Abstract
In eukaryotes, although the Mcm2-7 complex is a key component of the replicative DNA helicase, its association with Cdc45 and GINS (the CMG complex) is required for the activation of the DNA helicase. Here, we show that the CMG complex is localized to chromatin in human cells and describe the biochemical properties of the human CMG complex purified from baculovirus-infected Sf9 cells. The isolated complex binds to ssDNA regions in the presence of magnesium and ATP (or a nonhydrolyzable ATP analog), contains maximal DNA helicase in the presence of forked DNA structures, and translocates along the leading strand (3′ to 5′ direction). The complex hydrolyses ATP in the absence of DNA; unwinds duplex regions up to 500 bp; and either replication protein A or Escherichia coli single stranded binding protein increases the efficiency of displacement of long duplex regions. Using a 200-nt primed circular DNA substrate, the combined action of human DNA polymerase ε and the human CMG complex leads to the formation of products >10 kb in length. These findings suggest that the coordinated action of these replication complexes supports leading strand synthesis.
Eukaryotic DNA replication requires the assembly of the prereplication complex (pre-RC) during the G1 phase of the cell cycle. The pre-RC is formed following the ordered recruitment of the origin replication complex (ORC), Cdc6, Cdt1, and the Mcm2-7 complex at DNA origins (1). At the G1/S transition, two S-phase–promoting kinases, the cyclin-dependent (CDK) and Cdc7–Dbf4 (DDK) complexes (2), act jointly with multiple protein factors [which in yeast minimally include Sld2, Sld3, Dpb11, Mcm10, Ctf4, and DNA polymerase (Pol) ε] to promote the association of Cdc45 and GINS (heterotetramer of Sld5, Psf1, Psf2, and Psf3) with Mcm2-7 (3). The latter complex, containing Cdc45, Mcm2-7, and GINS (CMG complex), constitutes the eukaryotic replicative DNA helicase that unwinds origins and contributes to the recruitment of the replicative Pols essential for the synthesis of leading and lagging strands.
In yeasts, Dpb11, Sld2, and Sld3 play critical roles in the origin binding of Cdc45 and GINS. In budding yeast, Sld2 and Sld3 are the only replicative proteins that must be phosphorylated by CDK for the initiation of replication (4, 5). These modifications promote their specific interaction with Dpb11 that facilitate production of the CMG complex by an unknown process. In higher eukaryotes, homologs related to these proteins, RecQL4 (Sld2), Treslin (Sld3), and TopBP1 (Dpb11), are likely to play similar roles. Treslin was shown to be essential for replication, and specific interactions between TopBP1 and Treslin were reported (6). In vivo studies in HeLa cells using bimolecular fluorescence complementation assays revealed that interactions between Cdc45, Mcm2-7, and GINS occurred at the G1/S transition and required CDK and DDK activities (7). Stable interactions between the CMG components required a number of proteins that in yeast were shown to be essential for the formation of the yeast replication progression complex (RPC) (8). These findings suggest that the assembly of initiation proteins in higher eukaryotes and yeasts may be similar.
A number of reports have documented that key components of the replicative DNA helicase include Cdc45, Mcm2-7, and GINS. These components were coassociated at sites where fork progression was halted artificially by a streptavidin–biotin complex in the Xenopus cell-free replication system (9). Kanemaki and Labib (10) isolated a large complex from budding yeast (the RPC) that was formed at the preinitiation complex stage, which moved with the fork. This movement required the specific association of GINS, Cdc45, and the Mcm2-7 complex, and the targeted removal of any of these components immediately halted fork progression and the association of other components of the complex. Botchan et al. (11) isolated a complex from Drosophila embryos that contained stoichiometric levels of the CMG complex as well as the Drosophila CMG (dCMG) complex following expression of these proteins using the baculovirus Sf9 system (12). Both dCMG preparations contained 3′ → 5′ DNA helicase activity.
