in msh6 cells, that number is only about 50% in wild-type cells. As the model in Fig. 1B indicates, if TS transformants were also subject to random MMR action, some potential TS transformants could be eliminated and that appears to be the case with TS reversion in wild-type strains.
Transformation of 8-oxodGTP Is Subject to MMR Activity.
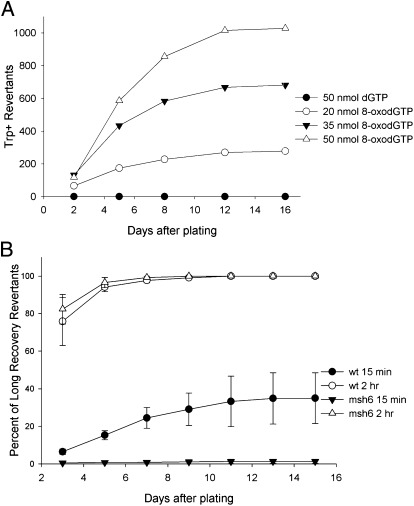
To determine if the MMR activity we observed was a peculiarity of oligo transformation, we studied the transformation of trp5 G148T mutants by 8-oxodGTP. As 8-oxoG can pair with either a C or A, mispairing of 8-oxoGTP incorporated during replication should lead to G148T revertants. We found that these strains could be transformed to Trp+ in a dose-dependent manner by electroporation with 8-oxodGTP, but not by dGTP (Fig. 4A). In this case, the 8-oxodGTP would transform by incorporation into the NTS, opposite an A, and upon replication in the next cell cycle, if replicated with a C opposite the 8-oxoG would create a Trp+ strain (Fig. S1). We would thus expect this transformation to be dependent on recovery time after electroporation, and therefore examined the effect of recovery time on transformation of both MMR-proficient and deficient strains. Because we had observed late-arising transformants (Fig. 4A), we also determined the number of transformants as a function of incubation time on plates. For historical reasons we used a recovery time of 15 min rather than 45 min for the short recovery time, but subsequent experiments revealed essentially no difference between a 15-min and 45-min recovery time. In msh6 strains, we observed almost no transformants with a short recovery time, but substantial transformants with a 2-h recovery time (Fig. 4B and Tables S3–S5) in accordance with our oligo experiments. In MMR-proficient cells, there were substantial numbers of transformants with a short recovery time (about 6% after 3 d on plates of the number of revertants obtained with a long recovery time observed after 2 wk on plates), and a much larger number with a longer recovery time (76%).
Fig. 4.
The 8-oxodGTP transformation displays what appears to be adaptive mutation in wild-type strains and a strict requirement for a second round of replication in msh6 strains. (A) Transformation of trp5 G148T wild-type cells with dGTP or 8-oxodGTP plated after a short recovery time (15 min). Shown is the number of Trp+ revertants counted after the indicated days of incubation on selective plates. (B) Transformation of trp5 G148T strains with 50 nmol 8-oxodGTP. One-half of each transformation was plated after a 15-min recovery time and one-half plated after a 2-h recovery time. The number of colonies on each plate was determined multiple times on the indicated days. Shown at each time point is the percent of transformants obtained for the indicated incubation and recovery time compared with the number of transformants obtained in that experiment after 15 d of incubation for the 2-h recovery time. Error bars indicate the SD from four experiments for each strain.
We had not observed any late-arising transformants with oligo transformation; however, in MMR-proficient strains, but not MMR-deficient strains, Trp+ transformants derived following electroporation of 8-oxodGTP and a short recovery time continue to arise on plates up to 2 wk of incubation, increasing to 35% of the number of revertants obtained when plated after a long recovery time (Fig. 4B and Tables S3–S5). Similar results were obtained in strains with the TRP5 gene in inverted orientation (Tables S6–S8). The continuous appearance of new revertants in wild-type strains on plates lacking tryptophan over a period of 2 wk appears to be “adaptive mutation” in which mutations arise in response to selection (8, 9). Trp− cells are viable on plates lacking tryptophan for long periods (Table S9) but do not replicate their DNA. The late-arising revertants were not inherently slow growing and were true revertants. Both the appearance of Trp+ revertants plated after a short recovery time in MMR-proficient but not MMR-deficient strains and the delayed appearance of Trp+ revertants in MMR-proficient strains can be explained by a model similar to that of Fig. 1B (Fig. S1). An 8-oxoG inserted during replication opposite the A at position 148 in the trp5 G148T mutant would result in an 8-oxoG-A mismatch and would produce only Trp− mRNA. There is no homolog of mutY (a glycosylase removing an A opposite 8-oxoG) in S. cerevisiae (10); therefore, in the absence of MMR this mismatch should not produce a sequence capable of producing Trp+ mRNA until a second round of replication, as is observed in msh6 strains. It is known that MMR recognizes 8-oxoG-A as a mispair (11); therefore, canonical MMR would remove the 8-oxoG inserted opposite A during replication. Thus, for any mutations to be induced by incorporation of 8-oxoG opposite an A, the initial mismatches would have to escape MMR and then template a C in the next cell cycle. However, recognition of the 8-oxoG-A mismatch by MMR postreplication with subsequent repair without a proper strand discrimination signal would, 50% of the time, lead to a transcribed strand containing wild-type sequence that could produce tryptophan and lead to subsequent cell growth.
