Abstract
TLS/FUS (TLS) is a multifunctional protein implicated in a wide range of cellular processes, including transcription and mRNA processing, as well as in both cancer and neurological disease. However, little is currently known about TLS target genes and how they are recognized. Here, we used ChIP and promoter microarrays to identify genes potentially regulated by TLS. Among these genes, we detected a number that correlate with previously known functions of TLS, and confirmed TLS occupancy at several of them by ChIP. We also detected changes in mRNA levels of these target genes in cells where TLS levels were altered, indicative of both activation and repression. Next, we used data from the microarray and computational methods to determine whether specific sequences were enriched in DNA fragments bound by TLS. This analysis suggested the existence of TLS response elements, and we show that purified TLS indeed binds these sequences with specificity in vitro. Remarkably, however, TLS binds only single-strand versions of the sequences. Taken together, our results indicate that TLS regulates expression of specific target genes, likely via recognition of specific single-stranded DNA sequences located within their promoter regions.
Expression of protein-coding genes in eukaryotes involves a number of tightly regulated steps, each of which is controlled by various proteins to ensure transcripts are appropriately expressed and processed. Some proteins are known to regulate more than one step to integrate the various events (1), and one candidate for linking transcription and pre-mRNA splicing is the protein TLS/FUS (translocated in liposarcoma or fused in sarcoma; here referred to as TLS). As the name suggests, the TLS gene was originally found at the breakpoint of a characteristic translocation in human liposarcomas (2). More recently, mutations in TLS have been implicated in both familial and sporadic amyotrophic lateral sclerosis (3, 4). TLS is structurally related to Ewing's sarcoma (EWS) and TATA-binding protein-associated factor 15 (TAF15), both of which are also involved in translocations that result in cancer-related fusion proteins. These three proteins comprise the TET (TLS, EWS, and TAF15) family of proteins.
TET proteins have been implicated in RNA polymerase (RNAP) II transcription by their association with the general transcription factor TFIID and with RNAP II itself (5). Proteins associated with TFIID can activate or repress transcription of specific genes both by directly recognizing and binding to core promoter sequences and by association with stimulatory or repressive factors and complexes. Each of the TET proteins copurifies with distinct and substoichiometric fractions of TFIID (6), perhaps influencing activation or repression of certain groups of genes. TLS interacts directly with the TATA-binding protein (TBP) and can enhance transcription by RNAP II in vitro (7).
Although TLS has been shown to bind DNA (8), RNA (2), and proteins involved in transcription (6), little is known about which RNAP II genes are directly regulated by TLS. TLS may activate transcription of certain response genes by interacting with the DNA-binding domain of various nuclear hormone receptors (9). Furthermore, the glutamine-rich amino termini of TET proteins can function as transcriptional activation domains when fused to a DNA-binding domain (10). TLS also associates with RNAP III-transcribed genes and represses their transcription both in vitro and in vivo (7).
TLS has also been linked to splicing. It contains an RNP-type RNA-binding domain and associates directly with SR protein splicing factors (11). TET proteins have been detected in spliceosomes (12), and TLS was found associated with RNAP II and snRNPs in a transcription and splicing complex in vitro (13). It is unclear whether and how TLS recruits splicing factors to sites of active transcription, but one possibility is through its interaction with TBP and the TFIID complex.
Here we provide insight into TLS regulation of RNAP II-transcribed genes. We used ChIP followed by promoter microarray analysis to identify putative TLS target genes, and confirmed that several of them are indeed associated with TLS. Furthermore, we detected changes in mRNA levels of several of these transcripts after siRNA-mediated knockdown or overexpression of TLS, indicating that TLS can both activate and repress target genes. Using bioinformatics to analyze the microarray data, we found specific sequences enriched in the DNA fragments immunoprecipitated by TLS, defining possible recognition motifs. Unexpectedly, these sequences were bound specifically as ssDNA by purified TLS in vitro. Together, our data establish TLS as an unusual transcriptional regulator with the potential to activate or repress target genes via specific ssDNA sequences.
Results
ChIP–Chip Analysis Identifies Possible TLS Target Genes.
