Abstract
While conservation of ATP is often a desirable trait for microbial production of chemicals, we demonstrate that additional consumption of ATP may be beneficial to drive product formation in a nonnatural pathway. Although production of 1-butanol by the fermentative coenzyme A (CoA)-dependent pathway using the reversal of β-oxidation exists in nature and has been demonstrated in various organisms, the first step of the pathway, condensation of two molecules of acetyl-CoA to acetoacetyl-CoA, is thermodynamically unfavorable. Here, we show that artificially engineered ATP consumption through a pathway modification can drive this reaction forward and enables for the first time the direct photosynthetic production of 1-butanol from cyanobacteria Synechococcus elongatus PCC 7942. We further demonstrated that substitution of bifunctional aldehyde/alcohol dehydrogenase (AdhE2) with separate butyraldehyde dehydrogenase (Bldh) and NADPH-dependent alcohol dehydrogenase (YqhD) increased 1-butanol production by 4-fold. These results demonstrated the importance of ATP and cofactor driving forces as a design principle to alter metabolic flux.
Keywords: biofuel, malonyl-CoA, metabolic engineering, synthetic biology
Biological production of chemicals and fuel from renewable resources is an attractive approach to a sustainable future. In particular, 1-butanol has received increasing attention because it is a potential fuel substitute and an important chemical feedstock. 1-Butanol can be produced by two distinct routes: the coenzyme A (CoA)-dependent pathway (1, 2) and the keto acid pathway (3–5). The CoA-dependent pathway follows the chemistry of β-oxidation in the reverse direction, in which acetyl-CoA is condensed to form acetoacetyl-CoA and then reduced to 1-butanol. The keto acid pathway utilizes 2-ketobutyrate as an intermediate, which goes through keto acid chain elongation to 2-ketovalarate using the leucine biosynthesis enzymes. 2-Ketovalarate is then decarboxylated and reduced into 1-butanol. In each case, the pathway can be extended to produce 1-hexanol and other higher alcohols (6–8).
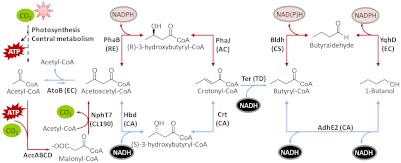
The CoA-dependent reverse β-oxidation is a natural fermentation pathway used by Clostridium species (9–11) and has been transferred to various recombinant heterotrophs, resulting in 1-butanol titers ranging from 2.5 mg/L to 1.2 g/L with glucose as the substrate (12–16). One of the challenges in transferring this pathway to other organisms lies in the hydrogenation of crotonyl-CoA to butyryl-CoA catalyzed by the butyryl-CoA dehydrogenase/electron transferring flavoprotein (Bcd/EtfAB) complex. Bcd/EtfAB complex is difficult to express in recombinant systems, is presumably oxygen sensitive (12, 17), and possibly requires reduced ferredoxin as the electron donor (18). This difficulty was overcome by expressing trans-2-enoyl-CoA reductase (Ter) (19, 20), which is readily expressed in Escherichia coli and directly reduces crotonyl-CoA using NADH. This modified 1-butanol pathway (Fig. 1; outlined in blue) is catalyzed by five enzymes: thiolase (AtoB), 3-hydroxybutyryl-CoA dehydrogenase (Hbd), crotonase (Crt), Ter, and bifunctional aldehyde/alcohol dehydrogenase (AdhE2). Simultaneously expressing these enzymes and engineering NADH and acetyl-CoA accumulation as driving forces, 1-butanol production with a high titer of 15 g/L and 88% of theoretical yield has been achieved using E. coli in flasks without product removal (19). This result demonstrates the feasibility of transferring the CoA-dependent pathway to nonnative organisms for high-titer 1-butanol fermentation from glucose.
Fig. 1.
Variations in the CoA-dependent 1-butanol pathway. The fermentative CoA 1-butanol pathway is in blue. Alternative routes are in red. EC, E. coli; RE, R. eutropha; CA, C. acetobutylicum; AC, A. caviae; TD, T. denticola; CS, C. saccharoperbutylacetonicum N1-4; CL190, Streptomyces sp. strain CL190.