Discussion
Mispairs that are formed during replication but that escape MMR will in general produce mutations in the next generation. In most cases, it would be difficult to distinguish unrepaired mismatches destined to create mutations in the next cell cycle from those that were repaired without proper strand discrimination. The near absolute dependence of our strains on added tryptophan for growth, coupled with an extremely low reversion rate, allows us to observe what would ordinarily be very rare events. The main indicator of MMR activity acting in a noncanonical way is the result of transformation with NTS oligos and 8-oxodGTP. In both cases, in cells lacking MMR there are few or no transformants if cells are plated before they have a chance to replicate, but there are large numbers of transformants if cells are allowed to replicate before plating. However, with NTS oligo transformation in MMR-proficient cells, there are substantial numbers of revertants when plated after a short recovery time and the number increases only about twofold when a round of replication is allowed with a long recovery time (Fig. 3) (46% of NTS revertants are observed with a short recover time). The number of revertants observed when plated after a long recovery time is a reflection of the maximum number of revertants that have escaped MMR. The observation that half of that number is seen after a short recovery time may reflect the fact that if those potential transformants are acted on randomly by MMR, only half should be repaired in the direction that would create revertants. Similarly, one observes for the TS transformation in Fig. 3 approximately one-half of the revertants in short—compared with long—recovery plating in wild-type MMR-proficient strains. The difference between these numbers can also be explained by MMR acting without proper strand discrimination and removing some of the mismatches that would otherwise form TS-induced revertants.
Transformation with 8-oxodGTP demonstrates even more clearly the effect of MMR in nondividing cells. In this case, there are almost no Trp+ revertants when cells are plated after a short recovery time in msh6 strains, whereas there are a substantial number of revertants in MMR-proficient strains under the same conditions (Fig. 4B). In addition, in MMR-proficient but not MMR-deficient strains, revertants continue to appear upon further incubation when plated after a short recovery time but not when plated after a long recovery time. If the late-arising colonies were because of some residual replication on the plates, one would expect to see late-arising colonies in the msh6 strains or in MMR-proficient strains plated after a long recovery period, but no substantial increase is observed under those conditions. [After 5 d on plates, 97% of transformants are observed in msh6 strains and 94% in MMR-proficient strains plated after 2 h of recovery, but only 15% of an eventual 35% in wild-type strains plated after 15 min of recovery (Fig. 4B and Tables S3–S5).] Because the 8-oxodGTP is incorporated during replication, normal MMR activity that would recognize the 8-oxoG-A mismatch would remove the 8-oxoG and thus prevent reversion. The noncanonical MMR-dependence of the 8-oxodGTP–induced reversion indicates that MMR must be occurring after the loss of any strand discrimination signals. The delayed appearance of some 8-oxodGTP–induced revertants indicates that 8-oxoG-A mismatches can be recognized and “repaired” in a replication-strand–independent manner in a time period of days after formation.
Why are late arising revertants observed with 8-oxodGTP and not with oligo transformation? One of the major differences between these experiments and the oligo transformation experiments is that the oligos create one additional mismatch in the cell, whereas one would expect incorporation of 8-oxodGTP to create a large number of 8-oxoG-A mismatches in the cell. The amount of MutSα in cells is low [one estimate is 1,230 molecules of Msh2p and 5,330 molecules of Msh6p per cell (12)]; thus, the large number of 8-oxodGTP generated mismatches likely saturates the MMR system such that some 8-oxoG-A mismatches in the TRP5 gene are only recognized by MutSα after long periods, thus accounting for the delayed appearance of Trp+ revertants on the plate. In addition, it appears from the number of revertants obtained with NTS oligos, creating a G-A mismatch, compared with TS oligos creating a C-T mismatch, that the G-A mismatch is recognized more poorly by MMR than the C-T mismatch (Table S10). The 8-oxoG-A mispair appears to be even less well recognized, as the final numbers of 8-oxodGTP–induced revertants is not much higher in msh6 strains than in wild-type strains (Table S3), indicating that there could also be significant delays in recognition of, and action on, the 8-oxoG-A mispairs. It has recently been found that mismatches accumulate many copies of MutLα apart from replication factories (5), and it may be that for persistent mismatches only upon that accumulation would any attempts at repair occur.