In this study, we describe the isolation and characterization of the human CMG (hCMG) complex. Like the dCMG complex, the human complex required ssDNA, ATP, and magnesium for its loading onto DNA. hCMG exhibited DNA helicase activity by migrating 3′ to 5′ on ssDNA, but was capable of moving through duplex DNA as well. Maximal hCMG helicase activity required forked DNA structures (containing both 5′ and 3′ DNA tails) as well as an oligo(dT) region at the 3′-loading site. The unwinding of duplex regions extended to 500 bp and replication protein A (RPA) or Escherichia coli single stranded binding protein (SSB) increased the efficiency of displacement of long duplex regions up to 1 kb. Using a 200-nt primed minicircle, we show that the combined action of hDNA polymerase ε and the hCMG helicase resulted in the production of long DNA chains >10 kb, in keeping with the synthesis of a leading DNA strand.
Results
hCMG Complex Is Associated with Chromatin in HeLa Cells.
We investigated whether the CMG complex could be detected in mammalian cells. For this purpose, soluble and chromatin fractions from HeLa cells were prepared and immunoprecipitated with Cdc45 antibodies. As shown in Fig. 1, subunits of the CMG complex were detected both in the soluble (SN) and chromatin fractions (C; lanes 1 and 2); the bulk of Cdc45 and Sld5 proteins (representing the GINS complex) was present in the soluble fraction, whereas Mcm2 (representing Mcm2-7) was distributed almost equally between the two fractions. Importantly, Cdc45 antibodies coimmunoprecipitated Cdc45 with the other subunits of the CMG complex only from the chromatin fraction, as shown by immunoblots using antibodies to Mcm2 and Sld5 subunits (Fig. 1, lane 4). Orc2 (representing the ORC complex), which is localized mostly on chromatin throughout the cell cycle, was used as a quality control of the lysate fractionation (Fig. 1, compare lanes 1 and 2). These findings indicate that the hCMG complex only stably forms on chromatin, although its subunits are abundantly present in chromatin-free fractions. Identical results were obtained from unsynchronized and synchronized cells. We also established that the CMG complex containing DNA helicase activity could be isolated from human 293 cells (Fig. S1).
Fig. 1.
The hCMG complex is associated with chromatin. Soluble (SN) or chromatin (C) fractions (500 μg) isolated from HeLa cells were incubated with Cdc45 antibodies (lane 3 and 4) or nonspecific (GST) antibodies (lane 5 and 6), as indicated at the top of the immunoblots; specific interactions were detected by Western blotting using antibodies to Mcm2, Cdc45, Sld5, or Orc2. Input represents 10% of the lysate used for immunoprecipitation (lanes 1 and 2). The IgG bands in the Cdc45 immunoblot (lanes 3–6) are indicated by an asterisk.
Isolation of the hCMG Complex.
Ample levels of the hCMG complex required for biochemical studies were generated using the baculovirus expression system. Sf9 cells were infected with 11 different viruses, each expressing a unique subunit of the CMG complex. After several purification steps, a protein complex that sedimented in glycerol gradients with a size similar to thyroglobulin (669 kDa) was isolated (Fig. 2A and enlargement of the peak fraction shown in Fig. 2B). In addition to the expected protein bands, slightly faster-migrating bands associated with Mcm7 and Cdc45 (doublets; Fig. 2B) were noted possibly due to proteolysis. The cosedimentation of the protein components of the hCMG complex was verified by Western blot analysis (Fig. S2). Furthermore, a peak of DNA unwinding activity cosedimented with the hCMG protein complex (Fig. 2C). Helicase assays were carried out using M13 DNA annealed to labeled oligonucleotides containing a 5′-dT40 tail. We also examined ATP hydrolysis using the peak hCMG glycerol gradient fraction. The complex hydrolyzed ATP (Km = 625 μM; Fig. S3) and its rate of hydrolysis was unaffected by the presence of ss or dsDNA. The rate of ATP hydrolysis by the hCMG complex was 181 molecules per minute per molecule of CMG (Kcat), comparable to that observed with the dCMG complex (12).
Fig. 2.
Purification of hCMG complex. (A) Silver-stained gel of glycerol gradient fractions of purified hCMG complex. Each gradient fraction (15 μL) was loaded onto a 4–20% gel (Invitrogen). The position of standard proteins (GE Healthcare Life Sciences) following sedimentation is indicated above the fractions (669 kDa, thyroglobulin; 150 kDa, aldolase; and 75 kDa, conalbumin). (B) Enlargement of the silver-stained fraction 7 shown in A. Protein bands are labeled at the right of the gel. (C) Cosedimentation of DNA helicase activity with the hCMG complex. Helicase assays were performed with aliquots (0.5 μL) of each fraction as described in Materials and Methods. The structure of the helicase substrate containing a 39-mer duplex region and a 5′-dT40 (M13 annealed to labeled oligonucleotide 3) is shown on the left side of the gel. B, boiled substrate.