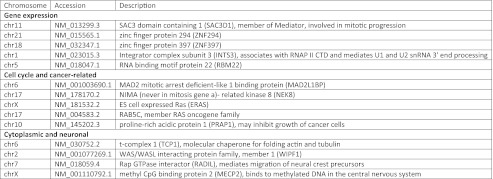
Important questions regarding TLS function include the nature of its role in RNAP II transcription and whether it regulates certain types or classes of genes. To identify RNAP II promoters that are bound, directly or indirectly, by TLS, we performed ChIP using antibodies directed against TLS, then amplified and labeled the DNA for hybridization to the Affymetrix Human Promoter 1.0 microarray chip (Materials and Methods). This tiling microarray contains 25-mer probes, with a gap of ∼10 bp between probes. The promoter region encompasses ∼7.5 kb upstream and ∼2.45 kb downstream of the transcription start site at >25,000 genes, yielding >4.6 million probes. We used a mock (no antibody) ChIP as a control for nonspecific immunoprecipitation and as a measure of noise. Subtraction of the mock from the TLS signal and comparison with a null model yielded a P value for each probe on the microarray. We found that, depending on the significance threshold chosen, 1,161 (P < 0.05) and 48 (P < 0.01) promoter regions were occupied by TLS (the latter are listed in Fig. S1; the former are available upon request). The corresponding genes could be grouped into general categories, many corresponding to processes in which TLS has been implicated (5). Putative target genes involved in gene expression; cell cycle and cancer; and cytoplasmic or neuronal functions are presented in Fig. 1.
Fig. 1.
TLS microarray candidate genes. TLS was enriched (P < 0.01) at the promoter of genes involved in gene expression; cell cycle and cancer,; cytoplasmic; and neuronal proteins. Accession information and a brief description are given for each target.
TLS Associates with Candidate Gene Promoter Regions.
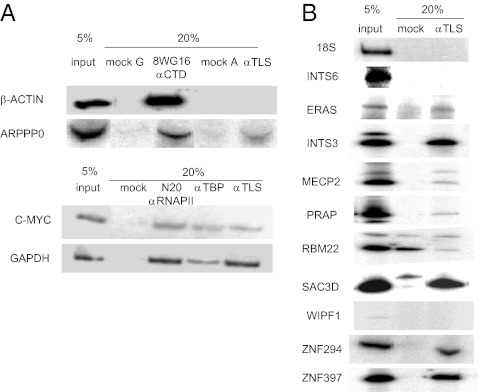
We next wished to verify that TLS associates with targets identified from the microarray. To this end, we designed primers specific to regions identified by microarray and used these for gene-specific ChIP. We also examined various additional genes, including the constitutively expressed genes β-actin, c-MYC, GAPDH, and the highly expressed gene encoding acidic ribosomal phosphoprotein P0 (ARPP P0). TLS was not detected at the β-actin promoter and only weakly at the ARPP P0 promoter (Fig. 2A). The microarray data showed low levels (P > 0.05) of TLS binding at both the c-MYC and GAPDH promoters, and we did detect TLS at these promoters (Fig. 2A). We also used primers for 18S rRNA genes, which are not expected to be recognized by TLS, and TLS occupancy was undetectable (Fig. 2B).
Fig. 2.
Confirmation of TLS target genes by ChIP. (A) ChIP assays were performed using antibodies to RNAP II, TBP, TLS, or mock (no antibody). DNA fragments were amplified by PCR. (B) ChIP assays were performed using antibodies against TLS or mock (no antibody). DNA fragments were then amplified using primers specific to the genes indicated on the left.
We next examined representative genes identified by ChIP and subsequent microarray with P < 0.01. We detected strong association with DNA fragments representing the promoter regions of ERAS, INTS3, MECP2, PRAP, SAC3D1, ZNF294, and ZNF397 in samples immunoprecipitated by TLS antibody but not in mock immunoprecipitation (Fig. 2B), confirming that TLS associates with these genes in vivo. In one case, RBM22, we did not detect enrichment in the TLS ChIP sample compared with mock immunoprecipitation, and in two cases, WIPF1 (Fig. 2B) and MAD2L1BP, the primers showed only weak amplification. Together, we analyzed eight genes from the microarray successfully, and seven gave robust ChIP signals.
TLS showed strong binding to the INTS3 gene. The protein product of this gene is part of the integrator complex that mediates 3′-end processing of snRNAs (14). We tested whether TLS associates with the gene encoding another component of the integrator complex, INTS6, which was not identified as a putative TLS target gene in the microarray, and did not detect TLS occupancy at this promoter (Fig. 2B), further confirming the specificity of the microarray results. Given that TLS is an RNA-binding protein, we also tested whether TLS occupancy at these promoters was dependent on RNA. Addition of RNase A before immunoprecipitation had no effect on TLS occupancy.
