Abstract
Neutrophil serine proteases (NSPs) in cytoplasmic granules of neutrophils are regarded as important antimicrobial defense weapons after engulfment and exposure of pathogens to the content of primary granules. Despite intensive studies on neutrophils during the last three decades, only three active serine proteases, neutrophil elastase (NE), cathepsin G (CG), and proteinase 3 (PR3) have been identified in these short-lived cells. Here, we report on the identification of a fourth serine protease (NSP4) with 39% identity to NE and PR3, but arginine specificity, yet sharing features like propeptide processing by dipeptidyl peptidase I, storage, and release as an active enzyme with the three active proteases. We established monoclonal antibodies against NSP4, excluded cross-reactivity to human granzymes, NE, CG, PR3, and azurocidin, and screened for NSP4 protein expression in various human tissues and blood leukocyte populations. Only granulocyte precursors and neutrophil populations from peripheral blood were positive. The content of NSP4 in neutrophil lysates, however, was about 20-fold lower compared with CG. Upon neutrophil activation, NSP4 was released into the supernatant. Profiling its specificity with peptide libraries from Escherichia coli revealed a preference for arginine in P1; it cleaved Tyr-Arg-Phe-Arg-AMC and Ala-Pro-Nva-thiobenzyl esters. NSP4 was inhibited by α1-proteinase inhibitor (α1–antitrypsin), C1 inhibitor, and most efficiently by antithrombin-heparin, but not by elafin, secretory leukocyte protease inhibitor, α1–antichymotrypsin, and monocyte-neutrophil elastase inhibitor. Functional specialization and preferred natural substrates of NSP4 remain to be determined to understand the biological interplay of all four NSPs during neutrophil responses.
Keywords: tissue distribution, serpins, cleavage site, proteomics, evolution
Recruitment of neutrophils to the site of infection is one of the earliest cellular defense reactions and is indispensable to stop the proliferation and dissemination of microbes within the host. To successfully kill microbes after their phagocytosis and in the pericellular tissue environment, neutrophils use a broad arsenal of efficient weapons, such as reactive oxygen species, hypochloric acid, antimicrobial peptides, and a unique set of serine proteases, namely neutrophil elastase (NE; encoded by ELANE), cathepsin G (CG), and proteinase 3 (PR3; encoded by PRTN3). Although intracellular killing of pathogens by neutrophil serine proteases (NSPs) is a beneficial function for the host (1–3), tissue damage is also mediated by these three proteases during infections and chronic inflammatory diseases (4). Furthermore, there is strong evidence that NSPs act as important modulators for a variety of regulatory functions in inflammation (e.g., by processing cytokines, chemokines, and cell surface receptors) (5–8).
From an evolutionary point of view and on the basis of their exon-intron organization, NSPs can be assigned to the sixth separate class (subtree) of serine protease genes (9). This class encompasses the five lymphocyte granzymes (Gzms) A, B, H, K, and M, the catalytically inactive serine protease homolog azurocidin (AZU1), the mast cell chymase, and the complement factor D (CFD). All members play important roles in immune defense reactions, such as in the elimination of virally infected cells (Gzms) or bacteria (NSPs), or they amplify an innate immune response by various mediator-dependent mechanisms (CFD, mast cell chymase). Among the 120 human gene sequences encoding serine protease homologs (10), we identified an as yet uncharacterized additional class 6 member, the gene of which is located in close proximity to the GZMM gene on chromosome 19p13.3 (11). The protein predicted from this sequence has so far not been detected in human tissues nor characterized at the biochemical and functional level.
Here we report the development and identification of specific monoclonal antibodies to this unique serine protease and its restricted expression in neutrophilic granulocytes and bone-marrow precursor cells. Like NE, CG, and PR3, it is converted into an active protease by dipeptidyl peptidase I (DPPI), is stored as a catalytically active serine protease in neutrophils, and released into the peri- and extracellular space upon neutrophil activation. In view of these similarities with known neutrophil serine proteases, we named it neutrophil serine protease 4, abbreviated as NSP4 in the following.
Results
NSP4 Is Most Similar to NSPs and CFD.
Genomic sequences for NSP4 have been deposited in GenBank and EMBL databases more than two decades ago. Expressed sequence tags derived from this serine protease gene were missing for a long time. The very few sequences deposited in GenBank subsequently did not give any clue about its tissue-specific expression and catalytic function. The respective gene sequence has been annotated by automated computer algorithms as PRSS57. NSP4 is found in the telomeric region on the short arm of human chromosome 19 (19p13.3) outside of the AZU1-PRTN3-ELANE-CFD cluster and is transcribed in opposite direction, compared with these genes. In view of four introns with phases 1-2-0-0, respectively, at identical position as in Gzms and NSPs, NSP4 can be regarded as the twelfth member of class-six serine proteases (9).