Other properties of the hCMG helicase were investigated. In the presence of a fixed level of magnesium acetate (10 mM), maximal unwinding was observed at low ATP levels (0.2–0.5 mM; Fig. S4A). In contrast, when the hCMG complex was replaced with the hMcm4/6/7 complex, higher ATP levels (∼10 mM) were required for its maximal helicase activation (Fig. S4B). Although the budding yeast Mcm2-7 complex was shown to possess weak DNA helicase activity in the presence of high levels of acetate or glutamate (13), we failed to detect helicase activity with the hMcm2-7 under these conditions (Fig. S5). These results indicate that similar to findings with the dMcm2-7 complex, activation of the hMcm2-7 helicase activity required its association with Cdc45 and GINS.
Properties of hCMG Helicase Activity.
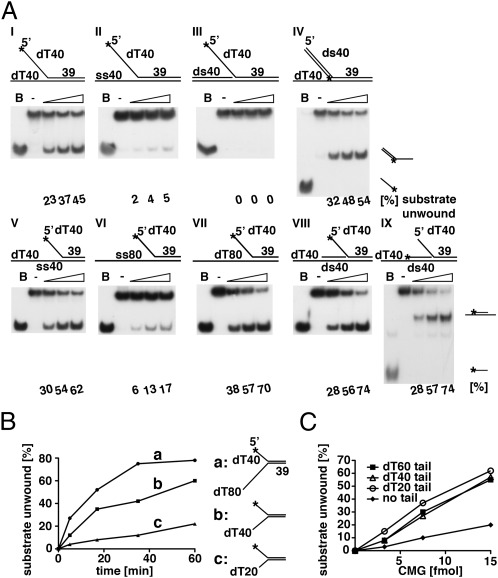
We examined the DNA helicase activity of the hCMG complex with oligonucleotide substrates containing the same 39-bp duplex sequence but different 3′- and 5′-tailed regions (Fig. 3A). Forked substrates possessing a 3′-tailed dT40 strand were displaced efficiently whether the 5′-tail was ss (designated I in Fig. 3A) or dsDNA (IV; Fig. 3A). However, unwinding was not detected when the 3′-tail was converted to a dsDNA structure (III; Fig. 3A), in keeping with the notion that the CMG complex unwinds DNA by translocating in the 3′ to 5′ direction on ssDNA. When the 3′ dT40 tail was changed to a random sequence, the unwinding efficiency was reduced ∼10-fold (compare activities observed with I and II in Fig. 3A), suggesting that the nucleotide sequence at the 3′-tail influenced the activity of the hCMG complex. The influence of the 3′-tail sequence on the helicase activity was examined using substrates with either oligo(dT20), oligo(dA20), oligo(dC20), or a random 20-nt sequence (Fig. S6). Although 3′-tailed oligonucleotides of this length are used less efficiently than longer 3′-tailed substrates, the 3′-oligo(dT20) substrate was unwound far more efficiently than those containing other 3′-tailed sequences. When the 3′ 40-nt ssDNA tail of the substrate described in Fig. 3A, II, was increased to 80 nt with either dT (V) or a random sequence (VI), the unwinding efficiency was increased ∼12-fold and ∼threefold, respectively. Extending the 3′-oligo(dT) tail from 40 to 80 nt increased the unwinding efficiency only ∼1.6-fold (compare unwinding of substrates I and VII in Fig. 3A). The efficiency of unwinding of an oligonucleotide containing a duplex region between the end of the 3′-ssDNA tail and the fork (shown in substrate VIII in Fig. 3A) was comparable to that devoid of the duplex region (substrate V). Interestingly, the unwinding reaction occurred without affecting the upstream duplex (shown with substrate IX; compare the labeled strand displaced in reactions using substrates IX and VIII; Fig. 3A). These findings indicate that the hCMG complex can translocate along dsDNA.
Fig. 3.