TLS Depletion or Overexpression Changes Expression of Target Genes.
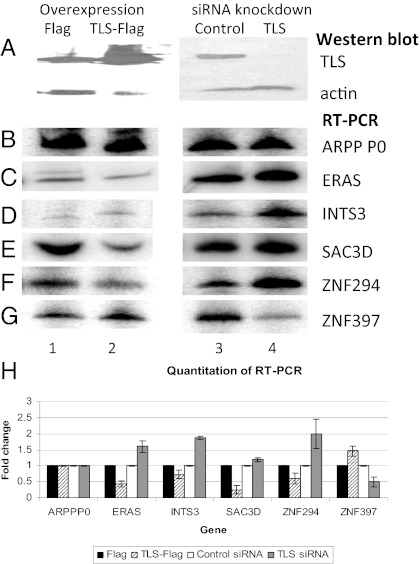
We next investigated whether TLS affects expression of any of the verified microarray target genes. To this end, we examined mRNA levels of several of these genes after altering TLS levels in HeLa cells. We either reduced TLS levels by using anti-TLS siRNAs, or increased TLS levels by transiently overexpressing Flag-tagged TLS, as described previously (7). Changes in TLS levels were verified by Western blotting (Fig. 3A). To assay effects on expression levels, we performed RT-PCR with gene-specific primers. Results with radioactive PCR are shown, but were confirmed by quantitative real-time PCR. Altering TLS levels did not affect expression of ARPP P0 mRNA (Fig. 3 B and H), consistent with weak occupancy of TLS at this gene (Fig. 2A), or the expression of β-actin and GAPDH. However, mRNA levels of six putative TLS target genes, ERAS, INTS3, SAC3D1, MECP2, ZNF294, and ZNF397, were all found to be sensitive to TLS levels.
Fig. 3.
Levels of TLS protein affect target gene expression. HeLa cells were transfected with plasmids encoding Flag or TLS-Flag, or with siRNA targeting luciferase (control) or TLS. (A) TLS and actin protein levels were analyzed by Western blot. (B–G) Reverse transcription using random hexamer primers followed by PCR analysis of the genes indicated on the right. (H) Graph of quantified mRNA amounts. At least three replicates of each experiment were quantified. Error bars depict SD.
TLS acts as a negative regulator of several target genes. For example, an increase in TLS levels decreased ERAS mRNA, and, conversely, a decrease in TLS protein resulted in increased levels of this mRNA (Fig. 3C; quantitation in Fig. 3H). A similar effect was seen for the INTS3 gene (Fig. 3 D and H). Likewise, SAC3D1 mRNA was decreased after TLS overexpression, and modestly increased after TLS knockdown (Fig. 3 E and H). Finally, ZNF294 mRNA levels were decreased upon TLS-Flag overexpression and almost doubled by knockdown of TLS (Fig. 3 F and H). Taken together, these results indicate that TLS has a repressive role in expression of several target genes.
TLS had the opposite effect on two tested genes. Overexpression of TLS increased the levels of ZNF397 mRNA, whereas a reduction in TLS levels resulted in decreased levels of ZNF397 mRNA (Fig. 3B; quantitation in Fig. 3H). TLS also has a positive effect on MECP2; decreased TLS protein levels resulted in less MECP2 mRNA, and increased TLS led to higher levels of MECP2 mRNA (Fig. S2). Our results thus indicate that TLS can influence target gene expression both positively and negatively.
MatrixREDUCE Identifies Sequence Motifs Preferentially Found in TLS Target Promoters.
We next used the microarray data to identify possible DNA sequences that were preferentially recognized by TLS. To this end, the raw microarray data were processed using model-based analysis of tiling arrays (MAT) (15). The MAT algorithm corrects for probe sequence bias and copy number to improve the signal-to-noise ratio. MAT also reduces false positives by performing a robust, trimmed mean that removes outliers and averages noise across all normalized probe intensities within a window of 600 bp, the average postsonication dsDNA fragment size. The final MAT enrichment score was used as a measure of TLS occupancy and affinity. We next applied the MatrixREDUCE algorithm (16) to the normalized occupancy scores. In general, the algorithm yields one or more position-specific affinity matrices (PSAMs), which quantify the relative affinity of a DNA-binding protein for each nucleotide relative to the optimal binding sequence at each position of the motif, under the assumption that each nucleotide position contributes independently to the overall affinity of the binding site. PSAM parameters are estimated by performing nonlinear regression (optimization) seeded by the K-mer with the highest Pearson correlation with normalized intensities. We restricted our analysis to 1,000 probes with the highest MAT score, which cover a wide range of binding affinities. The nucleotide sequence associated with each probe was taken to be the 600-bp window centered on the genomic location of the probe.