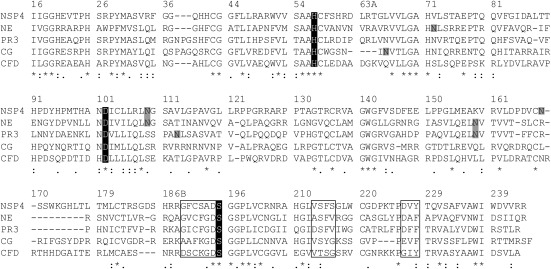
Fig. 1 presents the mature protein sequence of NSP4 in comparison with all catalytically active NSPs and CFD. The alignment and numbering of sequences is based on topological similarity between these proteases and chymotrypsinogen. The number and position of predicted N-linked glycosylation sites vary among these proteases, NE has three, NSP4 and PR3 two, and CG only one N-linked glycosylation site. Amino acid residues determining the size and shape of the active site in NE (189–195, 213–216, and 226–228) (12) are quite similar with those in NSP4. Of particular note for substrate specificity is the Gly189 at the bottom of the S1 pocket [nomenclature of Schechter and Berger (13)], which is shared by NSP4, NE, and PR3, and the Asp226, which is also present in NE and PR3, where it reduces the pocket size (12). In view of these structural features, we initially assumed an elastase-like activity for NSP4.
Fig. 1.
Sequence comparison between all four catalytically active human neutrophil serine proteases and complement factor D. Topologically aligned amino acid residues (single letter code) are numbered according to chymotrypsinogen. The catalytic triad of His57, Asp102, and Ser195 is highlighted in black. NSP4 carries two N-linked glycosylation sites marked in gray. Residues that shape the substrate binding pocket in NE are boxed (189–195, 213–216, 226–228) for all proteases and are similar between NSP4, NE, and PR3. An asterisk (*) below the alignments indicates positions which agree completely; a colon (:) or a period (.) indicate conservation of residues with strong or weak similarities (scoring > 0.5 or = < 0.5 in the Gonnet PAM 250 matrix), respectively.
Development of mAbs Specific for NSP4.
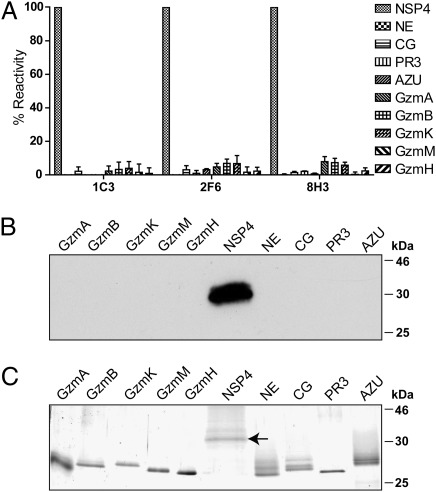
Reliable and specific detection of NSP4 in the presence of abundant, structurally related serine proteases can be achieved with well-characterized mAbs. For immunization of rats, larger quantities of NSP4 were produced as inclusion body (IB) material and solubilized in 6 M guanidinium hydrochloride. Fourteen hybridomas secreted antibodies that recognized both solubilized IB material and native NSP4. Native NSP4 was expressed in HEK 293 cells with an S-tag and an enterokinase cleavage site at the mature amino terminus. To identify potential cross-reactivities to other closely related serine proteases, all NSPs and the five Gzms (A, B, H, K, and M) were tested for binding to these monoclonal antibodies by ELISA and immunoblotting (Fig. 2). In this way, three antibodies (1C3, 2F6, and 8H3) were shown to be highly specific for NSP4 (Fig. 2A).
Fig. 2.
Identification of NSP4 specific rat monoclonal antibodies. (A) Only 3 of 14 monoclonal antibodies obtained from hybridomas after immunization of rats, named mAb 1C3, 2F6, and 8H3, were specific for NSP4. NSP4, the human Gzms (A, B, K, M, and H), and all known human NSPs were coated on microtiter plates. Supernatants of hybridoma clones secreting rat antibodies were analyzed in an ELISA using a secondary anti-rat HRP-conjugated polyclonal Ab. Percent reactivity was calculated by setting the mean reactivity of mAbs with NSP4 to 100%. Data are the mean from three independent experiments (n = 3, ± SD). (B) NSP4, the five human Gzms and human NSPs were separated by SDS/PAGE, transferred to Hybond ECL transfer membrane, and then probed with rat anti-NSP4 mAb 1C3, 2F6, or 8H3 and secondary HRP-labeled Ab. (C) Similar amounts of all tested proteins used as in A and B are shown after reducing SDS/PAGE and subsequent silver-staining. A horizontal arrow points to the glycosylated NSP4, carrying an amino-terminal extension (i.e., S-tag and an enterokinase cleavage peptide sequence).
NSP4 Expression in Polymorphonuclear Leukocytes.
