Abstract
Sequence exchange between PMS2 and its pseudogene PMS2CL, embedded in an inverted duplication on chromosome 7p22, has been reported to be an ongoing process that leads to functional PMS2 hybrid alleles containing PMS2- and PMS2CL-specific sequence variants at the 5′-and the 3′-end, respectively. The frequency of PMS2 hybrid alleles, their biological significance, and the mechanisms underlying their formation are largely unknown. Here we show that overall hybrid alleles account for one-third of 384 PMS2 alleles analyzed in individuals of different ethnic backgrounds. Depending on the population, 14–60% of hybrid alleles carry PMS2CL-specific sequences in exons 13–15, the remainder only in exon 15. We show that exons 13–15 hybrid alleles, named H1 hybrid alleles, constitute different haplotypes but trace back to a single ancient intrachromosomal recombination event with crossover. Taking advantage of an ancestral sequence variant specific for all H1 alleles we developed a simple gDNA-based polymerase chain reaction (PCR) assay that can be used to identify H1-allele carriers with high sensitivity and specificity (100 and 99%, respectively). Because H1 hybrid alleles harbor missense variant p.N775S of so far unknown functional significance, we assessed the H1-carrier frequency in 164 colorectal cancer patients. So far, we found no indication that the variant plays a major role with regard to cancer susceptibility.
Keywords: PMS2, PMS2CL, pseudogene, nonhomologous recombination, crossover, gene conversion hybrid allele, paralogous sequence exchange, HNPCC, Lynch Syndrome, DNA mismatch repair
Introduction
The mismatch repair (MMR) gene PMS2 (MIM# 600259), an important caretaker tumor-suppressor gene, forms a heterodimer with MLH1, termed MutLα. Loss of MutLα activity results in MMR deficiency, resulting in accumulation of somatic mutations manifested typically by microsatellite instability. Heterozygous inactivating mutations primarily in the MMR genes, MLH1 and MSH2, cause Lynch syndrome, a dominantly inherited cancer syndrome, predisposing mainly to early-onset colorectal cancer and other tumors such as endometrial cancer in women [Lynch et al., 2009]. The role of PMS2 in Lynch syndrome may have been underestimated in the past [Senter et al., 2008; Truninger et al., 2005]. Furthermore, homozygous or compound heterozygous mutations affecting PMS2 are responsible for the majority of cases of constitutional mismatch repair deficiency (CMMR-D) syndrome, a childhood cancer syndrome characterized by hematological malignancies, brain tumors, colorectal adenomas, and carcinomas, as well as other malignancies [Wimmer and Etzler, 2008]. Counseling and medical care of patients with these hereditary cancer syndromes as well as their families depends on reliable mutation analysis of all four mismatch repair genes.
The PMS2 gene has 15 exons, maps to chromosomal band 7p22, and encompasses ~37 kb of genomic sequence. In the human genome, a partial duplication of the PMS2 gene is located 700 kb proximal to the PMS2 gene. This partial PMS2 duplication spans 16 kb and is part of a larger repeat element of 100 kb. The PMS2 gene duplication is located in inverted orientation compared with the bona fide PMS2 gene. A transcribed pseudocopy of PMS2, termed PMS2CL, results from this duplication and harbors PMS2 exons 9 and 11–15, but lacks exon 10 due to an Alu-mediated 2.4-kb deletion [De Vos et al., 2004]. Sequence exchange between the inverted duplicons during evolution has led to considerable sequence homogenization affecting also the paralogues PMS2 and PMS2CL [Hayward et al., 2007]. As a consequence, the PMS2 reference sequences (NM_000535.5 and NT_007819.17) do not reliably allow distinguishing PMS2 from PMS2CL. Pseudogene specific variants (PSVs) according to the reference sequence are also found in the exonic and intronic regions of the PMS2 gene, where they could represent either allelic variants [Etzler et al., 2008; Hayward et al., 2007] or pathogenic alterations in patients [Auclair et al., 2007]. Vice versa, gene specific variants (GSVs) arealso found in some PMS2CL alleles. We will refer to such PMS2 and PMS2CL alleles, carrying variants derived from their respective paralogous sequence, as hybrid alleles. These hybrid alleles can severely compromise gDNA-based PMS2 mutation analysis in the 3′-region of the gene both by allelic drop-out and pseudogene coamplification.
To circumvent such problems we developed an RNA-based mutation analysis strategy based on the selective amplification of transcripts of the functional PMS2 gene followed by direct sequencing of the resulting reverse transcription-polymerase chain reaction (RT-PCR) products. We have shown that this assay does not only reliably detect mutations in syndromic cancer patients but also allows for the amplification and identification of PMS2 hybrid alleles [Etzler et al., 2008]. Hence, it can be used to assess the frequency and nature of PMS2 hybrid alleles, which have not been systematically studied so far. Using a similar RNA-based strategy Hayward et al. [2007] found PMS2 hybrid alleles with PSVs in the exons 13–15 in 3 of 16 individuals. Taken together with the small number of cases of our previous study, we estimated a PMS2-hybrid-allele frequency of ~0.1. A comparable frequency can be deduced from PSVs found in 41 patients with suspected Lynch syndrome who were analyzed for PMS2 mutations by a nested PCR strategy that uses gene-specific primers for the long-range PCR reaction [Clendenning et al., 2006]. However, a higher frequency of hybrid alleles is expected when considering MLPA dosage alterations that were found in at least 15% of individuals analyzed by Hayward et al. [2007].