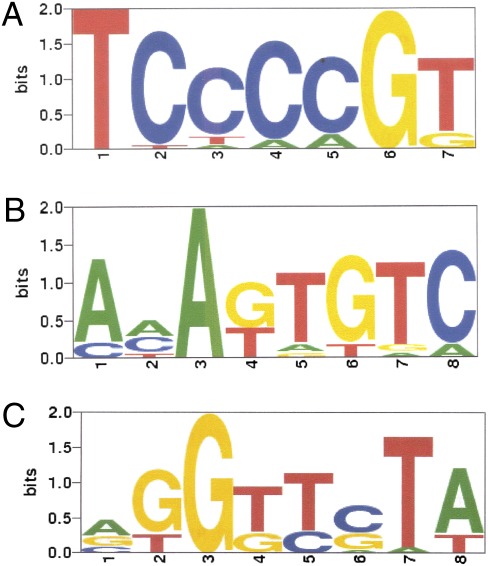
MatrixREDUCE identified three distinct PSAMs for TLS (Fig. 4) by recursively fitting to the residuals after each nonlinear optimization (Fig. 4). The first motif, with consensus sequence TCCCCGT and absolute conservation of T at position 1 and G at position 6, yielded a high R2 value—defined as the fraction of the variance in ChIP signal explained by the PSAM—of 0.17 (P value 2.54e-42). The second and third PSAMs, with consensus sequences AAAGTGTC and AGGTTCTA, also showed highly significant R2 values of 0.18 and 0.08, respectively (P values 2.2e-45 and 3.4e-20). Remarkably for a DNA-binding factor, the correlation of the first two sequences (but not the third) was direction dependent. Forward incidences of the motif, relative to the direction of transcription, correlated well with TLS enrichment, but incidence of the reverse complement motif did not. This directionality suggests that TLS might have a preference for binding to only one strand of DNA.
Fig. 4.
TLS-binding motifs modeled as PSAMs determined by MatrixREDUCE. (A) The first PSAM that best explains the variance in the normalized ChIP enrichment (MAT) scores using only the forward strand (R2 0.17, P value 2.54e-42). The height of each letter is proportional to its corresponding nucleotide's relative affinity at each position, and the letters are sorted in descending frequency order. The height of the entire stack at each position is then adjusted to signify the information content (in bits) of that position. (B) The second PSAM that best explains the variance in the residuals from the fit with the first PSAM, again using only the forward strand (R2 0.18, P value 2.2e-45). (C) The third PSAM that best explains the variance in the residuals from the fits with the first and second PSAMs, using both strands (R2 0.08, P value 3.4e-20).
TLS Binds with Specificity to Single-Stranded Motif Sequences.
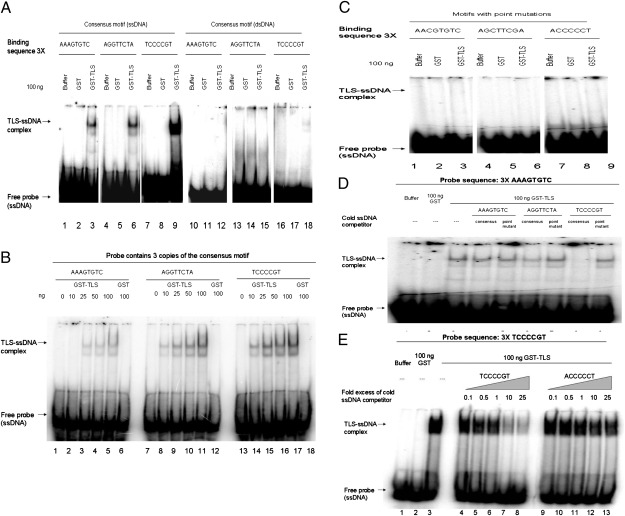
We next investigated whether TLS binds directly to the motifs predicted by MatrixREDUCE. To this end, we performed gel shift assays using 32P-labeled ssDNA and dsDNA probes containing three tandem repeats of the enriched sequences and purified GST-TLS (TLS). Strikingly, TLS (but not GST) bound strongly to ssDNA containing three copies of the TCCCCGT, AAAGTGTC, or AGGTTCTA sequences (Fig. 5A), and binding was concentration dependent (Fig. 5B). TLS showed strongest binding to TCCCCGT, followed by AGGTTCTA and AAAGTGTC. We did not detect binding to dsDNA containing three copies of the AAAGTGTC or AGGTTCTA sequences, and observed only very weak binding to dsDNA containing three copies of TCCCCGT (Fig. 5A).