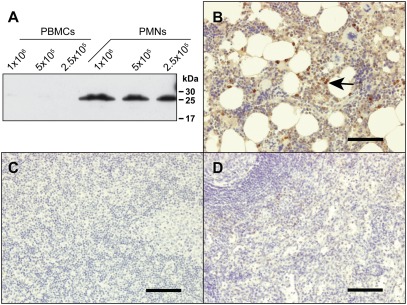
Given the structural similarity between NSP4 and other serine proteases of hematopoietic cell lineages, various leukocyte subsets were analyzed for NSP4 expression. Western blot analysis of total cell lysates using rat anti-NSP4 mAbs revealed the presence of NSP4 in polymorphonuclear leukocytes (PMNs), but not in other subsets of white blood cells (Fig. 3A). Restricted expression of NSP4 in the myelomonocyte cell lineage was ascertained by immunohistochemistry. NSP4 was only detected in neutrophils and neutrophil precursors of bone-marrow tissue (Fig. 3B), but not in lymph node, spleen (Fig. 3 C and D), neural network, pancreas, prostate, or arteries (Fig. S1). Hence, NSP4 exhibits a tissue-restricted expression pattern very similar to NE, CG, and PR3.
Fig. 3.
NSP4 is present in granulocytes. (A) Total cell lysates of peripheral blood mononuclear cells (PBMCs) and PMNs, which are almost exclusively neutrophil granulocytes, were analyzed by Western blotting using anti-NSP4 mAbs and secondary anti-rat HRP-labeled Ab. Natural NSP4 of PMNs runs lower than recombinant S-tag-NSP4 (Fig. 2B) because it is N-terminally processed and has shorter carbohydrate chains. (B–D) Staining of different human tissue samples using anti-NSP4 mAbs and the Ultravision LP staining kit. NSP4 was detected in neutrophil granulocytes and precursors in bone marrow tissue (arrow, B). NSP4 was not detected in lymph node (C) or spleen tissues (D). (Scale bars, 100 μm.)
Low Abundance of NSP4 in Neutrophils.
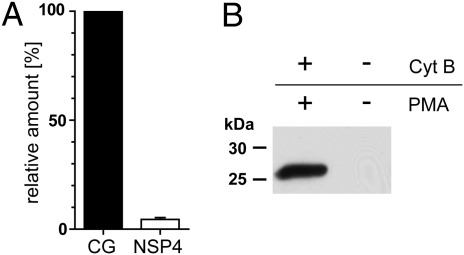
Neutrophil granulocytes store large amounts of NE, CG, and PR3 in granules during development (14) and are commonly used to purify these proteases. In contrast, we suspected that the content of NSP4 in neutrophils was low, and that this was the reason why NSP4 was overseen in the past. We performed semiquantitative Western blotting with PMN lysates using purified NSP4 and CG as a protein standard. To compare the expression of CG with NSP4 in human neutrophils, we calculated the content of NSP4 as a percentage of CG. These results are summarized in Fig. 4A. NSP4 in PMNs amounts to only 5% of the CG content.
Fig. 4.
Total amount of NSP4 in PMNs is 20-fold lower than that of CG; nevertheless, NSP4 is detected in supernatants of stimulated PMNs. (A) NSP4 and CG in total cell lysates of PMNs were compared with known amounts of purified CG and NSP4 by semiquantitative Western blotting using monospecific anti-CG antibodies and anti-NSP4 mAbs, respectively. Content of NSP4 in PMNs is given as a percentage of CG present in PMNs. Data were pooled from three independent experiments (n = 3, ± SD). (B) PMNs were incubated at 37 °C in the presence or absence of 5 μg/mL cytochalasin B (Cyt B) and 200 ng/mL PMA for 30 min. Cell-free supernatants were analyzed by Western blotting with anti-NSP4 mAbs.
Release of NSP4 from Activated Neutrophils.
NE, CG, and PR3 are released into the pericellular environment in response to neutrophil activation (15). To see if NSP4 is likewise released from cytoplasmic storage granules, PMNs were activated with the phorbolester phorbol-12-myristate-13-acetate (PMA) in the presence of cytochalasin B (cytB). The presence of NSP4 in the cell-free supernatant was determined by Western blotting. NSP4 was only detected in the supernatants after PMA treatment, but not in supernatants of neutrophils that were treated without cytB and PMA (Fig. 4B). Hence, NSP4 behaves like the other three serine proteases of PMNs and most likely acts in concert with these NSPs.
Biosynthetic Processing of NSP4 by DPPI.
NSPs and Gzms are synthesized as inactive precursors with a signal peptide and a propeptide at the amino terminus (16). Removal of the signal peptide by signal peptidase occurs during translocation into the endoplasmatic reticulum, and N-terminal dipeptides are removed by DPPI, also known as cathepsin C, before the mature enzymes are stored in cytoplasmic granules. To study biosynthetic processing, the full-length cDNA sequence of natural NSP4 with a C-terminal 6xHis-tag was expressed in HEK 293. The resulting protein was found to be secreted into the culture supernatant and purified by Ni-NTA affinity chromatography. N-terminal residues of the secreted protein were identified as Ala-Gln-Ile-Ile by stepwise Edman degradation. To test if the propeptide Ala-Gln can be removed by DPPI, the purified NSP4 precursor was incubated with DPPI and resequenced. The sequence of the DPPI-treated NSP4 started with Ile-Ile-Gly-Gly, indicating that NSP4 is posttranslationally processed to its mature form like the other NSPs.