The MMR activity we observe with NTS oligos and 8-oxodGTP transformation is taking place in nondividing cells; because of the presumably low levels of tryptophan, most of the cells cannot divide even once without tryptophan, and tryptophan can only be made if MMR acts randomly on the oligo or 8-oxoG–induced mismatches, creating a wild-type template strand for mRNA synthesis. However, could this random MMR activity take place in normally growing cells? Canonical MMR has been shown in a variety of both in vitro and in vivo assays to be strongly replication-strand directed (1–3), so it would be surprising to find that it would function efficiently, but randomly, in the absence of replication-strand signals. Our data suggest that a prolonged period of growth cessation may be important for activating random MMR, because some TS revertants appear to be lost in MMR-proficient but not MMR-deficient strains when plated after a short recovery time (Fig. 3).
The idea that a system such as MMR might in some contexts operate in a random manner is not new. As early as 1964, pure Lac− colonies induced by 5-BrUra in Escherichia coli were hypothesized to be the result of a system operating on mismatches persisting after replication (13). Early studies on MMR in yeast showed that a mismatch contained in a duplex plasmid could be repaired in either direction (14), although it is not clear whether individual repair events were directed by adventitious nicks in the plasmid DNA. Very recently, using a purified in vitro system, it was demonstrated that MutLα endonuclease activity could be induced on a closed circular plasmid in a strand-independent manner (15), indicating a possible mechanism for replication strand-independent MMR.
Another example of random MMR activity occurring outside of replication comes from studies of gene-conversion gradients that form as the result of recombination between homologous chromosomes in yeast meiosis. In this process gene conversions result from the repair of mismatches (using the donor template) in heteroduplex DNA that form during the repair of meiotically induced double-strand breaks (DSBs). At loci that display high levels of these DSBs, a gradient is observed where high frequencies of gene conversion are seen near the DSB and lower levels are seen at markers located further away. An explanation for how the gradient forms is that mismatches forming near a DSB are repaired by MMR using the donor chromosome as a template, but those occurring further away are repaired without such a bias (16–18). Another study examined the gene-conversion gradient at ARG4 in meiosis and found that the gradient was essentially flat in a low activity allele of mlh1, which was different from the gradient found in either wild-type cells or a null allele of MLH1 (19). In that work, the steep conversion gradient in the wild-type strain was ascribed to directed MMR close to the DSB and random repair distant from the DSB. The effect of the low-activity allele was explained by a delayed repair process that would be random at positions both close and distal to the initiating DSB because of loss of proper strand discrimination signals. Recent experiments have demonstrated two MMR pathways: one in which MutSα is associated with replication factories, and one in which MutSα recognition is independent of a PCNA association (5). This second pathway is likely the one of interest here.
An explanation of our results that is consistent with the other experiments cited above is that some mispairs present at the replication fork will inevitably escape MMR. In growing cells, those mispairs may accumulate some MMR proteins, but will likely persist until the next round of replication, in which case a mutation will be produced in one of the daughter duplexes. However, when mispairs escape replication, or are formed in other ways, and the cell stops growth, the mispair is subject to MMR recognition in a context that has lost proper strand-discrimination signals. With enough delay in this state, mispairs could be randomly corrected, being directed by a random nicking activity of MutLα (15), or by random transient nicks in the DNA. There may also be some cases, such as in meiosis, when random MMR activity is more common.
The existence of a random MMR activity has broad implications. In 1995, drawing on evidence from a variety of systems, MacPhee hypothesized that randomly acting MMR could be responsible for cancers that arise in nondividing cells (20). In particular, MacPhee imagined DNA damage in resting cells that would be repaired in part by error-prone systems that would leave mismatches subject to random MMR activity. Given a lag time that can be measured in decades, even low levels of random MMR activity could result in significant numbers of mutations. This random MMR activity could also be an additional explanation for stress-induced mutagenesis in growth-arrested cells (21). A related process might occur in somatic hypermutation. It was found in 1998 that contrary to expectations, somatic hypermutation was decreased in the absence of MMR (22) and that mutations at A-T base pairs were particularly sensitive to the presence of MMR (23). Although the mutations observed at C-G sequences in somatic hypermutation and class-switch recombination can be explained by activation-induced cytidine deaminase (24, 25), it has been more difficult to understand the MMR-dependent mutagenesis at A-T base pairs. One possibility is that such mutations could be the result of error-prone repair of U-G mismatches by polymerase (Pol) η, leading to mismatches created at flanking DNA sequences that are corrected in a random manner by MMR. Although these cells are not in a nondividing state, the presence of a localized concentration of mismatches may attract a sufficient number of MMR proteins that could facilitate correction, albeit without proper strand discrimination.