Fig. 5.
TLS binds to single-strand recognition motifs. (A) GST or GST-TLS was added to 32P-labeled ssDNA or dsDNA containing three tandem repeats of TLS motifs. (B) GST or increasing amounts (0–100 ng) of GST-TLS was added to 32P ssDNA containing three tandem repeats of TLS motifs. (C) GST or GST-TLS was added to 32P ssDNA encoding three tandem repeats of a mutated TLS motif. (D) GST and GST-TLS was added to 32P ssDNA containing three copies of AAAGTGTC. Cold competitor ssDNA containing three copies of consensus or mutated TLS-binding motifs was added as indicated. (E) GST or GST-TLS was added to 32P ssDNA containing three copies of TCCCCGT. Increasing amounts of cold competitor ssDNA was added as indicated. In all cases, complexes were resolved by native PAGE.
We next mutated key nucleotides in the binding sites to test the validity of the predicted TLS-binding motifs. Strikingly, point mutations of invariant nucleotides in each of the PSAMs abolished binding (Fig. 5C), suggesting TLS binds ssDNA with specificity and that the motifs identified by MatrixREDUCE are critical for recognition. For example, binding was abolished when the three AAAGTGTC sequences were altered to AACGTGTC. Likewise, mutating AGGTTCTA to AGCTTCGA, or TCCCCGT to ACCCCCT, prevented TLS binding to ssDNA containing three copies of these altered sequences. TLS also did not bind to dsDNA consisting of three copies of the mutated sequences or to ssDNA containing the reverse complement of the consensus motifs (Fig. S3).
To provide additional evidence that TLS binds sequence specifically, we used cold competitor ssDNA encoding consensus or mutated motifs. TLS binding to labeled AAAGTGTC ssDNA was modestly reduced by addition of an equal amount of cold competitor ssDNAs containing three copies of either AAAGTGTC or AGGTTCTA, but was essentially abolished by an equivalent amount of unlabeled ssDNA containing three copies of TCCCCGT (Fig. 5D), consistent with the strong affinity of TLS for this sequence. Cold competitor ssDNA containing the above-described mutations did not have a significant effect on TLS (Fig. 5D). Likewise, TLS binding to labeled AGGTTCTA- or TCCCCGT-containing ssDNA was decreased ≤85% upon addition of a twofold excess of unlabeled ssDNA consisting of any of the three consensus motifs, but much less so or not at all by mutant derivatives (Fig. S4 A and B). Competition was concentration dependent, because increasing amounts of unlabeled ssDNA containing three copies of TCCCCGT decreased TLS binding ≤90% to the same labeled DNA, whereas the unlabeled ACCCCGT mutant, even in 25-fold excess, had little effect on TLS binding (Fig. 5E). Finally, TLS binding to the ssDNAs containing the three consensus motifs was not affected by dsDNA containing either consensus or mutant sequences (Fig. S4C). Together, our data indicate that TLS binds sequence specifically and in a concentration-dependent manner to ssDNA consensus sequences derived from TLS target promoters.
Discussion
Our results show that TLS plays a significant role in expression of a number of RNAP II transcribed genes. Our findings define a group of genes regulated by TLS and identify putative TLS response elements in target gene promoters. Below we discuss properties of several of the TLS target genes, the significance of the ssDNA recognition elements, and how TLS, like several other RNA-binding proteins, binds ssDNA recognition sequences.