Principally, two processes associated with recombination, that is, gene conversion or reciprocal crossover, can cause sequence exchanges between the paralogues leading to formation of hybrid alleles. Both processes are initiated by a double-strand break and are the different outcomes of double-strand break repair [Chen et al., 2007]. Intra- or interchromosomal gene conversion, that is, the nonreciprocal sequence transfer from a donor to an acceptor, would lead to exchange of small sequence tracts of usually ~55–290 nucleotides [Jeffreys and May, 2004]. Such an event has been postulated to be responsible for the occurrence of two adjacent PSVs in exon 11 that lead to a pathogenic PMS2 allele [Auclair et al., 2007]. Intrachromosomal recombination with crossover between the duplicons would lead to reciprocal exchange of the entire sequences downstream of the breakpoint and to inversion of the intervening sequence that has been observed as polymorphic microinversion in at least 5% of normal chromosomes [Feuk et al., 2005; Szamalek et al., 2006]. However, so far it has not been shown whether the latter mechanism accounts also for the formation of PMS2 and PMS2CL hybrid alleles. Therefore, this study is aimed at assessing the frequency of PMS2 hybrid alleles and determining the recombination mechanisms that have formed them. We were particularly interested in hybrid alleles with pseudogene-derived exon 14 sequences since they harbor the pseudogene specific amino acid substitution p.N775S with possible biological significance. Clendenning et al. [2006] found the PSV p.N775S in 7/41 (17%) of colorectal cancer patients with PMS2-expression loss in the tumor. Here we assessed the frequency of PMS2 hybrid alleles with this missense variant in sporadic colorectal cancer patients and compared it to a control cohort.
Material and Methods
Individuals Analyzed
Forty-two individuals of European origin and further 150 individuals, comprising each 50 individuals of Caucasian, Hispanic, and African-American background were included in the study. Additionally, 22 PMS2-deficient and 142 sporadic colorectal cancer patients and 267 unrelated healthy control individuals were included. The project was approved by the ethics review board of the Medical University Vienna.
Reference Sequences and Alignments
Single nucleotide polymorphisms (SNPs) distinguishing paralogous and orthologous sequences refer to the human and chimpanzee (Pan troglodytes) gene and pseudogene reference sequences as published by Hayward et al. [2007]. All sequence variants are described in accordance with the recommendations of the human genome variation society (http://www.hgvs.org/mutnomen/). The reference sequence used for the human PMS2 mRNA is NM_000535.5 and for the PMS2 intron 12 NT_007819.17. Sequence alignments were performed with the programs SeqScape v2.5 (Applied Biosystems, Foster City, CA) and Clustal W v1.83 [Thompson et al., 1994].
Genotype Analysis of PMS2 and PMS2CL Transcripts in Exons 11 to 15
cDNA sequencing of PMS2 and PMS2CL transcripts was performed in exons 11 to 15. PMS2 RT-PCR conditions have been published previously [Etzler et al., 2008]. PMS2CL transcripts were amplified with the unspecific reverse primer (PMS2B_R) together with a primer (PMS2CLB_F) spanning the 3′ border of exon 9 and the 5′ border of exon 11. The PMS2 and PMS2CL RT-PCR products were sequenced with nested primers PMS2A_R, PMS2_6, PMS2_7 located in exon 11 and PMS2_8 spanning the exon 12 and 13 border. Primer sequences are listed in Supp. Table S1.
Haplotype Analysis for the Breakpoint Characterization of PMS2– PMS2CL Sequence Exchange in Three H1-Hybrid-Allele Carriers
Breakpoint analysis was initially performed in three Caucasian H1-hybrid-allele carriers. The primers LRPCR4_Fwd (located in intron 12) and PMS2c2253G_R (specific for PSV c.2253C) were used to amplify a 2,820-bp fragment expected to contain the 5′ breakpoint. Primer pair PMS2in12_PDS2CF und PMS2c2253A_R was used to amplify the same region on the presumed reciprocal H1 product on PMS2CL. The 3′-breakpoint region was amplified as a 1,792-bp fragment using the primers PMS2_*17CF and LRPCR4_Rev. All sequencing primers used are listed in Supp. Table S1.
Development of an H1 Hybrid Allele-Specific Multiplex PCR
The specificity of possible marker SNPs for the H1 hybrid allele in intron 12 was tested in 42 individuals of European origin with primers specific for the orthologue-discriminating SNPs c.21741+1857G>A (ODS1) and c.2175–1739A>G (ODS2). PMS2in12_ODS1AF together with primers PMS2in12_8F and PMS2in12_8R were used in a multiplex PCR to test for the presence of ODS1. ODS2 which was present in all H1 hybrid allele carriers as well as in an additional 10 of 42 individuals was specifically amplified with primer PMS2in12_ODS2GR. To directly allocate ODS2 to the gene or the pseudogene this primer was used together with PMS2in12_PDS2GF and PMS2in12_PDS2CF, which are gene and pseudogene specific, respectively.