Proteolytic Specificity of NSP4.
Enzymatic activity of NSP4 was evaluated with commercially available synthetic peptide substrates. Cleavage of the thiobenzyl ester Boc-Ala-Pro-Nva-4-chloro-SBzl (Fig. 5A), a typical substrate for NE and PR3, suggested an elastase-like specificity. Sixfold higher concentrations of NSP4, however, were required to achieve a similar activity as with NE.
Fig. 5.
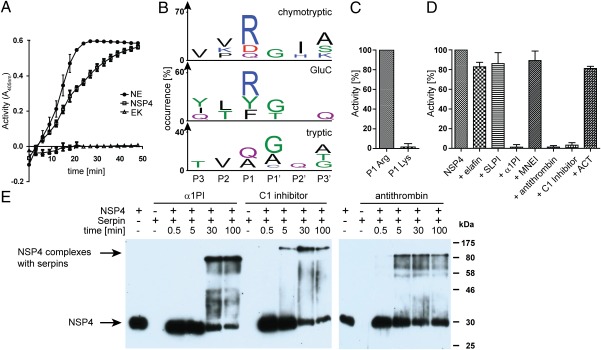
NSP4 preferably cleaves after arginine residues and is inhibited by α1PI, C1 inhibitor, and antithrombin. (A) After removing the propeptide extension with enterokinase (EK), proteolytic activity of recombinant NSP4 was measured using the thiobenzyl ester substrate Boc-Ala-Pro-Nva-4-chloro-SBzl and DTNB. NSP4 was used at a concentration of 3 × 10−6 M. NE and the convertase EK served as positive and negative controls at concentrations of 1 × 10−7 M and 3 × 10−2 U/μL, respectively. Substrate cleavage was determined by absorbance measurements at 405 nm. Data are results from three experiments, ± SD. (B) PICS specificity profile for NSP4 with chymotrypsin, GluC, and trypsin libraries generated from E. coli. Sequence logos were generated with IceLogo (26). (C) Enzymatic activity of 0.25 μM NSP4 was measured using AMC substrates with either Arg or Lys in P1 [H-Tyr-Arg-Phe-(Arg/Lys)-AMC] at 1 mM substrate concentration. Percent activity was calculated by setting the mean activity of NSP4 with P1 Arg to 100%. Data are the mean from three independent experiments (n = 3, ± SD). (D) 0.25 μM NSP4 was preincubated with 10 μM elafin, 10 μM SLPI, 25 μM α1PI, 3.3 μM MNEI with 1 mM DTT, 5.6 μM antithrombin with 300 U/mL heparin, 2.5 μM C1 inhibitor, or 5.6 μM ACT. Proteolytic activity was determined using H-Tyr-Arg-Phe-Arg-AMC. Percent activity was calculated by setting the mean activity of NSP4 without inhibitor to 100%. Data are the mean from three independent experiments (n = 3, ± SD), except for MNEI and ACT (n = 2, ± SD). (E) 0.8 μM NSP4 was incubated at 37 °C with 40 μM α1PI, 4 μM C1 Inhibitor or 5 μM antithrombin with 300 U/mL heparin and analyzed after different timepoints by Western blotting with anti NSP4 mAbs. For comparisons, NSP4 and the serpins alone were treated similarly.
To characterize potential cleavage sites of NSP4 in peptide substrates, a recently developed method called proteomic identification of protease cleavage sites (PICS) (17) was applied. Three proteome-derived peptide libraries generated by either chymotrypsin, GluC, or trypsin digestion of total Escherichia coli proteins were incubated with NSP4 and the resulting C-terminal cleavage products were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The corresponding N-terminal cleavage products are derived from database searches. These peptides are listed in Tables S1–S3 and represent 23, 45, and 59 nonredundant cleavage sites in the chymotrypsin, GluC, or trypsin generated libraries, respectively.
To our surprise, NSP4 displayed a strong P1 preference for arginine with both GluC and chymotrypsin PICS libraries (Fig. 5B). A minor enrichment of aromatic residues in P1 was only observed with the GluC PICS library, but not corroborated by the tryptic PICS library. In P1′, clear preference for glycine was detected with all three PICS libraries.
Because endopeptidolytic cleavages after arginine cannot occur in trypsin-generated peptide mixtures, additional P1 specificities, Gln and Ala, were disclosed with the trypsin library. Specificity of NSP4 for aliphatic side chains in P1 is consistent with its activity toward a thiobenzyl ester with norvaline in P1 (Fig. 5A). These data and the cleavage products obtained from the tryptic PICS library illustrate that arginine in P1 is not an absolute requirement. NSP4 can cleave, to some extent, peptides lacking a P1 arginine.