The functions of a number of the genes we identified as TLS target genes are of interest. The INTS3 gene, which is down-regulated by TLS, is known to be amplified in hepatocellular carcinomas (17). INTS3 is a component of the integrator complex, which mediates 3′-end processing of snRNAs (14). Down-regulation of U1 and U2 genes by TLS through INTS3 protein levels could suggest a role for TLS in U snRNA gene expression, because TLS also negatively regulates transcription by RNAP III of the U6 snRNA gene (7). TLS regulation of U1 and U2 expression through INTS3 and of U6 transcription could provide a mechanism for global control of RNA splicing.
We also found TLS at the promoter regions of several cell cycle-related genes, including RAS family genes, such as ERAS and RAB5C. TLS and the splicing factor SRSF2 have previously been found to stimulate alternative splicing of H-ras pre-mRNA (18). Thus, TLS may negatively regulate cell-cycle progression both through repressing transcription of RAS family genes and through alternative splicing that produces isoforms that delay cell-cycle progression. TLS was also found at other genes involved in cell-cycle regulation, including PRAP and SAC3D1. PRAP is down-regulated in hepatocellular carcinoma, and overexpression of this gene in cancer cell lines resulted in growth inhibition and decreased colony formation (19). SAC3D1, like INTS3, was also found associated with the integrator complex but is not stably associated with RNAP II (14). SAC3D1 is involved in centrosome duplication and cell-cycle progression in mammalian cells (20). These target genes suggest that TLS may be involved in regulating cell-cycle progression.
ZNF294, also known as LISTERIN and RNF160, is a potentially important disease-relevant TLS target; it was found mutated in colon cancer cell lines and encodes a protein that contains a RING finger domain that can function as an E3 ubiquitin ligase (21). Interestingly, a mutant mouse model indicates a role for ZNF294 in neurodegeneration, possibly including motor neuron dysfunction and specifically ALS (22). This finding is intriguing in light of the role of TLS mutants in ALS, and deregulation of ZNF294 expression may play some role in ALS.
ZNF397 encodes a protein that localizes to centromeres and may repress transcription of noncentromeric genes (23). TLS transcriptional control of ZNF397 could then regulate a cascade of other centromeric genes and segregation of chromosomes. TLS and EWS were previously shown to be involved in pairing autosomal and sex chromosomes, respectively, during meiosis (24, 25). Defects in this process could lead to the increased genomic instability observed in TLS knockout mice (24).
TLS positively regulates MECP2, which encodes methyl CpG-binding protein 2. Mutations in MECP2 cause Rett syndrome, a neurodevelopmental disorder, and initial experiments suggested that MECP2 acts as a transcriptional repressor (26). MECP2 may also have roles in RNA splicing, chromatin organization, and L1 retrotransposition in neurons (27). Another DNA- and RNA-binding protein, TAR DNA-binding protein 43 (TDP-43), is also associated with ALS (28). Interestingly, TDP-43 binds MECP2 in neurons (29), suggesting that TLS and TDP-43 regulate a common pathway in neurodegeneration.
All of the target genes we have discussed contain multiple TLS-binding motifs generated by MatrixREDUCE in their promoter regions. These TLS-binding motifs include the highest-affinity sequences as well as sequence variants that contain one or two tolerated nucleotide substitutions, as specified in the TLS affinity matrix generated by MatrixREDUCE (16). For example, INTS3 contains three TCCCCGT motif variants, one copy of AAAGTGTC, and two AGGTTCTA variants in the 5.6-kb promoter region upstream of its transcription start site. In contrast, the 7.5-kb β-actin promoter region, which is not occupied by TLS, does not contain any copies of TLS-binding motifs or variants, whereas the ARPP P0 promoter region contains only two copies of a tolerated variant, AGGTTGTA. Together, this result suggests that multiple copies of at least one TLS recognition motif are present in TLS target gene promoter regions. Given the presence of each motif in genes that are positively and negatively regulated by TLS, it is difficult to conclude that a specific motif is associated with activation or repression.
Unexpectedly, the reverse complement of two of the three identified TLS-binding motifs did not correlate with its genomic variation in occupancy, which suggested that the binding motif is ssDNA. A possible explanation for the existence of three apparently disparate binding motifs is that TLS contains multiple distinct nucleic acid-binding domains, which may function independently or in combination. Because TLS did bind these regions only as ssDNA in vitro, there is support for the idea that directional binding is indicative of ssDNA motifs. For the third sequence, AGGTTCTA, which had the lowest R2 value, the reverse complement was found enriched in the TLS ChIP microarray data, but this sequence was also bound by TLS only as ssDNA in vitro. Our data are consistent with previous studies in which TLS was found to have greater nonspecific affinity for ssDNA than dsDNA (8), and to bind to ssDNA but only weakly to dsDNA consisting of human telomeric sequences (30). The telomere sequence TTAGGG is not related to any of the consensus sequences we have described, and how and if binding to this sequence is related to the binding we have described here is not known.