The specificity of SNP c.*17G>C for the H1 hybrid allele was tested by an allele-specific multiplex PCR with primer PMS2_*17CF and the universal primers PMS2_15_1F and PMS2B_R. The variants ODS1, ODS2, and c.*17G>C were used to develop an H1 allele-specific multiplex assay. Primers PMS2_17CF and PMS2B_R generated a c.*17G>C-specific PCR product of 164 bp, the primers PMS2in12_ODS1AF and PMS2 in 12_ODS2GR were used to amplify an ODS1-specific PCR product of 573 bp. An unspecific PCR product of 696 bp was amplified with the primers PMS2in14_1F and PMS2B_R as internal control.
For all PCR reactions standard conditions with Taq DNA Polymerase of New England Biolabs (Frankfurt, Germany) or Invitrogen (Carlsbad, CA) were used. For sequencing, PCR products were purified with the ExoSAP-IT PCR clean-up kit (GE Healthcare, Vienna, Austria) and subsequent sequencing was performed using the BigDyeTerminator V3.1Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Primersequences used for PCR reactions and sequencing are listed in Supp. Table S1.
Interphase FISH Analysis for the Analysis of Inversion
FISH analysis was performed with genomic clones as previously described [Szamalek et al., 2006]. In brief, 1 μg of the DNA of BAC RP11-425P5 and BAC RP11-1275H24 was labeled with biotin-16-dUTP (Roche-Diagnostics, Mannheim, Germany), whereas 1 μg of PAC RP4-810E6 was labeled with digoxygenin-11-dUTP (Roche-Diagnostics). The cohybridizations were detected with antibodies coupled with Texas-red (Dianova, Hamburg, Germany) as well as FITC-avidin and biotinlyated antiavidin (Vector, Burlingame, CA). Approximately 50 interphase nuclei from each individual were investigated.
Analysis of H1-Hybrid-Allele Haplotypes in IVS12
Haplotypes of all 38 H1 hybrid alleles were determined around the breakpoint of PMS2– PMS2CL sequence exchange by sequencing PCR products generated with the ODS1-specific forward primer (PMS2in12_ODS1AF) and a reverse primer specific for the PSV c.2253C (PMS2c2253G_R). The PCR-product that covers 2393 bp of IVS12 was amplified using Taq DNA Polymerase from Invitrogen (Carlsbad, CA) and standard conditions. Sequencing primers covering almost the entire sequence (2277 bp) are listed in Supp. Table S1.
Phylogenetic Network Analysis and Statistics
Phylogenetic network analysis was performed by means of the median-joining-network method [Bandelt et al., 1999] using Network 4.5.1.0. available at www.fluxus-technology.com.
The chi-squared test was used to compare allele frequencies in patients and controls.
Results
Genotyping of 42 Individuals of European Origin Identifies at Least Three Different PMS2 Hybrid Alleles
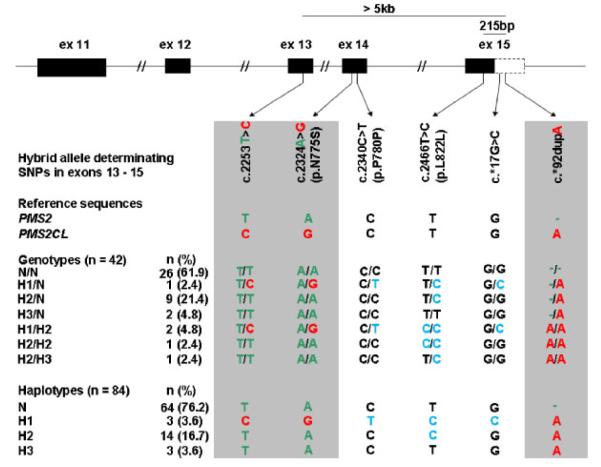
To determine the frequency of PMS2– PMS2CL hybrid alleles we first genotyped a cohort of 42 individuals of European origin for PMS2 exons 11 to 15 by sequencing of PMS2 transcripts. PSVs were found in PMS2 transcripts of 16 individuals either in the heterozygous or homozygous state (Fig. 1). Thirteen individuals harbored only the PSV c.*92dupA in exon 15. Three individuals carried PSVs (c.2253T>C, c.2324A>G and c.*92dupA) in PMS2 exons 13, 14, and 15. These three individuals also carried c.2340C>T in exon 14 and c.*17G>C in exon 15 not reported by Hayward et al. [2007] as paralogue-discriminating SNPs (PDSs). Because these two latter variants were observed only in these three individuals they were allocated to the hybrid alleles. Furthermore, variant c.2466T>C in exon 15 was present once in a heterozygous and twice in a homozygous state in these three individuals. It was also observed in 11 individuals carrying only PSV c.*92dupA, but in none of the 26 individuals without PSVs. Thus, c.2466T>C was also allocated to the hybrid alleles. No individual was found with PSVs in exon 11. Three different hybrid-allele haplotypes, termed H1 to H3, can be deduced from the six genotypes observed in the 16 individuals (Fig. 1). Hybrid allele H1 harbors PSVs in all three exons 13, 14, and 15. Alleles H2 and H3 reveal PSVs only in exon 15 and the variant c.2466C>T distinguishes both alleles. In total, 20 of 84 (24%) analyzed PMS2 alleles were hybrid alleles. The deduced allele frequencies of 0.04 for H1 and H3 alleles and 0.17 for H2 alleles were in Hardy-Weinberg equilibrium with the observed genotypes.