PICS libraries only contain peptides with dimethylated lysine residues. Hence, these libraries are not suited to judge about the lysine specificity of the S1 pocket and the role of lysines in substrate recognition (17). Fortunately, two custom-made tetrapeptide AMC substrates [H-Tyr-Arg-Phe-(Arg/Lys)-AMC] with either Arg or Lys at the P1 site were at hand in our laboratory and suited to compare the binding of Arg and Lys in the S1 pocket. Cleavage of the Arg substrate, but not of the Lys substrate, was observed indicating a high selectivity of NSP4 for Arg residues at the P1 position (Fig. 5C).
Natural Inhibitors of NSP4.
Cleavage of H-Tyr-Arg-Phe-Arg-AMC by NSP4 was completely prevented by α1-antitrypsin (α1AT), heparin-accelerated antithrombin, or C1 inhibitor (Fig. 5D). Elafin, secretory leukocyte protease inhibitor (SLPI), monocyte-neutrophil elastase inhibitior (MNEI), and α1-antichymotrypsin (ACT) did not inhibit NSP4 activity (Fig. 5D).
To investigate complex formation with serpins, we analyzed the NSP4 by immunoblotting after various incubation times. In the presence of a 50-, 6.5-, or 5-fold molar excess of α1PI, heparin-activated antithrombin, or C1 inhibitor at concentrations found in human plasma, free NSP4 declined and covalent complexes with all three serpins of ∼80, 90, or 130 kDa, respectively, were identified (Fig. 5E). Complexes with antithrombin in the presence of heparin could be detected within 1 min and occurred most rapidly. Because NSP4 activity was completely abolished, but noncovalently linked NSP4 was still found after prolonged incubation times, we suggest that noncovalent interactions between these serpins and NSP4 contributed to the observed inhibitory activity. Similar noncovalent inhibition of target proteases by serpins have been reported in the past (18).
Discussion
Serine proteases of the immune defense system represent a conserved subfamily of serine protease genes that are characterized by a unique exon-intron organization and by their location in three specific chromosomal regions. Although the human Gzms form tandem clusters on human chromosome 5 (GZMA and GZMK) and chromosome 14 (GZMB and GZMH, together with mast cell chymase, CMA1, and neutrophil cathepsin G, CTSG), GZMM, the fifth Gzm is found on chromosome 19 in some distance away from the PRTN3, ELANE, and CFD cluster. The orphan reading-frame coding for NSP4 is embedded within this 300-kb segment. So far, nothing was known about this serine protease gene, and its relationships to the granzymes and neutrophil serine proteases was completely unclear. Expression profiling of its transcripts gave little clue to the site of expression and potential function. The Genome Novartis Foundation (GNF) database suggested expression of the mouse homolog in bone, bone marrow, and the thymus, but the respective data for the human gene were missing.
Although the PRTN3-ELANE-CFD gene cluster has previously been reported to be lacking in both zebra finch and chicken (19), we found clear evidence for the existence of a CFD ortholog in both bird species, but no directly clustered elastase-like neighbor. In birds, CFD is neighbored by the polypyrimidine tract binding protein 1 (PTPB1) on one side as in mammalian genomes. In opposite direction, a CFD-linked single copy equivalent of the CMA1, CTSG, GZMB, and GZMH genes can be noticed. This zebra finch gene has only six cysteine residues forming three disulfide bonds and an Asp-189 residue at the bottom of the S1 pocket, as seen in CFD homologs of vertebrates. Besides the loss of the fourth intramolecular cysteine bond, this feature of the S1 pocket excludes it as being an NSP4 or elastase-like equivalent.
Orthologs for the human NSP4 gene are not directly linked to the CFD locus in mammals. Two unrelated genes, FGF22 and POLRMT genes next to NSP4, however, were found to mark a well-conserved synteny group around the NSP4 locus in mammals, birds and bony fishes. CFD is about 100-kb apart from the NSP4-FGF22-POLRMT region in these vertebrates. In birds and bony fishes, NSP4-like genes are located in close proximity to the FGF22-POLRMT genes. Although this region of the zebra finch genome contains only one copy of NSP4, it has been expanded in fishes and harbors a cluster of several paralogs with predictable NSP4-like substrate specificity. In zebra finches the NSP4 homolog is the only class 6 serine protease that carries a Gly instead of an Asp residue at the bottom of the S1 pocket. In bony fishes, all paralogs show this feature of the S1 pocket. These findings strongly suggest that NSP4 preceded the emergence of NE, CG, and PR3 and emerged as a class 6 serine protease quite early during vertebrate evolution.