However, the success of the CoA-dependent pathway in E. coli is not directly transferrable to photoautotrophs. By expressing the same enzymes in cyanobacteria Synechococcus elongatus PCC 7942, photosynthetic 1-butanol production from CO2 was barely detectable (21). 1-Butanol production was achieved by this strain only when internal carbon storage made by CO2 fixation in light conditions was fermented under anoxic conditions (21). We hypothesized that both the acetyl-CoA and NADH pools in this organism under photosynthetic conditions may be insufficient to drive 1-butanol formation. Acetyl-CoA is the precursor for fermentation pathway and the TCA cycle, both of which are not active in light conditions. Furthermore, photosynthesis generates NADPH, but not NADH, and the interconversion between the two may not be efficient enough. Without a significant driving force against the unfavorable thermodynamic gradient, 1-butanol production cannot be achieved. The difficulty of direct photosynthetic production of 1-butanol is in sharp contrast to the production of isobutanol (450 mg/L) and isobutyraldehyde (1,100 mg/L) by S. elongatus PCC 7942 (22), which has an irreversible decarboxylation step as the first committed reaction to drive the flux toward the products. This difference suggests the importance of driving forces in altering the direction of metabolic flux.
We reason that instead of the acetyl-CoA pool, ATP may be used to drive the thermodynamically unfavorable condensation of two acetyl-coA molecules under photosynthetic conditions. Thus, we engineered the ATP-driven malonyl-CoA synthesis and decarboxylative carbon chain elongation used in fatty acid synthesis to drive the carbon flux into the formation of acetoacetyl-CoA, which then undergoes the reverse β-oxidation to synthesize 1-butanol. We further replaced the subsequent NADH-dependent enzymes with NADPH-dependent ones and successfully achieved 1-butanol synthesis under photosynthetic conditions. In theory, excess ATP consumption in the cell might cause a decrease in biomass. Thus, with notable exceptions (23–26), most metabolic engineering design do not choose to increase ATP consumption. Although many natural examples of microbes using ATP to drive reactions, most of them are highly regulated. Therefore, it is unpredictable whether it is feasible to use ATP consumption to push flux in a nonnative pathway, for which no regulation exists.
Results
Incorporating an ATP Driving Force in 1-Butanol Pathway Design.
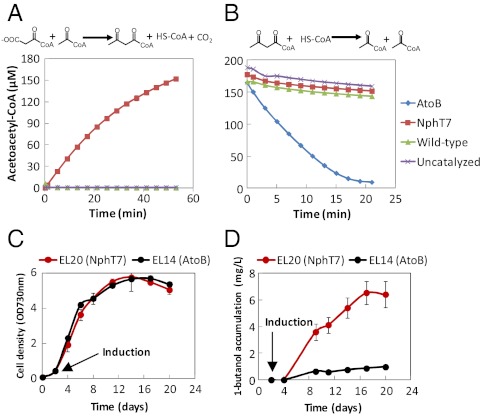
The thiolase-mediated condensation of two acetyl-CoA molecules is reversible but strongly favors the thiolysis of acetoacetyl-CoA. To examine the thermodynamic property of this reaction, we overexpressed and purified E. coli thiolase (AtoB) and used an in vitro assay to determine its equilibrium constant and ΔG∘′. The result showed that the condensation reaction is unfavorable (Fig. 2) with equilibrium constant (Keq) of (1.1 ± 0.2) × 10-5 at pH 8.0, within the optimum pH range for cyanobacteria (27). This experimentally determined Keq approximately corresponds to a ΔG∘′ of 6.8 kcal/mol, consistent with the literature reported Keq using partially purified thiolase from pig heart protein homogenate (28). Therefore, without a sufficiently large acetyl-CoA pool or an efficient product trap, there is no driving force for the formation of acetoacetyl-CoA. Although the irreversible hydrogenation of crotonyl-CoA catalyzed by Ter provides a driving force, it is insufficient to drive the reaction forward without large pools of acetyl-CoA and reducing equivalent (19).
Fig. 2.
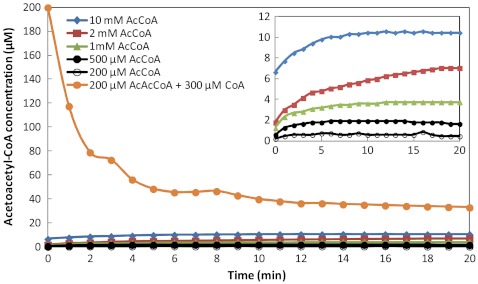
Determination of equilibrium concentrations for the thiolase (AtoB) mediated reaction. The equilibrium constant (Keq) was determined from the equilibrium concentrations. E. coli AtoB was cloned, purified, and used in an in vitro assay. AcCoA, acetyl-CoA; AcAcCoA, acetoacetyl-CoA; CoA, coenzyme A. Detailed conditions and methods are listed in SI Text.