Figure 1.
Genotypes of hybrid allele-determinating SNPs in exons 13 to 15 observed with cDNA sequencing in 42 European individuals and inferred haplotypes. In the upper part of the figure the location of the SNPs in the PMS2 gene map is shown. PDSs are indicated with gray underlay. The gene-specific variants are given in green and the pseudogene-specific variants in red letters. Variants c.2340C>T, c.2466T>C, and c.*17G>C are indicated in blue. Six different genotypes containing pseudogene-specific variants were found. Three different hybrid-allele haplotypes were deduced and named H1, H2, and H3. Alleles containing only gene-specific variants were named N ( = normal). The number of observed genotypes and alleles (n) and their frequency (%) is given. In H1 alleles, pseudogene-derived sequence variants were observed in a region that exceeds 5 kb at genomic DNA level, while they were detected within a 215-bp region in H2 and H3 alleles.
Reciprocal Hybrid Pseudogenes Indicate Intrachromosomal Recombination with Crossover as the Underlying Cause of the H1 Hybrid Allele
Hybrid allele H1 contains PSVs in the consecutive exons 13, 14, and 15. Together with the three additional variants c.2340C>T, c.2466T>C, and c.*17G>C, these PSVs characterize a haplotype that is likely to result from the insertion of more than 5 kb PMS2CL-derived sequence into PMS2. Because this 5-kb segment exceeds the usual length of sequences that are introduced into acceptor sequences by gene conversion, we assumed that intrachromosomal recombination with crossover may be the underlying cause of this hybrid-allele haplotype. This mechanism would also lead to the reciprocal sequence exchange in the donor sequence, in this case the PMS2CL pseudogene (Fig. 2). To test this possibility we specifically amplified PMS2CL derived transcripts in exons 11 to 15 from three individuals carrying heterozygous H1 alleles. Sequencing of the RT-PCR products showed that all three individuals carried the GSVs at positions c.2253 and c.2324 in exons 13 and 14 in a heterozygous state and two of three individuals also at position c.*92 in exon 15. This result strongly suggests that the PMS2CL hybrid alleles reciprocal to the H1 PMS2 hybrid alleles exist and that recombination with crossover is responsible for the formation of the H1 hybrid allele.
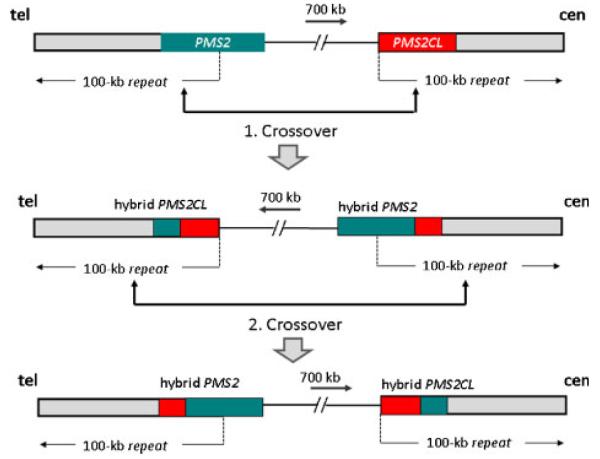
Figure 2.
Scheme showing PMS2– PMS2CL sequence exchange due to intrachromosomal recombination with crossover. The normal localization of the functional PMS2 gene and the PMS2CL pseudogene on chromosomal band 7p22 is shown in the top panel. The gray bar indicates the 100-kb repeat element duplicated in inverse orientation 700-kb centromeric. The dotted line indicates the start of the repeats and the arrow their orientation. The green bar corresponds to the PMS2 gene and the red bar to the PMS2CL pseudogene. The C-terminal part of the PMS2 gene is situated in the telomeric repeat. The PMS2CL pseudogene is located in the inverse orientation in the centromeric repeat. Recombination with crossing over would lead to the formation of the PMS2 hybrid allele, the reciprocal PMS2CL pseudogene hybrid allele, and inversion of the intervening sequence (middle panel). A second crossover occurring in the more central part of the duplicons either subsequently to or simultaneously with the first event would lead to the normal orientation of the duplicon with the hybrid alleles (bottom panel).
Intrachromosomal recombination with crossover in the inverted duplicon would lead to inversion of the intervening sequence. We tested this in our three European individuals using interphase FISH. The FISH probe order in interphase nuclei of the individuals was determined as being not inverted according to previous analysis performed with these probes in 11 human control individuals and different primate species [Szamalek et al., 2006] (data not shown). Hence, we conclude that the 700-kb sequence interval located between the 100-kb duplicons is not inverted in these three individuals. Therefore, the most likely explanation for the PSV pattern of hybrid allele H1 in these individuals would be a second crossover event in the more central part of the duplicon (Fig. 2) occurring either simultaneously with or subsequently to the first event.