Sequence comparisons of NSP4 homologs from vertebrate species (www.ensembl.org, gene name Prss57) point to a high conservation of the S1-specificity pocket. Although the critical residues Gly189 and Asp226, which determine the shape and electrostatic properties of the S1, are shared by NE and PR3, the cleavage specificity of NSP4 is unexpectedly unique and differs by its clear arginine preference from all other NSPs. In NE, however, the acidic side chain of Asp226 is fully shielded by the hydrophobic side chains of Val216 and Val190; in PR3 it is inaccessible and thus does not confer a negative character on the S1-specificity pocket in these two NSPs. The third well-known NSP, CG with a larger Glu226, strongly prefers phenylalanine or tyrosine residues at the P1 substrate position, but also cleaves substrates after arginine or lysine residues with lower efficacy (20); its Glu226 side chain extends into the S1 pocket and is fixed to Ala190. The carboxylate group of Glu226 may be protonated in this environment. The nonoptimal interactions of the S1 pocket with Lys and Arg residues and the higher flexibility of the pocket because of the lack of the active site Cys191-Cys220 bond may explain the low catalytic efficacy of CG. In contrast, all NSP4 homologs are stabilized by a fourth Cys191-Cys220 disulfide bond and possess a well-conserved Tyr228, the hydroxyl group of which may interact with the Asp226 carboxylate as in the rat anionic trypsin mutant D189G/G226D (21). The shorter side chain of Asp226 and a differently shaped S1 pocket may explain this unexpected selective preference of NSP4 for arginine residues.
Recently, murine NSP4 was selected for disruption as one of 472 target genes coding for secreted or transmembrane proteins using a lacZ-neomycin cassette (22). Although the lacZ-encoded β-galactosidase transcribed by the NSP4 promoter is a highly sensitive reporter enzyme for expression profiling, no lacZ activity was recorded in any of the tissues studied. Expression of mRNA in wild-type mice was found to be restricted to the thymus and spleen, but the cell type transcribing this gene has not been identified. Despite screening a broad panel of phenotypic parameters in this mouse line with a mixed genetic background, no specific biological effect of this gene, called Prss57, was detected. The authors investigated a total of four Prss57-deficient mice on a mixed 129/SvEvBrd × C57BL/6 hybrid background and observed no notable phenotype except for a marginal decrease in peripheral blood CD8 and NK cells, a slight increase in B cells, and slightly higher levels of TNF-α and monocyte chemotactic protein-1 in response to intraperitonal LPS injections. Because the altered values are within the broad range of variability and depend on the housing environment and the health status of the animals, the causative relationships of these alterations to the functions of NSP4 is questionable. In light of its cell type-restricted expression and sequestration in neutrophils, its functional similarity with other neutrophil serine proteases, and inhibition by abundant extracellular inhibitors, it is unlikely to have effects at distant sites and to mediate a broad range of physiological functions.
Our experiments with natural inhibitors clearly indicate that the proteolytic effects of NSP4 are controlled by natural serine protease inhibitors of the serpin-type and are most likely limited to the intracellular and pericellular microenvironment of activated neutrophils. Using highly specific rat mAbs to human NSP4, we were able to unequivocally demonstrate restricted expression and storage of NSP4 in human neutrophils, but not in other leukocyte populations, neither in CD8 T cells nor B cells of peripheral blood. This tissue distribution is fully consistent with mRNA expression (GNF) (22) in bone marrow and with our immunohistological findings showing strongest expression in myeloid cell populations of the bone marrow. Although NSP4 may be produced by additional specialized cells, in particular after specific stimulation or in response to microbial invasion, neutrophils are by far the most abundant source for this protease under physiological conditions. Nevertheless, this protease escaped its identification and biochemical characterization in the past for various reasons. Its molecular size, its proteolytic specificity, and cellular storage are very similar to NE and PR3. Besides these difficulties in identifying this protein, the relatively low abundance of NSP4 in neutrophils also makes purification much more difficult. Our semiquantitative comparison of NSP4 with CG suggests a 20-fold lower abundance of NSP4 in human neutrophils.
To demonstrate its functional integrity after recombinant expression and purification, we used a widely known elastase substrate, with Nva at the P1 position in the beginning of our studies. Sixfold higher concentrations of the enzyme were required to obtain a similar cleavage rate as with NE, suggesting a different type of cleavage specificity. Proteome-wide mapping of cleavage sites subsequently revealed a preference for Arg at the P1 site. Formation of a covalent complex with heparin-accelerated antithrombin and C1 inhibitor is a further indication for its Arg-specific activity as complex formation is associated with a cleavage after the P1 Arg residue in these serpins. Further comprehensive characterization of the fine specificity of NSP4 with human peptide libraries will be required to distinguish its activity from those of CG and other serine proteases. The low abundance in neutrophils and the strict conservation of this gene in vertebrates supports our suggestion that it is not just digesting bacterial proteins in the phagolysosome, but rather regulating neutrophil responses and innate immune reactions. As a secreted protease that may potentiate the proteolytic damage of tissues by activated neutrophils, NSP4 also appears to be an attractive therapeutic target that was completely overseen in the past.
Materials and Methods
Generation of mAbs Against NSP4.