Instead of using the direct condensation of acetyl-CoA, we contemplated an alternative route through the ATP-driven malonyl-CoA synthesis. Malonyl-CoA is synthesized from acetyl-CoA, 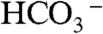 and ATP by acetyl-CoA carboxylase (Acc). The formation of malonyl-CoA is effectively irreversible due to ATP hydrolysis. In fatty acid synthesis, malonyl-CoA is then converted into malonyl-acyl carrier protein (malonyl-ACP) and acts as the carbon addition unit for fatty acid synthesis. For 1-butanol synthesis, malonyl-CoA can react with acetyl-coA in a decarboxylative condensation to form acetoacetyl-CoA, in a reaction analogous to ketoacyl-ACP synthase III (KAS III) that catalyzes the irreversible condensation of malonyl-ACP and acetyl-CoA to synthesize the four carbon intermediate 3-ketobutyryl-ACP.
and ATP by acetyl-CoA carboxylase (Acc). The formation of malonyl-CoA is effectively irreversible due to ATP hydrolysis. In fatty acid synthesis, malonyl-CoA is then converted into malonyl-acyl carrier protein (malonyl-ACP) and acts as the carbon addition unit for fatty acid synthesis. For 1-butanol synthesis, malonyl-CoA can react with acetyl-coA in a decarboxylative condensation to form acetoacetyl-CoA, in a reaction analogous to ketoacyl-ACP synthase III (KAS III) that catalyzes the irreversible condensation of malonyl-ACP and acetyl-CoA to synthesize the four carbon intermediate 3-ketobutyryl-ACP.
We note that the energy release from ATP hydrolysis (ΔG∘′ of -7.3 kcal/mol) would compensate for the energy required for condensation of acetyl-CoA into acetoacetyl-CoA. By combining the reaction catalyzed by thiolase with ATP hydrolysis (Fig. S1), the net reaction is thermodynamically favorable (ΔG∘′ < 0), which would reduce the need for high concentration of acetyl-CoA required to push the reaction forward. More importantly, CO2 released from the second step, decarboxylative chain elongation, shifts the reaction toward the formation of acetoacetyl-CoA. Fatty acid and polyketide syntheses have naturally evolved this mechanism to enable the thermodynamically unfavorable formation of 3-ketoacyl-ACP. By taking advantage of this mechanism, it is possible to push the carbon flux to the 1-butanol pathway without the acetyl-CoA driving force artificially constructed in E. coli (19).
Expression of Acetoacetyl-CoA Synthase Enables Photosynthetic Production of 1-Butanol.
Therefore, we bioprospected for a KAS III that utilizes malonyl-CoA rather than malonyl-ACP for condensation with acetyl-CoA. Because both ACP and CoA carry phosphopantetheine, which forms thioester bond with the malonyl moiety, KAS III and KAS III-like enzymes may also react with malonyl-CoA. We cloned a variety of KAS III and KAS III-like enzymes from different organisms and examined their expression in E. coli (Fig. S2). After His-tag purification, we assayed their activity for condensing malonyl-CoA with acetyl-CoA (Table 1). Among the enzymes tested, NphT7 (29) was the most active. Other enzymes such as Bamb6244, GOX0115, and PAE-FabH2 were also active while the rest showed no detectable activity. The condensation reaction (Fig. S3) catalyzed by NphT7 using malonyl-CoA and acetyl-CoA is irreversible and accumulates acetoacetyl-CoA as the product. At low starting concentrations of malonyl-CoA, conversion yield to acetoacetyl-CoA is higher than high starting substrate concentrations. This result is likely due to the fact that NphT7 also catalyzes malonyl-CoA self condensation and is particularly useful for 1-butanol synthesis, because both malonyl-CoA and acetyl-CoA pools are expected to be low in S. elongatus PCC 7942.
Table 1.