DNA helicase activity of the hCMG complex on various DNA substrates. (A) Increasing levels of CMG (7.5, 15, and 30 fmol) were incubated with oligonucleotide substrates (see Table S1 for description of oligonucleotides used in this assay). The first two substrates (I and II; Upper) were formed by annealing oligonucleotide 3 (labeled) with 9 or 11, respectively. Substrate III was prepared by annealing oligonucleotide 3 (labeled) with 11 (bottom strand) and 14. Substrate IV was made by annealing oligonucleotide 15 (labeled) with 5 and 9 (bottom strand). The three substrates (V, VI, and VII; Lower) were synthesized by annealing oligonucleotide 3 (labeled) with oligonucleotide 12, 13, or 10, respectively. Substrate VIII was made by annealing oligonucleotide 3 (labeled) with oligonucleotides 12 (bottom strand) and 14. Substrate IX was generated by annealing oligonucleotide 14 (labeled) with oligonucleotides 12 (bottom strand) and 3. (B) Helicase assays with oligonucleotide substrates containing different 3′-tail lengths. Substrates containing the 3′-oligo(dT40) or 3′-oligo(dT80) tails are the same as I and VII described in A. Substrates containing 3′-oligo(dT20) tails was made by annealing oligonucleotide 3 (labeled) with 8. CMG (15 fmol) was incubated with substrates for varying incubation periods, and the substrate unwound (%) plotted against the time of incubation. (C) Helicase assays with M13 substrates containing a 39-mer duplex region and different 5′-dT tail lengths (0, 20, 40, and 60 nt) prepared by annealing labeled oligonucleotides 1, 2, 3, or 4 with M13. The unwound substrate formed (%) was calculated and plotted against the level of CMG complex added.
As shown in Fig. 3B, the rate of unwinding observed at early time points increased ∼5- to 10-fold as the length of the 3′-dT tail of oligonucleotide substrates was increased from 20 to 80 nt. When the 5′-dT tail length of M13 templates was increased from 0 to 20, 40, or 60 nt, helicase activity increased only when chains were extended from 0 to 20 nt (Fig. 3C). The hCMG complex readily displaced short duplex M13 substrates devoid of a 5′-tail but failed to displace oligonucleotide substrates that lacked a 5′-tail. This discrepancy may be due to an increased loading of the hCMG complex on the long ssDNA regions present in M13 substrates.
Collectively, these findings indicate that the hCMG complex unwinds duplex DNAs by translocating in the 3′ to 5′ direction on ssDNA, and its movement is stimulated by 3′-tailed dT sequences and fork structures. Though the significance of the movement through duplex regions is unclear, this property could permit the complex to move through short duplex regions that cannot be displaced. A more detailed analysis of this type of translocation, previously noted with other replicative helicases (14, 15), may help to define its importance.
Processivity of the hCMG Helicase.
The processivity of the hCMG helicase activity was examined with substrates prepared by annealing a 5′ 32P-labeled oligonucleotide containing a 5′-oligo(dT40) tail to M13 DNA. The duplex region was extended by Sequenase as previously described (16). The extent of unwinding by the hCMG complex was examined with two substrates, one containing a duplex that varied from 39 to ∼500 bp and the other with duplex regions that were >500 bp. The length of the DNA duplex in these substrates was controlled by the level of dideoxy CTP (ddCTP) added during the extension reaction. The hCMG complex displaced duplex regions extending up to 500 bp but failed to unwind longer regions under the conditions used (Fig. 4A), similar to findings reported with the dCMG complex (11).
Fig. 4.
Processivity of the CMG helicase activity. (A) Substrates used for the processivity studies were prepared as described in Materials and Methods. For the preparation of the short duplex DNA substrate (39–500 bp) used in lanes 1–5, the annealed oligonucleotide was extended in the presence of dNTPs containing ddCTP. For the preparation of the long duplex substrate (>500 bp), described in lanes 6–10, the annealed oligonucleotide was extended in the absence of ddCTP. Increasing levels of CMG (12.5, 25, and 50 fmol) were incubated with DNA substrates as described in Materials and Methods. (B) Comparison of the processivity in the presence and absence of RPA or E. coli SSB. Substrates containing duplex regions, as shown, were preincubated with 40 fmol the hCMG complex in the presence of 0.05 mM ATP and then supplemented with 0.45 mM ATP and RPA (1.7 pmol) or E. coli SSB (0.27 pmol) followed by further incubation for 30 min.