Eighty micrograms of the NSP4 IB material was injected intraperitoneally and subcutaneously into LOU/C rats using incomplete Freund's adjuvant supplemented with 5 nmol CpG 2006 (TIB MOLBIOL). After a 6-wk interval, a final boost with 50 μg NSP4 IB material and CpG 2006 was given intraperitoneally and subcutaneously 3 d before fusion. Fusion of the myeloma cell line P3 × 63-Ag8.653 with the rat immune spleen cells was performed according to standard procedures. Hybridoma supernatants were tested in a solid-phase immunoassay with the NSP4 IB material or an irrelevant C-His fusion protein coated to ELISA plates. Antibodies from tissue culture supernatant bound to NSP4 were detected with biotin-conjugated mAbs against the rat IgG isotypes (TIB173 IgG2a, TIB174 IgG2b, TIB170 IgG1, all from ATCC; R-2c IgG2c homemade), thus avoiding mAbs of IgM class, and HRP-streptavidin (Vector). HRP was visualized with OPD (Sigma). The immunization of rodents was approved by the local government of Upper Bavaria.
Determination of the Amount of NSP4 Stored in PMNs.
PMN total cell lysates, CG, and NSP4 were serially diluted and analyzed in parallel by immunoblotting. For this we used mouse anti-CG 1:60 (ab50845; Abcam), rat anti-NSP4 mAb 2F6 1:30, goat anti-mouse IgG+IgM 1:2,500 (31444; Thermo Scientific), and goat anti-rat IgG+IgM 1:10,000 (Jackson Immunoresearch) antibodies, the latter two peroxidase-conjugated. With the use of this semiquantitative comparison, the total amounts of CG and NSP4 in PMNs were classified.
Degranulation of PMNs.
After isolation, PMNs were resuspended in RPMI (Invitrogen) at a concentration of 1.7 × 105 cells/μL. CytB was added to a final concentration of 5 μg/mL and after 5-min incubation at 37 °C, PMA was added to a final concentration of 200 ng/mL. Because cytB and PMA were both solved in DMSO, DMSO alone was added to the control sample. After another 30 min at 37 °C, PMNs were pelleted for 10 min at 500 × g and resulting supernatants were analyzed by Western blotting.
PICS.
NSP4 specificity profiling using proteome-derived peptide libraries was performed as described previously (17). Tryptic, GluC, and chymotryptic peptide libraries were generated from E. coli. Enzyme-to-library ratios were 1:200 (wt/wt). Incubation was at 37 °C for 16 h in 50 mM Hepes, pH 7.5, 300 mM NaCl. For LC-MS/MS, the peptides were injected onto a 15 cm × 75 μm ProteoPep 2 PicoFrit column (New Objectives) connected to an LTQ-OrbiTrap XL mass spectrometer (Thermo Scientific). Peptide sequences were identified by X!Tandem (23), in conjunction with PeptideProphet (24) at a confidence level > 95%. Mass tolerance was 10 ppm for parent ions and 0.5 Da for fragment ions. Nonenzyme constraint searches were applied. Static modifications were cysteine carboxyamidomethylation (+57.02 Da), lysine dimethylation (+28.03 Da), and thioacylation of peptide amino-termini (+88.00 Da). A Web-based PICS service (25) was used to derive nonprime sequences. Sequence logos were generated with iceLogo (26).
Activity Assays.
Enzymatic activity was assessed using 1 mM Boc-Ala-Pro-Nva-4-chloro-SBzl (Bachem) and 0.5 mM 5′,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) or 1 mM H-Tyr-Arg-Phe-[Arg/Lys]-AMC (Novabiochem). Substrate hydrolysis was recorded with a spectrofluorometer (BMG Labtech).
Inhibition Assays.
Enzymatic inhibition of NSP4 (0.25 μM) from HEK 293 cells was assayed after 45 min of incubation at 37 °C using the following inhibitors and concentrations, elafin, 10 μM (Proteo Biotech); SLPI, 10 μM (R&D Systems); α1PI, 25 μM (Athens Research); MNEI (Creative Biomart) with 1 mM DTT, 3.3 μM; antithrombin (Athens Research) with 300 U/mL heparin (Sigma), 5.6 μM; C1 Inhibitor, 2.5 μM (Athens Research); or ACT, 5.6 μM (Athens Research). Subsequent activity measurements were carried out as described above using H-Tyr-Arg-Phe-Arg-AMC. For comparisons, NSP4 without inhibitor was treated similarly. To test whether NSP4 forms covalent complexes with serpins, 0.8 μM NSP4 was incubated at 37 °C with 40 μM α1PI, 5 μM antithrombin and 300 U/mL heparin or 4 μM C1 inhibitor in PBS. Samples of 2 μL were taken after 0.5, 5, 30, and 100 min and analyzed by SDS/PAGE and immunoblotting with anti-NSP4 mAbs, as described in SI Materials and Methods.
Ethical Statement.
All human samples were obtained and used in accordance with the procedures approved by the Institutional Review Board of the Ludwig Maximilian University, Munich.