Characterization of the Breakpoints of PMS2– PMS2CL Sequence Exchange Identifies SNPs Highly Specific for the H1 Allele
Because the H1 hybrid allele likely results from two recombination events with crossover, we aimed at characterizing the breakpoints of these PMS2– PMS2CL sequence exchanges. Moreover, by characterizing the breakpoints of crossover we aimed to develop a simple assay to test for the presence of the H1 hybrid allele harboring the pseudogene-derived missense variant p.N775S.
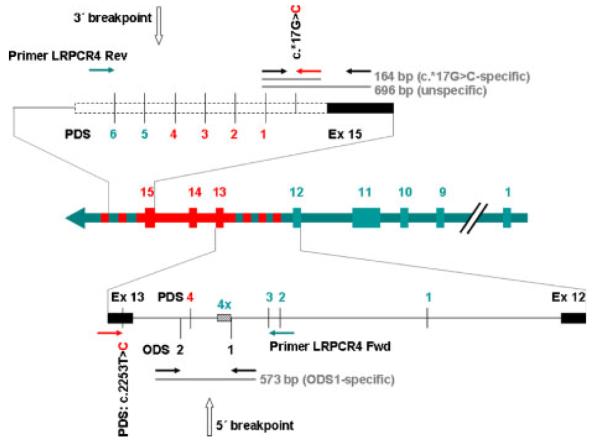
First we developed H1-allele-specific PCR reactions spanning the breakpoint regions of PMS2– PMS2CL sequence exchange with primers specific for the PMS2 sequence at one end and for the PMS2CL sequence at the other end. Such primer pairs are expected to generate products only from H1 hybrid alleles. Five patients analyzed by Clendenning et al. [2006] performing their gene-specific long-range nested PCR-approach obviously carried H1 hybrid alleles (lack of variant c.2253T>C in these alleles is explained by allelic drop-out due to stringent gene-specific nested primers for exon 13). This indicated that H1 hybrid alleles are gene-specific at the PMS2-specific long-range forward primer LRPCR4_Fwd in intron 12. Consequently, the 5′ breakpoint of sequence exchange was expected between this primer site and the PSV c.2253C in exon 13. Therefore, we used a primer specific for c.2253C and LRPCR4_Fwd to amplify the breakpoint-containing sequence in intron 12 (Fig. 3). This 2,820-bp region harbors 27 PDSs, 26 of which were analyzed (Supp. Fig. S1) and a 24-bp minisatellite repeated four and eight times in the gene and pseudogene reference sequences, respectively. All three H1 alleles investigated shared the same haplotype in which the seven first PDSs at the 5′-end of this intronic region as well as the paralogue-discriminating 24-bp repeat were gene specific and the analyzed 19 PDSs at the 3′-end with one exception (PDSxiv, Supp. Fig. S1) carried the pseudogene-specific variants. Thus, the crossover breakpoint is located in an Alu sequence between the 24-bp minisatellite and PDS c.2175–1802T>C. This PDS was named PDS4 because it is the fourth PDS of intron 12 that distinguishes also the pan troglodytes (pt) paralogues and, hence, predates the separation of the lineages (Fig. 3 and Supp. Fig. S1).
Figure 3.
Scheme of the breakpoints of sequence crossover mapping into intron 12 and the 3′ UTR of the H1 hybrid allele. The gene map of the H1 hybrid allele is shown in the middle panel. The arrow indicates the orientation. Exons are shown as boxes and introns as lines between the boxes. Exons with gene-derived SNPs are depicted in green and exons with pseudogene-derived sequences in red. The scheme of the 3′ breakpoint region is enlarged in the top panel and the scheme of the 5′ breakpoint in the bottom panel. Primers used to amplify PCR products spanning the breakpoints of H1 hybrid alleles are indicated by horizontal arrows. Green arrows indicate gene-specific, red arrows pseudogene-specific, and black arrows unspecific primers. The gene-specific primer LRPCR4_Rev was used together with the specific primer PMS2_*17CF to amplify a 1,792-bp product spanning the 3′ breakpoint. The gene-specific primer LRPCR4_Fwd was used together with the pseudogene-specific primer PMS2c2253G_R to amplify a 2,820-bp product spanning the 5′ breakpoint. Paralogue-discriminating SNPs (PDS) and orthologue-discriminating SNPs (ODS) are numbered as in Supp. Table S3. A green number indicates the gene-specific and a red number the pseudogene-specific variant at the respective PDS. A 24-bp minisatellite (shaded square) is repeated four times in H1 hybrid alleles that corresponds to the reference sequence of the PMS2 gene. The breakpoints are indicated by vertical arrows. PCR products generated in the H1 hybrid allele-specific multiplex PCR are indicated as gray horizontal lines and their size is given.
Among the sequence variants determining the haplotype of the H1 hybrid allele in intron 12 were two variants, that is, c.21741 1857G>A and c.2175–1739A>G, which were unexpected in the human sequence, because they were described as chimpanzee specific at orthologous-discriminating SNPs (ODSs). To test whether these variants denoted ODS1 and ODS2 (Fig. 3 and Supp. Fig. S1) could serve as marker SNPs specific for the H1 hybrid allele we assessed their presence in all 42 individuals of our cohort. Although ODS2 was not only associated with the H1 haplotype, but also with one other gene- and one pseudogene-specific haplotype present in one and nine individuals, respectively, the variant ODS1 was exclusively associated with the H1 haplotype (data not shown). We, therefore, concluded that ODS1 can serve as a surrogate marker for H1 alleles.