The ability of TLS to bind specific ssDNA sequences raises interesting questions. Are these sequences relevant to RNA binding? Arguing against this, TLS has been reported to bind a GGUG motif in RNA, with relatively low affinity (31). Additionally, the ssDNA binding we described is not likely to reflect RNA binding in vivo, because the sequences enriched in the promoter microarray were from upstream promoter regions, making it unlikely that they reflect interaction with RNA. Do other putative RNA-binding proteins recognize ssDNA? At least five proteins containing RNA-binding domains have been shown to be capable of binding ssDNA: polypyrimidine tract binding protein (PTB), the FUSE-binding protein (FBP), hnRNP K, hnRNP A1, and, most relevantly, EWS.
How might TLS and other ssDNA-binding proteins recognize what would normally be dsDNA? Though PTB binds with specificity to pyrimidine-rich ssDNA (32), an intriguing possibility is that DNA binding by the other proteins involves G-quadruplex structures. HnRNP A1 is known to bind such structures in telomeric DNA (33) and the KRAS promoter (34). Both FBP and hnRNP K recognize and bind ssDNA regions of the c-MYC promoter through their K homology domains, resulting in transcription activation (35, 36). Interestingly, hnRNP K binds the pyrimidine-rich strand of the CT element of c-MYC, a region that consists of four imperfect repeats of the sequence CCCTCCCA (37). This sequence bears a resemblance to TCCCCGT, the sequence for which TLS has greatest affinity. The CT element is hypersensitive to nucleases, indicative of ssDNA, and the purine-rich strand can form a G-quadruplex structure (38). Notably, a number of proto-oncogenes have promoter sequences that can form G-quadruplex structures (39), raising the possibility that recognition of the complementary strand by ssDNA/RNA-binding proteins is important for expression of these genes. Likewise, the promoter regions of many of the genes we identified here are predicted to contain G-quadruplex–forming sequences, based on analysis with the program Quadfinder (40). EWS has also recently been reported to bind G-rich DNA in a G-quadruplex structure (41), and the high degree of similarity between TLS and EWS supports the view that TLS does bind to G-quadruplex–containing DNA. Indeed, the strongest of the three TLS consensus motifs we identified, TCCCCGT, could become single stranded as a result of G-quadruplex formation by the complementary strand.
In summary, we have identified RNAP II promoters occupied by TLS and have confirmed that at least some of these target genes are regulated by TLS. We identified TLS recognition elements in the promoter regions of these genes, and showed that TLS binds these as ssDNA. This finding adds to the mechanisms by which TLS, and likely other TET family proteins, can modulate transcription. Likewise, the functions of TLS target genes indicate a role for TLS in regulating processes as diverse as transcription; cell-cycle progress; DNA repair and genomic stability; and neurodegeneration.
Materials and Methods
ChIP on Chip.
ChIP DNA was amplified as described in the Affymetrix Chromatin Immunoprecipitation Assay Protocol. DNA was purified with Affymetrix cDNA cleanup columns, and then subjected to fragmentation and labeling using GeneChip WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix). Labeled DNA was hybridized to GeneChip Human Promoter 1.0R Array (Affymetrix) in the Columbia University Cancer Center microarray facility. Data were analyzed using Partek Genomics Suite and Affymetrix GeneChip Operating Software, and genes were identified using the University of California–Santa Cruz Genome Browser. Microarray data has been deposited in the EBI Express Database under accession no. E-MEXP-3568.
Bioinformatics Analysis.
Data from the microarray (.cel files) were standardized using MAT (15), then analyzed using MatrixREDUCE (16). The human hg18 assembly, released in March 2006, was used to extract 600-bp sequences centered at each probe start position in the human genome. The MAT algorithm was used to model probe sequence effects, sequence copy number, and windowed, trimmed-mean averaging to remove noise and standardize the signal. MatrixREDUCE was then used to find motifs within the 600-bp sequences with the highest linear correlation with the MAT standardized enrichment scores. The human hg18 assembly was also used to align the 7,158 isoforms of the 1,000 genes that contained the top MAT enrichment scores. The same analysis was performed on all 96,576 gene isoforms currently known in the hg18 assembly.