Specific activities of acetoacetyl-CoA synthases (μmol/ min /mg)
| Enzyme | Specific activity | Enzyme | Specific activity |
| Burkholderia ambifaria BAMB6244 | 0.0116 ± 0.0002 | Pseudomonas aeruginosa PAE-FabH2 | 0.0140 ± 0.0010 |
| Gluconobacter oxydans GOX0115 | 0.0099 ± 0.0011 | Streptomyces avermitilis SAV-FabH4 | ND |
| Helicobacter pylori HP0202 | ND | Streptomyces coelicolor SCO5858 | ND |
| Listeria monocytogenes LMO2202 | ND | Streptomyces sp. strain CL190 NphT7 | 6.02 ± 0.25 |
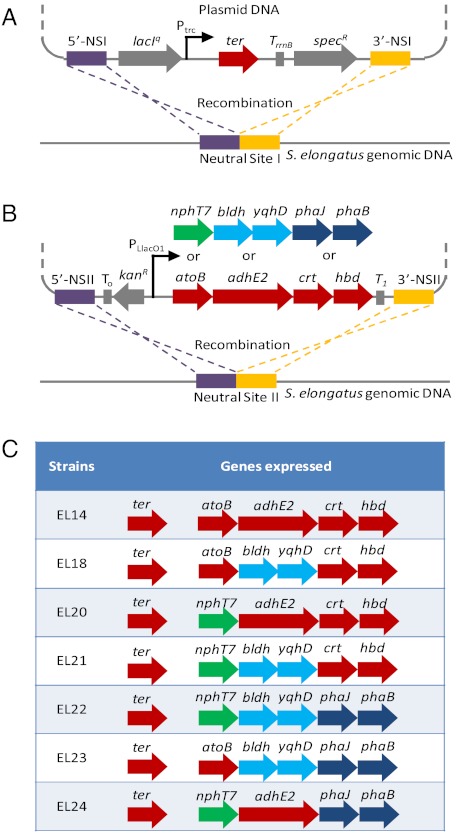
Next, we constructed a plasmid harboring genes nphT7, hbd, crt, and adhE2 under an IPTG inducible promoter PLlacO1 with Neutral Site II (NSII) recombination sequences flanking the genes and kanamycin resistance marker (Fig. 3). By DNA homologous recombination, we integrated these genes into the genome of S. elongatus strain EL9 [expressing ter at Neutral Site I (NSI) under the control of another IPTG inducible promoter Ptrc] at NSII and selected for successful transformant on kanamycin containing BG-11 plates. The successful transformant strain EL20 was then analyzed for in vitro enzyme activity and 1-butanol production. As shown in (Fig. 4A), crude extract from strain EL20 catalyzed the formation of acetoacetyl-CoA by condensation of malonyl-CoA and acetyl-CoA and did not catalyze the thiolysis of acetoacetyl-CoA (Fig. 4B). On the other hand, crude extract from strain EL14 expressing atoB along with hbd, crt, ter, and adhE2 catalyzed thiolysis (Fig. 4B) much more efficiently than the condensation reaction (Fig. 4A). The two strains EL20 and EL14 exhibited nearly identical growth rate (Fig. 4C). However, Strain EL20 produced 6.5 mg/L (Fig. 4D) of 1-butanol while Strain EL14 produced only barely detectable amounts (detection limit of about 1 mg/L) of 1-butanol (Fig. 4D). This result indicated that ATP-driven acetoacetyl-CoA formation is required for photosynthetic production of 1-butanol using the CoA-dependent pathway.
Fig. 3.
Schematic representation of recombination to integrate (A) ter at NSI, (B) atoB, adhE2, crt, and hbd at NSII in the genome of S. elongatus. Different combinations of alternative genes nphT7, bldh, yqhD, phaJ, and phaB can replace their counterpart enzymes to recombine into NSII. (C) List of strains with different combinations of overexpressed genes used in this study.
Fig. 4.
(A) In vitro assay for the synthesis of acetoacetyl-CoA using crude extracts of wild-type S. elongatus PCC 7942, strain EL14 and EL20. (B) In vitro assay for the thiolysis of acetoacetyl-CoA using crude extracts of wild-type S. elongatus PCC 7942, strain EL14 and EL20. (C) Cell density and (D) 1-butanol accumulation as a function of time of strain EL14 and EL20. 1-Butanol production by strain EL14 was near detection limit of about 1 mg/L.
Substitution of NADPH Utilizing Enzymes Aids 1-Butanol Production.
Another useful driving force in 1-butanol synthesis is the reducing equivalent (19). Cyanobacteria produce NADPH as the direct result of photosynthesis. Intracellular NAD+ and NADP+ levels exist in a ratio of about 1∶10 (30) in S. elongatus PCC 7942. Thus NADH utilizing pathway may be unfavorable in cyanobacteria. To synthesize 1 mol of 1-butanol from acetoacetyl-CoA requires 4 mol of NADH. Therefore, changing the cofactor preference to NADPH may aid the production of 1-butanol.
As depicted in Fig. 1 (outlined in red), we identified enzymes that utilize NADPH or both NADPH and NADH by bioprospecting. Acetoacetyl-CoA reductase (PhaB) (31) was used to replace Hbd. PhaB from Ralstonia eutropha is an enzyme found in the poly-hydroxyalkanoate biosynthetic pathway for reducing 3-ketobutyryl-CoA to 3-hydroxybutyryl-CoA using NADPH. However, PhaB produces the (R)-stereoisomer of 3-hydroxybutyryl-CoA instead of the (S)-stereoisomer produced by Hbd. As a result, Crt cannot be used for the subsequent dehydration to produce crotonyl-CoA. Therefore, a different crotonase is necessary for dehydration of (R)-3-hydroxybutyryl-CoA. (R)-specific enoyl-CoA hydratase (PhaJ) (32) is found in Aeromonas caviae and dehydrates (R)-3-hydroxybutyryl-CoA into crotonyl-CoA. Together, PhaB and PhaJ can replace Hbd and Crt, respectively, to change the cofactor preference of 3-ketobutyryl-CoA reduction to NADPH.