Preparation of NSP4 IB material, production of native NSP4 (S-tag NSP4), PR3, the Gzms A, B, K, M, H, NSP4 precursor, ELISA, isolation of different blood cell populations, preparation of total cell lysates, SDS/PAGE, immunoblotting, immunohistochemistry, conversion of NSP4 precursor by DPPI, and Edman sequencing are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Elisabeth Stegmann and Bettina Mayer for excellent technical assistance; Hartmut Wekerle for his continuous interest in the project; and Debora Bonenfant (Novartis) for excellent LC-MS/MS analysis. This work was supported in part by the German Research Council (SFB 571); O.S. is supported by an Emmy-Noether grant of the Deutsche Forschungsgemeinschaft (DFG SCHI 871/2) and a starting grant of the European Research Council (Programme “Ideas” ERC-2011-StG 282111-ProteaSys).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200470109/-/DCSupplemental.
References
- 1.Belaaouaj A. Neutrophil elastase-mediated killing of bacteria: Lessons from targeted mutagenesis. Microbes Infect. 2002;4:1259–1264. doi: 10.1016/s1286-4579(02)01654-4. [DOI] [PubMed] [Google Scholar]
- 2.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams R. Killing controversy. J Exp Med. 2006;203:2404. doi: 10.1084/jem.20311fta. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss SJ. Tissue destruction by neutrophils. N Engl J Med. 1989;320:365–376. doi: 10.1056/NEJM198902093200606. [DOI] [PubMed] [Google Scholar]
- 5.Kessenbrock K, Dau T, Jenne DE. Tailor-made inflammation: How neutrophil serine proteases modulate the inflammatory response. J Mol Med (Berl) 2011;89:23–28. doi: 10.1007/s00109-010-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 7.Nathan C. Neutrophils and immunity: Challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 8.Pham CT. Neutrophil serine proteases: Specific regulators of inflammation. Nat Rev Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 9.Jenne DE. Structure of the azurocidin, proteinase 3, and neutrophil elastase genes. Implications for inflammation and vasculitis. Am J Respir Crit Care Med. 1994;150:S147–S154. doi: 10.1164/ajrccm/150.6_Pt_2.S147. [DOI] [PubMed] [Google Scholar]
- 10.Puente XS, Sánchez LM, Overall CM, López-Otín C. Human and mouse proteases: A comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 11.Pilat D, et al. The human Met-ase gene (GZMM): Structure, sequence, and close physical linkage to the serine protease gene cluster on 19p13.3. Genomics. 1994;24:445–450. doi: 10.1006/geno.1994.1651. [DOI] [PubMed] [Google Scholar]
- 12.Bode W, Meyer E, Jr, Powers JC. Human leukocyte and porcine pancreatic elastase: X-ray crystal structures, mechanism, substrate specificity, and mechanism-based inhibitors. Biochemistry. 1989;28:1951–1963. doi: 10.1021/bi00431a001. [DOI] [PubMed] [Google Scholar]
- 13.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 14.Campbell EJ, Silverman EK, Campbell MA. Elastase and cathepsin G of human monocytes. Quantification of cellular content, release in response to stimuli, and heterogeneity in elastase-mediated proteolytic activity. J Immunol. 1989;143:2961–2968. [PubMed] [Google Scholar]
- 15.Bentwood BJ, Henson PM. The sequential release of granule constitutents from human neutrophils. J Immunol. 1980;124:855–862. [PubMed] [Google Scholar]
- 16.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev. 2010;62:726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- 18.Shieh BH, Potempa J, Travis J. The use of alpha 2-antiplasmin as a model for the demonstration of complex reversibility in serpins. J Biol Chem. 1989;264:13420–13423. [PubMed] [Google Scholar]
- 19.Quesada V, Velasco G, Puente XS, Warren WC, López-Otin C. Comparative genomic analysis of the zebra finch degradome provides new insights into evolution of proteases in birds and mammals. BMC Genomics. 2010;11:220. doi: 10.1186/1471-2164-11-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wysocka M, et al. New chromogenic substrates of human neutrophil cathepsin G containing non-natural aromatic amino acid residues in position P(1) selected by combinatorial chemistry methods. Mol Divers. 2007;11:93–99. doi: 10.1007/s11030-007-9063-7. [DOI] [PubMed] [Google Scholar]
- 21.Perona JJ, Tsu CA, McGrath ME, Craik CS, Fletterick RJ. Relocating a negative charge in the binding pocket of trypsin. J Mol Biol. 1993;230:934–949. doi: 10.1006/jmbi.1993.1211. [DOI] [PubMed] [Google Scholar]
- 22.Tang T, et al. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 23.Craig R, Beavis RC. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 24.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 25.Schilling O, auf dem Keller U, Overall CM. Factor Xa subsite mapping by proteome-derived peptide libraries improved using WebPICS, a resource for proteomic identification of cleavage sites. Biol Chem. 2011;392:1031–1037. doi: 10.1515/BC.2011.158. [DOI] [PubMed] [Google Scholar]
- 26.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.