Additional materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank P. Richard and E. Rosonina for assistance with ChIP; Y. Zhang for performing computational identification of bound promoter regions; Y. Feng and other J.L.M. laboratory members for discussions; X.-J. Lu for implementing the MatrixREDUCE software; and L. Tora for TLS antibody used in early experiments. A.Y.T. was partially funded by a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada. This work was supported by grants from the National Institutes of Health to J.L.M. (R01 GM048259) and to H.J.B. (R01HG003008 and U54CA121852).
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarray data reported in this paper have been deposited in the European Bioinformatics Institute Array Express Database, http://www.ebi.ac.uk/arrayexpress/ (accession no. E-MEXP-3568).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203028109/-/DCSupplemental.
References
- 1.Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 3.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 4.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan AY, Manley JL. The TET family of proteins: Functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 7.Tan AY, Manley JL. TLS inhibits RNA polymerase III transcription. Mol Cell Biol. 2010;30:186–196. doi: 10.1128/MCB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baechtold H, et al. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 9.Powers CA, Mathur M, Raaka BM, Ron D, Samuels HH. TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- 10.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 13.Kameoka S, Duque P, Konarska MM. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foat BC, Morozov AV, Bussemaker HJ. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics. 2006;22:e141–e149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]
- 17.Inagaki Y, et al. CREB3L4, INTS3, and SNAPAP are targets for the 1q21 amplicon frequently detected in hepatocellular carcinoma. Cancer Genet Cytogenet. 2008;180:30–36. doi: 10.1016/j.cancergencyto.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Camats M, Guil S, Kokolo M, Bach-Elias M. P68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS ONE. 2008;3:e2926. doi: 10.1371/journal.pone.0002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, et al. The proline-rich acidic protein is epigenetically regulated and inhibits growth of cancer cell lines. Cancer Res. 2003;63:6658–6665. [PubMed] [Google Scholar]
- 20.Khuda SE, et al. The Sac3 homologue shd1 is involved in mitotic progression in mammalian cells. J Biol Chem. 2004;279:46182–46190. doi: 10.1074/jbc.M405347200. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene. 2007;26:2873–2884. doi: 10.1038/sj.onc.1210098. [DOI] [PubMed] [Google Scholar]
- 22.Chu J, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci USA. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey SL, et al. ZNF397, a new class of interphase to early prophase-specific, SCAN-zinc-finger, mammalian centromere protein. Chromosoma. 2008;117:367–380. doi: 10.1007/s00412-008-0155-7. [DOI] [PubMed] [Google Scholar]
- 24.Hicks GG, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 25.Li H, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117:1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 27.Hite KC, Adams VH, Hansen JC. Recent advances in MeCP2 structure and function. Biochem Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: The FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sephton CF, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahama K, Arai S, Kurokawa R, Oyoshi T. Identification of DNA binding specificity for TLS. Nucleic Acids Symp Ser (Oxf) 2009;53:247–248. doi: 10.1093/nass/nrp124. [DOI] [PubMed] [Google Scholar]
- 31.Lerga A, et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 32.Jansen-Dürr P, et al. The rat poly pyrimidine tract binding protein (PTB) interacts with a single-stranded DNA motif in a liver-specific enhancer. Nucleic Acids Res. 1992;20:1243–1249. doi: 10.1093/nar/20.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krüger AC, et al. Interaction of hnRNP A1 with telomere DNA G-quadruplex structures studied at the single molecule level. Eur Biophys J. 2010;39:1343–1350. doi: 10.1007/s00249-010-0587-x. [DOI] [PubMed] [Google Scholar]
- 34.Paramasivam M, et al. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: Implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duncan R, et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 36.Tomonaga T, Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 37.Braddock DT, Baber JL, Levens D, Clore GM. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21:3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González V, Hurley LH. The c-MYC NHE III(1): Function and regulation. Annu Rev Pharmacol Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 40.Scaria V, Hariharan M, Arora A, Maiti S. Quadfinder: Server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res. 2006;34(Web Server issue):W683–W685. doi: 10.1093/nar/gkl299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahama K, Kino K, Arai S, Kurokawa R, Oyoshi T. Identification of Ewing's sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 2011;278:988–998. doi: 10.1111/j.1742-4658.2011.08020.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.