To replace AdhE2, NADP-dependent alcohol dehydrogenase (YqhD) (33) from E. coli has been demonstrated to aid the production of higher chain alcohols (22). In addition, we needed a CoA-acylating butyraldehyde dehydrogenase (Bldh) to replace the aldehyde dehydrogenase function of AdhE2. We thus bioprospected for enzymes catalyzing reduction of butyryl-CoA to butyraldehyde. Bldh was found in high butanol producing Clostridium species including C. beijerinckii NCIMB 8052 (34), C. saccharobutylicum ATCC BAA-117, and C. saccharoperbutylacetonicum NI-4 (35). In particular, Bldh from C. beijerinckii has been purified and demonstrated activity in vitro with both NADH and NADPH as a reducing cofactor.
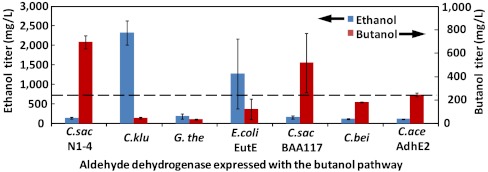
Using the sequence of C. beijerinckii Bldh, we searched by homology and cloned additional Bldh-like enzymes from various organisms including C. saccharoperbutylacetonicum NI-4, C. saccharobutylicum ATCC BAA-117, Geobacillus thermoglucosidasius, Clostridium kluyveri, and E. coli. We assessed the performance of these Bldh’s by 1-butanol production in recombinant E. coli. As shown in Fig. 5, the E. coli strain expressing C. saccharoperbutylacetonicum NI-4 Bldh along with rest of the CoA 1-butanol pathway produced the highest titer of 1-butanol, exceeding the 1-butanol produced by E. coli strain expressing AdhE2 by nearly threefold.
Fig. 5.
Production of 1-butanol and ethanol by recombinant E. coli strains JCL299 expressing CoA-dependent 1-butanol pathway with YqhD from E. coli and Bldh from different organisms. Dashed line represents the baseline production by using AdhE2. Detailed production procedure is listed in SI Text.
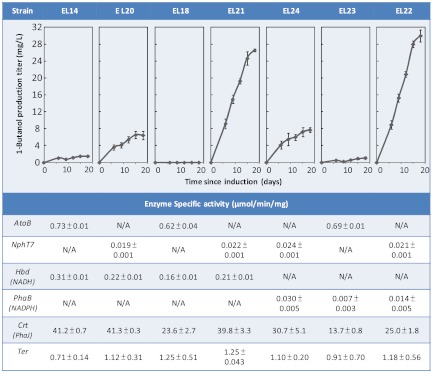
To test the effect of cofactor utilization, we constructed various combinations of different routes by overexpressing different genes in S. elongatus PCC 7942 (Fig. 3C). We constructed plasmids containing different genes and recombined them into the genome of S. elongatus strain EL9. We then assayed the activity of overexpressed enzymes to confirm expression (Fig. 6). Of the strains tested, strain EL22 expressing the NADPH utilizing enzymes produced the highest amount of 1-butanol (29.9 mg/L) exceeding that of EL20 (6.4 mg/L) by more than fourfold. This result reinforced the importance of cofactor as driving force.
Fig. 6.
1-Butanol production and enzyme activity of strains expressing different enzymes. Strain genotype is listed in Fig. 3C. Expression of nphT7 enables direct photosynthetic production of 1-butanol under oxygenic condition. Strains EL21 and EL22 expressing bldh and yqhD achieved the highest production.
Discussion
In a metabolic system involving multiple pathways, the direction and rate of each reaction are determined by kinetics, regulated by the enzyme expression levels and metabolite pool sizes. Typically, a reaction with a large positive ΔG∘′ is considered practically unfeasible in the forward direction because it requires high concentrations of the substrate pool to drive the reaction forward. The condensation of two molecules of acetyl-CoA to acetoacetyl-CoA is such an example. On the other hand, the reverse direction, thiolysis of acetoacetyl-CoA, is readily achievable and used as the last step in the β-oxidation. However, some fermentative organisms, such as Clostridium species, use direct acetyl-CoA condensation for 1-butanol synthesis. These organisms accomplish this thermodynamically unfavorable reaction presumably through a large pool of acetyl-CoA and high reducing equivalents that drive the subsequent reactions. This situation was recreated in E. coli expressing the enzymes for 1-butanol synthesis (19). Unfortunately, this strategy cannot be readily implemented in photosynthetic organisms for multiple reasons: Acetyl-CoA is a precursor for fermentative pathways and the oxidative TCA cycle, which are not active under photosynthetic conditions. Thus, the acetyl-CoA pool size is not expected to be high and is difficult to modulate. Instead of using high acetyl-CoA pool as a driving force, here ATP and the evolution of CO2 effectively drive the reactions toward 1-butanol synthesis. This strategy is used in fatty acid synthesis in nature, and is used to couple the fatty acid synthesis to the energy status in the cell.