In general, it is difficult to distinguish random MMR activity that would occur with mismatches postreplication, from mutations that would occur in the next round of replication. The lack of growth of the Trp− cells under selective conditions combined with a sensitive detection method has allowed us to observe what is ordinarily a rare event. Evidence indicates that such activity exists in all organisms from bacteria to humans.
Materials and Methods
S. cerevisiae Strains and Oligos.
The genotypes of strains can be found in Table S11. Oligos were 40 nt in length and differed by one centrally located nucleotide from the mutant chromosomal sequence and were gel purified (Eurofins MWG Operon); sequences of oligos are listed in Table S12.
Transformation with Oligos or Nucleotides.
Transformation by electroporation was modified from that used previously (6). An overnight culture of a strain was diluted 1:50 in YPAD (rich medium) (26), incubated with shaking at 30° to an A600 of 1.3–1.4, washed twice with cold H2O, and once with cold 1 M sorbitol. After the final centrifugation, all solution was removed from the cells and a volume of cold 1 M sorbitol equal to that of the cell pellet added to resuspend the cells. For a typical transformation, 200 pmol of an oligo was added to 200 μL of this cell suspension in a 2-mm gap electroporation cuvettete, and the mixture electroporated at 1.55 kV, 200 Ω, and 25 μF (BTX Harvard Apparatus ECM 630). Immediately after electroporation, the cell suspension was added to a volume of YPAD equal to that of the initial culture, and the cells incubated at 30° with shaking for various times. After this recovery period, cells were centrifuged, washed with H2O, and plated on synthetic dextrose medium lacking the appropriate amino acid (26) to select transformants. Transformation with dGTP or 8-oxodGTP (TriLink Biotechnologies) was performed similarly, using 300 μL of cell suspension.
Supplementary Material
Acknowledgments
We thank Rachelle Spell, Sue Jinks-Robertson, Natasha Degtyareva, Frank Gordon, and Paul Doetsch for helpful comments on the manuscript; and Josef Jiricny for suggesting the role of random MMR in somatic hypermutation. This work was supported by National Institutes of Health Grants R01 GM80754 (to G.F.C.) and P01 ES011163 (to Y.W.K. and G.F.C.), and Winship Cancer Institute Seed Grant funds (to Y.W.K. and G.F.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115361109/-/DCSupplemental.
References
- 1.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 2.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 3.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 4.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kow YW, Bao G, Reeves JW, Jinks-Robertson S, Crouse GF. Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands. Proc Natl Acad Sci USA. 2007;104:11352–11357. doi: 10.1073/pnas.0704695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams T-M, Fabbri RM, Reeves JW, Crouse GF. A new reversion assay for measuring all possible base pair substitutions in Saccharomyces cerevisiae. Genetics. 2005;170:1423–1426. doi: 10.1534/genetics.105.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster PL. Adaptive mutation in Escherichia coli. J Bacteriol. 2004;186:4846–4852. doi: 10.1128/JB.186.15.4846-4852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg SM. Evolving responsively: Adaptive mutation. Nat Rev Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- 10.Jansson K, Blomberg A, Sunnerhagen P, Alm Rosenblad M. Evolutionary loss of 8-oxo-G repair components among eukaryotes. Genome Integr. 2010;1:12. doi: 10.1186/2041-9414-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 12.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 13.Witkin EM, Sicurella NA. Pure clones of lactose-negative mutants obtained in Escherichia coli after treatment with 5-bromouracil. J Mol Biol. 1964;8:610–613. doi: 10.1016/s0022-2836(64)80017-6. [DOI] [PubMed] [Google Scholar]
- 14.Bishop DK, Andersen J, Kolodner RD. Specificity of mismatch repair following transformation of Saccharomyces cerevisiae with heteroduplex plasmid DNA. Proc Natl Acad Sci USA. 1989;86:3713–3717. doi: 10.1073/pnas.86.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pluciennik A, et al. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detloff P, White MA, Petes TD. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992;132:113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alani E, Reenan RAG, Kolodner RD. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reenan RAG, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: Evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argueso JL, et al. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol Cell Biol. 2003;23:873–886. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacPhee DG. Mismatch repair, somatic mutations, and the origins of cancer. Cancer Res. 1995;55:5489–5492. [PubMed] [Google Scholar]
- 21.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cascalho M, Wong J, Steinberg C, Wabl M. Mismatch repair co-opted by hypermutation. Science. 1998;279:1207–1210. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- 23.Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 24.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 25.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 26.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.