We examined whether RPA or E. coli SSB influenced the extent of unwinding of these substrates (Fig. 4B). The CMG complex was first preincubated with M13 DNA substrates in the presence of 0.05 mM ATP (conditions that favored binding rather than unwinding; Fig. S7) followed by the addition of more ATP (final 0.5 mM) and RPA or E. coli SSB. Both DNA-binding proteins stimulated displacement of longer duplex regions of the 39- to ∼500-bp duplex substrates preferentially (>200 bp) and supported the unwinding of regions up to 1 kb. These results suggest that the hCMG complex acts as a processive helicase and replication protein A (RPA; or E. coli SSB) sequesters the ssDNA generated during the unwinding reaction.
DNA-Binding Properties of the hCMG Complex.
Interactions between the hCMG complex and forked DNA oligonucleotides were examined using EMSA assays. In the presence of adenosine 5′-(γ-thio)triphosphate (ATPγS) and magnesium acetate, relatively stable interactions between hCMG and DNAs were detected, whereas no binding was observed when ATPγS or magnesium acetate was omitted (Fig. 5A). Although a significant amount of the DNA–helicase complex was detected as a band that barely entered the gel, substantial levels of the 32P-labeled DNA were distributed throughout the gel, suggesting that the DNA complex dissociated during the electrophoretic separation. When ATP rather than ATPγS was added, substrate unwinding and low levels of DNA binding were observed (Fig. 5A). In the presence of ATP, the level of substrate unwound (39%) was less than the amount of substrate bound in the presence of ATPγS (65%). This discrepancy may be due to some binding of the hCMG complex to the 5′-dT40 tail or to the instability of the DNA–helicase complex. The binding of hCMG to DNA was also observed when adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP) was used in lieu of ATPγS, whereas AMP and ADP failed to support this interaction.
Fig. 5.
DNA binding activity of the hCMG complex. EMSA assays were performed with DNA substrates under different conditions. Reactions contained 15 fmol of hCMG. (A) EMSA assays with fork-structured DNAs containing 5′- or 3′-oligo(dT40) tails prepared by annealing oligonucleotides 3 and 9 (Table S1). (Right) EMSA assays were performed with the nucleotides indicated at the top of the gel. The DNA products formed (substrate–protein complex, substrate, and unwound product) are illustrated. (B) EMSA assays with substrates shown at top of gels. The first three substrates used were prepared by annealing oligonucleotides 1 with 7, 3 with 7, and 1 with 9. The next two substrates were prepared with labeled oligonucleotides 3 and 9, and the last substrate indicated was identical to that described in A. (C) CMG binding to fork-structured substrates containing different 3′-tail lengths. The substrates used were the same as those described in Fig. 3B Right. The nucleoprotein complex formed (%) was calculated and plotted against the CMG complex added.
The interactions between different DNA substrates and hCMG complex were analyzed (Fig. 5B). The complex bound to ssDNA- and ssDNA-tailed duplexes but not to fully duplex DNA. In the presence of ATPγS, hCMG interacted more efficiently (twofold) with ssDNA containing a 3′-oligo(dT40) tail than with 5′-oligo(dT40)-tailed ss oligonucleotides. Perhaps the 3′-terminal location of the oligo(dT40) tail contributed to this increased efficiency. EMSA assays with forked DNA substrates containing different 3′-dT tail lengths (20-, 40-, and 80-mer), similar to those described in Fig. 3B, showed that the hCMG complex binding to DNA increased as the length of the 3′-tail increased (Fig. 5C). Thus, increasing the 3′-tail length of forked DNA structures supports a more efficient binding of the complex as well as an increased rate of unwinding.
CMG Complex and Pol ε Are Coupled on a Rolling Circle Substrate.