ATP consumption has been used by cells to drive various thermodynamically unfavorable reactions. In engineered E. coli, ATP consumption has been used to stimulate glycolysis by futile cycling (23, 24) or by deletion of membrane-coupling subunits in (F1F0)-ATP synthase (25). Increased ATP consumption by overexpressing enzymes that promote malonyl-CoA biosynthesis also increased production yield of compounds downstream of malonyl-CoA (26). However, additional ATP consumption in heterologous pathways may cause adverse effects in the cell and may result in reduced biomass formation. As such, most metabolic engineering design models have primarily focused on maximizing carbon yield and minimizing ATP expenditure. Here we provide a distinct example using ATP to alter the thermodynamics of the CoA-dependent 1-butanol pathway, which naturally does not require additional ATP consumption. By incorporating an ATP-dependent step into the CoA-dependent 1-butanol synthesis pathway, we demonstrated for the first time the direct photosynthetic production of 1-butanol from CO2.
Direct photosynthetic 1-butanol production from CO2 is desirable because it reduces the number of processing steps. S. elongatus PCC 7942 is naturally competent and therefore is an attractive model organism for engineering. The DNA recombination method used in this study has also been broadly practiced for engineering cyanobacteria for the production of various chemicals. By metabolic engineering, cyanobacteria has also enabled the production of Isobutyraldehyde (1,100 mg/L) (22), isobutanol (450 mg/L) (22), ethanol (550 mg/L) (36), ethylene (451 nl/nl/h/OD730) (37), isoprene (0.05 mg/g dry cell weight) (38), sugars (45 mg/L) (39), lactic acid (56 mg/L) (39), fatty alcohols (0.2 mg/L) (40), and fatty acids (194 mg/L) (41) from CO2. The pathways for the relatively high production of isobutyraldehyde, isobutanol and ethanol naturally involve a decarboxylation step as the first committed reaction. The loss of CO2 is considered irreversible and serves as a driving force to the product formation. Our result is also consistent with this phenomenon that decarboxylation aids in directing the carbon flux.
Reducing cofactor preference is another important aspect of pathway design. Depending on the production condition and organisms’ natural metabolism, changing cofactor preference is necessary to achieve high flux production. For example, changing NADPH-dependent enzymes into NADH-dependent increases the isobutanol productivity and yield under anaerobic condition in recombinant E. coli (42). Replacing the Bcd-EtfAB complex that requires an unknown electron donor with an NADH-dependent Ter is helpful in 1-butanol production (19, 20). In contrast, pathways utilizing NADPH are preferred in cyanobacteria because NADPH is more abundant. By utilizing NADPH-dependent enzymes, our 1-butanol production enhanced from 6.5 mg/L to 29.9 mg/L (Fig. 6).
Current limitation of our 1-butanol production using cyanobacteria may be the synthesis of malonyl-CoA. Compared to the high flux production of isobutanol and isobutyraldehyde in cyanobacteria, the carbon flux through our 1-butanol pathway is suboptimal. Malonyl-CoA biosynthesis is considered as the limiting step in fatty acid synthesis (43). Therefore, increasing carbon flux toward the synthesis of acetyl-CoA and malonyl-CoA may be necessary to increase 1-butanol production. Intracellular acetyl-CoA and malonyl-CoA supply may be increased by increasing CoA biosynthesis (44), overexpression of Acc (45–48), glycolytic enzymes such as phosphoglycerate kinase and glyceraldehyde-3-phosphate dehydrogenase (26), and inhibition of fatty acid biosynthesis (49). With these approaches, the 1-butanol production in cyanobacteria may be further improved.
Materials and Methods
For details, see SI Text.
Culture Medium and Condition.
All S. elongatus PCC 7942 strains were grown on modified BG-11 (1.5 g/L NaNO3, 0.0272 g/L CaCl2·2H2O, 0.012 g/L ferric ammonium citrate, 0.001 g/L Na2EDTA, 0.040 g/L K2HPO4, 0.0361 g/L MgSO4·7H2O, 0.020 g/L Na2CO3, 1,000× trace mineral (1.43 g H3BO3, 0.905 g/L MnCl2·4H2O, 0.111 g/L ZnSO4·7H2O, 0.195 g/L Na2MoO4·2H2O, 0.0395 g CuSO4·5H2O, 0.0245 g Co(NO3)2·6H2O), 0.00882 g/L sodium citrate dihydrate (50)) agar (1.5% w/v) plates. All S. elongatus PCC 7942 strains were cultured in BG-11 medium containing 50 mM NaHCO3 in 250 mL screw cap flasks. Cultures were grown under 100 μE/s/m2 light, supplied by four Lumichrome F30W-1XX 6500K 98CRI light tubes, at 30 °C. Cell growth was monitored by measuring OD730 with Beckman Coulter DU800 spectrophotometer.