To determine the 3′ breakpoint of sequence exchange, anticipated because of the normal orientation of the duplicon intervening 700-kb sequence in the three H1-allele carriers, a similar strategy was applied. The 3′-breakpoint was expected between variant c.*17G>C that, similar to ODS1, was highly specific for the H1 allele (data not shown) and c.*160+1603 the site of the gene-specific long-range reverse PCR primer (LRPCR4_Rev) used by Clendenning et al. [2006]. Therefore, we used LRPCR4_Rev together with a primer specific for variant c.*17G>C to amplify the H1 allele around the 3′ breakpoint of PMS2– PMS2CL sequence crossover. Sequence analysis of the six PDSs within the resulting PCR product showed in all three alleles the same haplotype and the 3′ breakpoint mapped between the PDSs c.*160+489_*160+490TG>CA and c.*160+979C>T, named PDS4 and PDS5 in Figure 3, which carry the pseudogene and gene specific variant, respectively.
Evaluation of a Multiplex PCR-Assay for the H1 Hybrid Allele in Individuals from Different Ethnic Backgrounds Reveals H1 Hybrid Allele-Diversity
Because the H1 hybrid allele carried the pseudogene-derived missense variant p.N775S of so far unknown biological significance, we aimed at developing a simple gDNA-based assay that could test for the presence of H1 alleles in retrospective patient cohorts. To this end we used SNPs ODS1 (c.2174+1857G>A) and c.*17G>C located close to the 5′- and 3′-breakpoint, respectively, and highly diagnostic for H1 to develop an H1-allele specific multiplex assay (Fig. 3). This assay was 100% specific and sensitive in our cohort of 42 Caucasian control individuals (data not shown). To evaluate the sensitivity and specificity of this assay in a larger cohort of control individuals of different ethnic backgrounds we analyzed further 150 control individuals of Caucasian (n = 50), Hispanic (n = 50) and African-American (n = 50) background. The results of the multiplex PCR were compared with the PMS2 genotypes of these individuals as determined by cDNA sequencing (Supp. Table S2). Thirty-three of 150 individuals were positive for the ODS1-specific PCR product. All but one of the ODS1-positive individuals carried PSVs in exons 13 and 14 including the missense variant p.N775S. Combined with the results in the 42 Caucasian individuals of our initial cohort, ODS1-specific PCR products were found in all 35 individuals carrying PSV p.N775S but in only 1 of 157 individuals without PSVs in PMS2 exons 13 and 14. Hence, we deduce that ODS1 may serve as surrogate marker for H1 hybrid alleles with the PSV p.N775S in the functional gene. Our multiplex PCR assay detects carriers with a sensitivity of 100% and a specificity of 99%.
The H1 hybrid allele frequency in the 150 individuals with different ethnic backgrounds was deduced from their genotypes and the multiplex assay results. It was higher in African-Americans (0.25) than in Caucasians (0.06) and Hispanics (0.04). H1 hybrid alleles in African-Americans showed also a higher diversity with respect to the presence of three variants c.2340C>T, c.2466T>C, and c.*17G>C that were found in almost all Caucasian H1 alleles (Supp. Table S2). Therefore, only a proportion of H1 alleles derived from individuals of African descent were positive for variant c.*17G>C in the multiplex assay (examples can be seen in Fig. 4). In contrast to H1, the H2-allele frequency was lowest in African-Americans (0.06) and substantially higher in Caucasians (0.18) and Hispanics (0.16). The H3 allele was found with allele frequencies of 0.03, 0.09, and 0.10 in Caucasians, Hispanics, and African-Americans, respectively.
Figure 4.
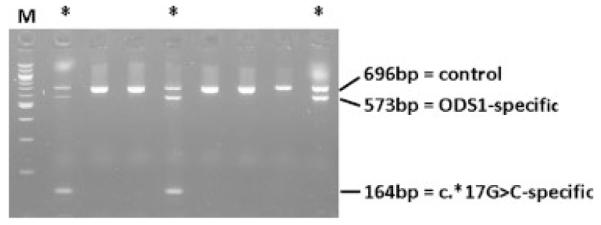
H1 hybrid allele-specific multiplex PCR assay. PCR products generated from genomic DNA of eight individuals by the multiplex PCR assay are shown. Besides the unspecific PCR product of 696 bp present in all individuals, lanes marked with a star show the ODS1-specific band of 573 bp, indicating the presence of an H1 hybrid allele and in two lanes also the c.*17G>C-specific band of 164 bp. M = 100 bp DNA ladder.