We investigated whether the hCMG helicase activity (moving 3′ → 5′) could be coupled with the replicative Pols (resulting in leading strand synthesis). For this purpose, we devised a rolling circle assay using a 200-nt minicircle containing an annealed oligonucleotide that provided a 5′-dC30 tail and a 20-nt duplex region. Fig. 6 and SI Materials and Methods describe the rolling circle substrate and order of addition of reagents. In the presence of hCMG, human Pol (hPol) ε, replication factor C (RFC), proliferating cell nuclear antigen (PCNA), and E. coli SSB, DNA fragments >10 kb were formed (Fig. 6A, lane 7). Formation of these products required hPol ε; the hCMG complex; hRFC and hPCNA; ATP (Fig. 6A, lanes 2–5, respectively); and E. coli SSB (lane 6), presumably to sequester the emerging ssDNA product. Low levels of the long DNA products were formed in the absence of the hCMG helicase, which we attribute to the limited displacement action of Pol ε (Fig. 6A, lane 3). The addition of hCMG increased the synthesis of long DNA chains ∼fivefold, compared with that observed with Pol ε alone (Fig. 6A, compare lanes 3 and 7; Fig. 6B, compare lanes 2 and 3 with lanes 8 and 9). When hPol δ was used in place of hPol ε, significantly shorter leading-strand products were formed in the presence and absence of hCMG (Fig. 6B, lanes 5 and 6, and 11 and 12). Both hPols were capable of fully extending singly primed M13 substrates, as previously reported (Fig. 6C) (17). For reasons that remain to be explained, RPA in contrast to E. coli SSB, failed to support the Pol ε-catalyzed synthesis of long DNA products. In addition, the reactions described in Fig. 6 yielded low levels of DNA synthesis (2 pmol of nucleotide incorporated in Fig. 6A, lane 7). However, these findings suggest that hPol ε acts more efficiently with the hCMG helicase than hPol δ to support the synthesis of long leading strands.
Fig. 6.
Rolling circle assay. The primed 200-nt minicircle was synthesized as described in SI Materials and Methods. (A) Requirements for Pol ε-catalyzed leading-strand synthesis. Reactions (15 μL) containing 20 mM Tris⋅HCl (pH 7.5), 10 mM magnesium acetate, 10 mM potassium glutamate, 1 mM DTT, 0.1 mg/mL BSA, 0.2 mM EDTA, 3.75% glycerol, 0.5 mM AMP-PNP, rolling circle DNA substrate (50 fmol), and hCMG complex (15 fmol) were incubated for 10 min at 37 °C; 0.12 mM dCTP, 0.12 mM, dGTP, and 0.03 mM [α-32P]dATP (specific activity 37,700 cpm/pmol) were added with RFC (20 fmol), PCNA (1 pmol), and hPol ε (70 fmol). After 5 min at 37 °C, 5 mM ATP was added and the reaction incubated for 10 min, after which E. coli SSB (0.5 pmol) was added and the mixture incubated for 60 min. Mixtures were adjusted to 10 mM EDTA and separated on an alkaline agarose gel (1%) at 15 W for 2.5 h. The gel was washed with water, dried, and audioradiographed at −80 °C. (B) Comparison of leading-strand synthesis by hPol ε and hPol δ. Reactions were as described in A with 50 mM potassium glutamate, 50 fmol CMG complex, and 35 and 70 fmol hPol ε or 35, 70, and 300 fmol of hPol δ, where indicated. After incubation, samples were treated with Proteinase K (0.1 mg/mL) in reactions containing 20 mM EDTA, 1% SDS, and 40 μg of yeast tRNA. Following ethanol precipitation, samples were subjected to alkaline agarose gel separation as described above. (C) Elongation of singly primed M13 by hPol ε and hPol δ. The activity of hPol ε (55 fmol) and hPol δ (44 fmol) observed with singly primed M13 (7 fmol) following incubation for 30 min at 37 °C was carried out as previously described (17).
Discussion
In this report, we described the isolation and characterization of the hCMG complex. Like the dCMG complex, the human complex contains DNA helicase activity and translocates in a 3′-to-5′ direction. Although the hMcm2-7 complex contains all of the essential helicase motifs, demonstrable unwinding activity required its association with Cdc45 and GINS. Similar to the dCMG complex, the human complex required ATP (or analogs such as ATPγS, AMP-PNP) and magnesium to bind to ssDNA and hydrolyzed ATP to ADP and Pi in the absence of DNA. hCMG displaced duplex regions extending to 500 bp, and the length was increased substantially by SSBs (RPA and E. coli SSB). We noted that the hCMG helicase activity is most active with forked DNA substrates containing oligo(dT) sequences on the 3′-strand. Both helicase and DNA-binding activities were increased significantly as the length of the 3′-ss tail was increased. In addition to translocating along ssDNA, the hCMG complex moved through dsDNA regions and displaced downstream 5′-ss tailed duplex regions, similar to observations noted with other DNA helicases (14, 15).