Production of 1-Butanol.
A loopful of S. elongatus PCC 7942 was used to inoculate fresh 50 mL BG-11. 500 mM IPTG was used to induce the growing culture at cell density OD730 nm of 0.4 to 0.6 with 1 mM IPTG as final concentration. 5 mL of growing culture was sampled for cell density and 1-butanol production measurements every 2 d for up to day 8 after which sampling time was switched to every 3 d. After sampling, 5 mL of fresh BG-11 with 500 mM NaHCO3, appropriate antibiotics, and IPTG were added back to the culture. Method for 1-butanol quantification is listed in SI Text.
Enzyme Assays.
Thiolase activity was measured via both condensation and thiolysis direction. The enzymatic reaction was monitored by the increase or decrease of absorbance at 303 nm. Acetoacetyl-CoA synthase activity was measured by monitoring the increase of absorbance at 303 nm, which corresponds to appearance of acetoacetyl-CoA. For details, see SI Text.
Alcohol Production by E. coli Expressing Butyraldehyde Dehydrogenase.
Transformed E. coli strain JCL299 (ΔadhE, ΔldhA, Δfrd, Δpta) expressing different Bldh and the CoA-dependent pathway were cultured in Terrific broth (TB; 12 g tryptone, 24 g yeast extract, 2.31 g KH2PO4, 12.54 g K2HPO4, 4 mL glycerol per liter of water) supplemented with 20 g/L glucose. Fermentation was conducted for 2 d, after which samples were taken for measurement of 1-butanol concentration. For details, see SI Text.
DNA Manipulations.
For details, see SI Text.
Plasmid Constructions.
For details, see SI Text.
Strain Construction and Transformation.
For details, see SI Text.
Protein Purification and SDS/PAGE.
For details, see SI Text.
1-Butanol Quantification.
For details, see SI Text.
Supplementary Material
Acknowledgments.
This research was supported by the Kaiteki Institute and partially supported by National Science Foundation Grant NSF MCB1221392. The authors would like to thank Dr. Hao Luo, Dr. Claire R. Shen, Yasumasa Dekishima, Dr. Hidevaldo Machado, and Dr. Kwang Myung Cho for their valuable support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200074109/-/DCSupplemental.
References
- 1.Papoutsakis ET. Engineering solventogenic clostridia. Curr Opin Biotechnol. 2008;19:420–429. doi: 10.1016/j.copbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Ezeji TC, Qureshi N, Blaschek HP. Bioproduction of butanol from biomass: From genes to bioreactors. Curr Opin Biotechnol. 2007;18:220–227. doi: 10.1016/j.copbio.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 4.Atsumi S, Liao JC. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl Environ Microbiol. 2008;74:7802–7808. doi: 10.1128/AEM.02046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen CR, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab Eng. 2008;10:312–320. doi: 10.1016/j.ymben.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Dekishima Y, Lan EI, Shen CR, Cho KM, Liao JC. Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J Am Chem Soc. 2011;133:11399–11401. doi: 10.1021/ja203814d. [DOI] [PubMed] [Google Scholar]
- 7.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476:355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 8.Zhang K, Sawaya MR, Eisenberg DS, Liao JC. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci USA. 2008;105:20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng. 2010;12:307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Sillers R, Chow A, Tracy B, Papoutsakis ET. Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab Eng. 2008;10:321–332. doi: 10.1016/j.ymben.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Yu MR, Zhang YL, Tang IC, Yang ST. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production. Metab Eng. 2011;13:373–382. doi: 10.1016/j.ymben.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Atsumi S, et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Inui M, et al. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol. 2008;77:1305–1316. doi: 10.1007/s00253-007-1257-5. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen DR, et al. Engineering alternative butanol production platforms in heterologous bacteria. Metab Eng. 2009;11:262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Berezina OV, et al. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl Microbiol Biotechnol. 2010;87:635–646. doi: 10.1007/s00253-010-2480-z. [DOI] [PubMed] [Google Scholar]