The Haplotypes of Different H1 Hybrid Allele Subtypes are Consistent with a Single Recombination Event with Crossover
In total, we found 35 individuals carrying 38 H1 hybrid alleles. Because all 38 H1 hybrid alleles harbor PSVs in exons 13 and 14 and are positive for variant ODS1 we assumed that, although showing differences particularly within exon 15, they trace back to a single recombination event, which caused the PMS2– PMS2CL crossover. This assumption is strongly supported by the results of the haplotype analysis in intron 12. All alleles showed the same pattern of gene and pseudogene-specific variants at 21 of 22 analyzed paralogue-discriminating sites within the ~2.4 kb analyzed (Supp. Table S3). Furthermore, all H1 hybrid alleles carried the variant ODS2 and a T at the CpG-dinucleotide c.2175–1786.
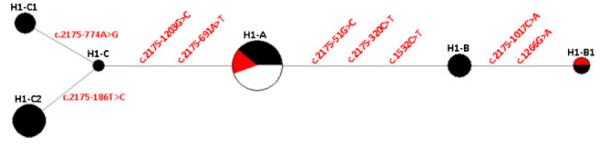
Seven variant SNPs (SNP2–SNP8 in Supp. Table S3 and Supp. Fig. S1) were found in the intronic sequences of the analyzed alleles. These variants were used to group the 38 characterized H1 alleles into six different H1 subhaplotypes (Supp. Table S3). The H1 subhaplotype without any of the variant SNPs was defined as H1-A. The remainder alleles fall into two main subhaplotypes denoted H1-B and H1-C, which can further be subdivided into H1-B and H1-B1 as well as H1-C, H1-C1, and H1-C2. Although the H1-A subhaplotype was present at nearly equal frequencies in Caucasians, Hispanics, and African-Americans, subhaplotypes H1-B and H1-C were found only in individuals of African descent. These latter subhaplotypes accounted for the substantially higher frequency of H1 haplotypes in African-Americans (Table 1). A phylogenetic network of the eight defined H1 subhaplotypes produced by split decomposition showed a fully tree-like structure (Fig. 5), which is in agreement with the assumption that all H1 subhaplotypes have evolved from a common ancestor that resulted from a single recombination event with crossover. Furthermore, it is also evident that subhaplotype H1-A present in all three ethnic groups predates the dispersal of modern humans out of Africa approximately 100,000 years ago.
Table 1.
Hybrid Allele Frequencies in Three Ethnic Groups
| Hybrid allele |
Hybrid allele frequencies |
|||
|---|---|---|---|---|
| H1 subhaplotypes | C (n=92) | H (n=50) | A (n=50) | Total |
| H1-A | 9 | 3 | 8 | 20 (0.05) |
| H1-B | 0 | 1 | 5 | 6 (0.02) |
| H1-C | 0 | 0 | 12 | 12 (0.03) |
| H1 total | 9 (0.05) | 4 (0.04) | 25 (0.25) | 38 (0.10) |
| H2 | 33 | 16 | 6 | 55 (0.14) |
| H3 | 6 | 9 | 10 | 25 (0.07) |
| H1-H3 total | 48 (0.26) | 29 (0.29) | 41 (0.41) | 118 (0.31) |
H1-hybrid-allele subhaplotypes are individually indicated. A = Afro-American; C = Caucasian; H = Hispanic; n = number of individuals analyzed. Seven alleles with PDSs in exon 11 only are not included in this table.
Figure 5.
Phylogenetic network analysis of H1-hybrid-allele subhaplotypes. Genealogical relationship of six PMS2-hybrid-allele haplotypes reconstructed through median-joining network. Each node represents a different PMS2-hybrid-allele haplotype with the size of the circle proportional to the haplotype frequency. Circles are color coded according to ethnic background of the individuals carrying the haplotype (black: African-Americans, red or gray: Hispanics, white: Caucasians). The differences between the H1 hybrid allele subhaplotypes listed also in Supp. Table S3 are indicated between the haplotypes.
Analysis of H1-Hybrid-Allele Frequency in Colorectal Cancer Patients Renders no Evidence for a Strong Role of PSV p.N775S in Cancer Susceptibility
All H1-hybrid-allele haplotypes carry PSV c.2324A>G in exon 14 that replaces Serine for Asparagine at residue 775 (p.N775S), which is conserved down to zebrafish and mosquito. Although this missense alteration is unlikely to play a role as pathogenic mutation because it was also found in patients with clearly truncating PMS2 mutations [Clendenning et al., 2006], it may still have some impact on the function of the resulting gene product. It is possible that it modulates cancer susceptibility in carriers. In a first attempt to test a possible role of p.N775S as cancer susceptibility allele, we assayed the frequency of H1 hybrid allele-carriers in a cohort (n = 142) of sporadic European colorectal cancer patients and small group of patients with isolated PMS2 loss in the tumor (n = 22) using our H1 hybrid allele-specific multiplex assay. We found no significant difference in the frequency with a similarly sized cohort of age and sex-matched controls (Supp. Table S4) that would provide evidence for a role of PSV p.N775S as a major cancer susceptibility variant.