We immunoprecipitated the hCMG complex from HeLa cells and demonstrated it was localized to chromatin. Using human 293 cells synchronized in S phase and constitutively expressing N-terminal Flag-tagged Cdc45, we also isolated the hCMG complex following glycerol gradient sedimentation and showed that helicase activity cosedimented with Mcm2 (reflecting the Mcm2-7 complex), Psf3 (reflecting the GINS complex), and Cdc45. These findings suggest that the hCMG complex is associated with chromatin, in keeping with the cell-cycle and stepwise pathway that leads to the formation of a replication fork (1). It is not clear whether production of the complex from overexpressed proteins (i.e., the baculovirus system) depends on or circumvents the biologically relevant pathway. The relatively low yield of the isolated human complex (∼35 pmol from 2 L of Sf9 cells) may be due to its specific formation on chromatin and/or the intrinsic difficulties in the coexpression of 11 proteins. The yield of the human complex was ∼20% of that reported for the dCMG complex (12). These findings may also be due to protein modifications required for CMG complex formation. Perhaps viral-infected insect cells modify Drosophila proteins more efficiently than human proteins, contributing to the differences in the level of complex production.
How the hMcm2-7 complex, which lacks helicase activity, is activated following its interactions with Cdc45 and the GINS complex is unclear. In vivo, Mcm2-7, Cdc45, and GINS are loaded separately onto chromatin. Before formation of the CMG complex, duplex DNA appears to pass through the central cavity of the Mcm2-7 complex (18, 19). In accord with this notion, the yeast Mcm2-7 complex was shown to roll along duplex DNA in the absence of ATP (18). Conversion of the Mcm2-7 complex to the helicase active CMG complex requires alterations in the duplex DNA-Mcm2-7 structure such that only an ssDNA strand is located within the central cavity of the CMG complex. Mutant studies with the yeast Mcm2-7 complex (13, 20) and EM studies with the dCMG complex (21) suggest that the circular Mcm2-7 complex contains an opening (gate) between the Mcm2 and Mcm5 subunits. EM studies indicate that Cdc45, GINS, and ATP may contribute to the dynamic opening and closing of this gap to generate an active helicase. The finding that the hCMG complex translocates through duplex DNA (Fig. 3A), presumably with duplex DNA located within its central cavity, suggests that the hCMG, like other replicative helicases, can translocate through both ss and dsDNA structures. Further biochemical and structural studies will be required to understand how this switch is carried out.
We initiated experiments to develop a eukaryotic replication fork to investigate whether the hCMG helicase activity could be coupled with the replicative DNA Pols. We constructed a 200-nt circular primed DNA template and found that in the presence of Pol ε, long (>10 kb) leading-strand DNA products were generated. Thus, by coupling the hCMG helicase to the action of Pol ε, the helicase displaced duplex regions >10 kb, much longer than the duplex region displaced in the absence of Pol ε (up to 1 kb). The extensive unwinding reaction required the processivity factors PCNA and RFC, both of which are essential for the in vitro replication of singly primed DNA templates by purified hPol ε (17). When Pol ε was replaced by Pol δ, long hCMG-dependent DNA chains were not observed. Although the reasons for this discrepancy remain unclear, in vitro studies with hPol δ and hPol ε revealed that the latter was a more processive polymerase (17). In vivo studies in Saccharomyces cerevisiae suggest that Pol ε synthesizes leading strand, whereas Pol δ supports lagging-strand synthesis (22). The mechanisms contributing to this strand specificity are presently unclear. In the SV40 replication system, both leading and lagging strands are synthesized by Pol δ, but Pol ε cannot be used in lieu of Pol δ (23–25). Whether this is due to the specific roles of SV40 T antigen (an origin binding protein as well as a helicase), RPA, and/or the Pol α-primase complex are unknown. We have carried out leading- and lagging-strand synthesis using the archaeal proteins Pol B, RFC, PCNA, and Mcm (3′ → 5′ DNA helicase), and the two-subunit DNA primase (26). Efforts to carry out similar reactions with the eukaryotic enzymes are underway.