- 16.Steen EJ, et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Fact. 2008;7 doi: 10.1186/1475-2859-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boynton ZL, Bennett GN, Rudolph FB. Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, et al. Coupled ferredoxin and crotonyl coenzyme a (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen CR, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol. 2011;77:2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bond-Watts BB, Bellerose RJ, Chang MCY. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7:222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 21.Lan EI, Liao JC. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab Eng. 2011;13:353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 23.Patnaik R, Roof WD, Young RF, Liao JC. Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. J Bacteriol. 1992;174:7527–7532. doi: 10.1128/jb.174.23.7527-7532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao YP, Liao JC. Alteration of growth yield by overexpression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in Escherichia coli. Appl Environ Microbiol. 1993;59:4261–4265. doi: 10.1128/aem.59.12.4261-4265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Causey TB, Zhou S, Shanmugam KT, Ingram LO. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: Homoacetate production. Proc Natl Acad Sci USA. 2003;100:825–832. doi: 10.1073/pnas.0337684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MAG. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern JR, Coon MJ, Delcampillo A. Acetoacetyl coenzyme-a as intermediate in the enzymatic breakdown and synthesis of acetoacetate. J Am Chem Soc. 1953;75:1517–1518. [Google Scholar]
- 29.Okamura E, Tomita T, Sawa R, Nishiyama M, Kuzuyama T. Unprecedented acetoacetyl-coenzyme A synthesizing enzyme of the thiolase superfamily involved in the mevalonate pathway. Proc Natl Acad Sci USA. 2010;107:11265–11270. doi: 10.1073/pnas.1000532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005;42:504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 31.Slater SC, Voige WH, Dennis DE. cloning and expression in Escherichia-coli of the alcaligenes-eutrophus h16 poly-beta-hydroxybutyrate biosynthetic-pathway. J Bacteriol. 1988;170:4431–4436. doi: 10.1128/jb.170.10.4431-4436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukui T, Shiomi N, Doi Y. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J Bacteriol. 1998;180:667–673. doi: 10.1128/jb.180.3.667-673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem. 2008;283:7346–7353. doi: 10.1074/jbc.M708846200. [DOI] [PubMed] [Google Scholar]
- 34.Yan RT, Chen JS. Coenzyme a-acylating aldehyde dehydrogenase from Clostridium-beijerinckii Nrrl-B592. Appl Environ Microbiol. 1990;56:2591–2599. doi: 10.1128/aem.56.9.2591-2599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosaka T, Nakayama S, Nakaya K, Yoshino S, Furukawa K. Characterization of the sol operon in butanol-hyperproducing Clostridium saccharoperbutylacetonicum strain N1-4 and its degeneration mechanism. Biosci Biotechnol Biochem. 2007;71:58–68. doi: 10.1271/bbb.60370. [DOI] [PubMed] [Google Scholar]
- 36.Dexter J, Fu PC. Metabolic engineering of cyanobacteria for ethanol production. Energy Environ Sci. 2009;2:857–864. [Google Scholar]
- 37.Takahama K, Matsuoka M, Nagahama K, Ogawa T. Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J Biosci Bioeng. 2003;95:302–305. doi: 10.1016/s1389-1723(03)80034-8. [DOI] [PubMed] [Google Scholar]
- 38.Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng. 2010;12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Niederholtmeyer H, Wolfstadter BT, Savage DF, Silver PA, Way JC. Engineering cyanobacteria to synthesize and export hydrophilic products. Appl Environ Microbiol. 2010;76:3462–3466. doi: 10.1128/AEM.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan XM, et al. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab Eng. 2011;13:169–176. doi: 10.1016/j.ymben.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Liu XY, Sheng J, Curtiss R. Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci USA. 2011;108:6899–6904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastian S, et al. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab Eng. 2011;13:345–352. doi: 10.1016/j.ymben.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Davis MS, Solbiati J, Cronan JE. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275:28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- 44.Vadali RV, Bennett GN, San KY. Cofactor engineering of intracellular CoA/acetyl-CoA and its effect on metabolic flux redistribution in Escherichia coli. Metab Eng. 2004;6:133–139. doi: 10.1016/j.ymben.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Leonard E, Lim KH, Saw PN, Koffas MAG. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zha WJ, Rubin-Pitel SB, Shao ZY, Zhao HM. Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab Eng. 2009;11:192–198. doi: 10.1016/j.ymben.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Miyahisa I, et al. Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol. 2005;68:498–504. doi: 10.1007/s00253-005-1916-3. [DOI] [PubMed] [Google Scholar]
- 48.Lu XF, Vora H, Khosla C. Overproduction of free fatty acids in E. coli: Implications for biodiesel production. Metab Eng. 2008;10:333–339. doi: 10.1016/j.ymben.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Santos CNS, Koffas M, Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng. 2011;13:392–400. doi: 10.1016/j.ymben.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 50.Bustos SA, Golden SS. Expression of the psbdii gene in synechococcus sp Strain-Pcc-7942 requires sequences downstream of the transcription start site. J Bacteriol. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.