In vivo DNA replication occurs on chromatin rather than naked DNA. Thus, it would be important to examine the interaction of the CMG complex with chromatin and determine whether the complex can unwind chromatinized DNA. It is likely that nucleosome chaperones and chromatin-remodeling complexes play important roles in the action of the CMG complex. In support of this notion, FACT was identified as a component of the yeast RPC (8). Other factors such as MCM10, Ctf4, Tim-Tipin, and Claspin are critical for DNA replication. How and whether these factors affect the action of the CMG complex in vitro remain to be examined.
Materials and Methods
Expression and Purification of hCMG, Mcm2-7, and Mcm4/6/7 Complexes.
The isolation of these complexes is described in SI Materials and Methods. The purity of Mcm2-7 and Mcm4/6/7 complexes is show in Fig. S8.
DNA Helicase Assay.
Unless indicated, helicase assays were carried out in reactions (20 μL) containing 25 mM Hepes-NaOH (pH 7.5), 2.5–9.75 mM NaCl, 0.5 mM ATP, 10 mM magnesium acetate, 1 mM DTT, 0.1 mg/mL BSA, and 2.5 fmol of DNA substrate. Reactions were incubated at 37 °C for 30 min, halted with 4 μL 6× stop solution [50 mM EDTA/pH 8.0, 40% (wt/vol) glycerol, 2% SDS, 0.8% xylene cyanol, 0.3% bromophenol blue], subjected to 10% PAGE separation at 150 V in 1× 89 mM Tris base, 89 mM boric acid, and 2 mM EDTA [Tris/boric acid/EDTA (TBE)] and dried on DEAE-cellulose paper and autoradiographed. The resolved DNA products were quantified using a PhosphorImager (Fujifilm). Processivity analysis of the CMG helicase activity in the presence of RPA or E. coli SSB was carried out as follows: the CMG complex was preincubated with the substrate for 10 min at 37 °C in the presence of ATP (0.05 mM) followed by the addition of more ATP (final 0.5 mM) and RPA or E. coli SSB and an additional incubation of 30 min. Reactions were stopped with 5 μL stop solution (100 mM EDTA and 0.5% SDS), deproteinized with 5 μg of Proteinase K (New England Biolabs) at 37 °C for 1 h and products separated by electrophoresis through a 2.5% agarose gel (12 × 22 cm) in 1× TBE at 200 V for 3.5 h.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant GMS R01 GM034559.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203734109/-/DCSupplemental.
References
- 1.Prasanth SG, Méndez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos Trans R Soc Lond B Biol Sci. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka S, Araki H. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma. 2010;119:565–574. doi: 10.1007/s00412-010-0291-8. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 5.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 6.Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Im JS, et al. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci USA. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 9.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 10.Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan DL, Davey MJ, O'Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 15.Shin JH, Jiang Y, Grabowski B, Hurwitz J, Kelman Z. Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J Biol Chem. 2003;278:49053–49062. doi: 10.1074/jbc.M308599200. [DOI] [PubMed] [Google Scholar]
- 16.Lee JK, Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc Natl Acad Sci USA. 2001;98:54–59. doi: 10.1073/pnas.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bermudez VP, Farina A, Raghavan V, Tappin I, Hurwitz J. Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. J Biol Chem. 2011;286:28963–28977. doi: 10.1074/jbc.M111.256289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochman ML, Schwacha A. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 ‘gate’. Nucleic Acids Res. 2010;38:6078–6088. doi: 10.1093/nar/gkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 24.Lee SH, Pan ZQ, Kwong AD, Burgers PM, Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J Biol Chem. 1991;266:22707–22717. [PubMed] [Google Scholar]
- 25.Nethanel T, Kaufmann G. Two DNA polymerases may be required for synthesis of the lagging DNA strand of simian virus 40. J Virol. 1990;64:5912–5918. doi: 10.1128/jvi.64.12.5912-5918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chemnitz Galal W, Pan M, Kelman Z, Hurwitz J. Characterization of the DNA primase complex isolated from the archaeon, Thermococcus kodakaraensis. J Biol Chem. 2012 doi: 10.1074/jbc.M111.338145. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.