Discussion
In this study we genotyped 192 individuals from different ethnic backgrounds in exons 11 to 15 of the PMS2 gene. Within the exonic sequences of the functional gene, 103 of 192 (54%) individuals carried, in a heterozygous or homozygous state, variants that are considered PSVs according to the reference sequences of PMS2 and PMS2CL [Hayward et al., 2007]. Variant c.1621A>G (p.K541E) in exon 11 was not taken into account for this calculation. According to Hayward et al. [2007], A at position c.1621 is considered gene specific, whereas G pseudogene specific. However, c.1621G is the ancestral (present also in the chimpanzee gene and pseudogene, which serve as out-groups) and more frequent variant also in the PMS2 gene of all ethnic groups. Therefore, we consider c.1621A occurring with an overall frequency of 0.18 a SNP in PMS2 and list this as c.1621G>A (p.E541K) in Supp. Table S2.
For simplicity we name PMS2 alleles with PSVs PMS2 hybrid alleles. Deduction of haplotypes from genotypes leads to an estimated overall PMS2-hybrid-allele frequency of 125 of 384 (33%). The majority (80 of 125 = 64%) of hybrid alleles carried PSVs only in the terminal exon 15. 38/125 (30%) of the deduced haplotypes contained PSVs in the exons 13 and 14 and 36/38 also in exon 15. These alleles were named H1 alleles. PMS2 exons 11 with PSVs were present in seven individuals of African-American background. We made no attempt to group these PSVs into haplotypes. Five of the seven individuals carried variant c.1488C>T and one variant c.1437C>G in a heterozygous state. One individual was heterozygous for both these variants, which were located in trans (data not shown). Because both PSVs represent the ancestral nucleotide also present in the chimpanzee gene and pseudogene that serve as out-groups, it is likely that they represent remainders of the common ancestor of the human PMS2 and PMS2CL sequences in an ancient PMS2 haplotype.
Depending on the population, 14% (Hispanics) to 60% (African-Americans) of the hybrid alleles were H1 alleles. We present here strong evidence that all H1 hybrid alleles trace back to a single crossover event:
The haplotypes of 38 H1 hybrid alleles show in intron 12 the same pattern of gene specific variants (GSVs) and PSVs with a distinct switch from gene to pseudogene-specific indicating that they all carry the same breakpoint of sequence exchange. Notably, there is a 24-bp minisatellite repeat close to the assumed breakpoint of sequence crossover. Such elements have been reported to be frequently associated with nonhomologous recombination hotspots [Lupski, 2004]. The only PDS that showed differences among the subhaplotypes was c.2175–691A>T (PDSxv in Supp. Table S3). All H1-C subhaplotypes carried the evolutionary new GSV, whereas the remaining subhaplotypes were pseudogene specific at this site. It is possible that the occurrence of the GSV in the otherwise pseudogene-derived sequence tract results from a recurrent mutation. Alternatively, the GSV may have been reintroduced by gene conversion into the H1-C subhaplotype. This would suggest that the diversity of the H1 hybrid allele partly results from gene conversion that occurred after the allele-forming crossover event and would represent an example of how gene conversion and recombination with crossover together lead to sequence homogenization of the paralogues. A similar mechanism may be at play for c.2340C>T and c.2466T>C in exons 14 and 15, respectively.
All H1-allele haplotypes harbored two variants at orthologue-discriminating SNPs that are chimpanzee specific according to the reference sequences. These two variants, ODS1 and ODS2, are also found in the PMS2 gene of the out-groups, gorilla and orangutan, indicating that these variants are the ancestral variants. Hence, it is likely that ODS2 and ODS1 are remainders of the ancestor sequence that can still be found in some ancient human alleles. The nearly exclusive occurrence of ODS1 in all H1-hybrid-allele subhaplotypes further corroborates their common origin. We have also sequenced intron 12 of the PMS2 allele giving rise to the only false positive ODS1-specific product in the multiplex PCR assay. Interestingly, this sequence carried two additional variants (c.2175–426_2175–429delCTCC, c.2175–407A>C) that appeared to be remainders of the common ancestor of man and chimpanzee, as well. It appeared to have the same breakpoint of PMS2 and PMS2CL sequence exchange as the H1 hybrid allele with four copies of the 24-bp minisatellite repeat and the PSV at the paralogue-discriminating SNP 4 (PDS4) (see individual H17 in Supp. Table S3). However, this haplotype contained the GSV c.2175–691A present also in the H1-C subhaplotype as well as the GSVs c.2175-969_2175-970AA, c.2175-947G, c.2175-926G, c.2175-836G, and c.2175-823G. Hence, there are striking similarities of the latter allele and the H1 allele, but currently we cannot fully deduce their relationship.
The detection of a PMS2CL hybrid allele that correlated to the presumptive H1 reciprocal product on PMS2CL renders further evidence for the assumed crossover event that formed the H1 hybrid allele. In all three H1 hybrid allele-carriers of our cohort of 42 Caucasians this PMS2CL hybrid allele was present. In one of these individuals (Ci) it was possible to specifically amplify the intronic region containing the presumptive reciprocal 5′ breakpoint of sequence exchange on PMS2CL by use of a primer pair specific for the pseudogene-specific variant at PDS2 in intron 12 and the gene-specific one at position c.2253 in exon 13. Sequence analysis of the resulting PCR-product showed that 6/7 PDSs before the 24-bp repeat element carried the pseudogene specific variant, whereas 14 of 17 analyzed PDSs after the repeat including PDS4 were gene specific indicating a breakpoint in the anticipated intron 12 region. In contrast to the expected eight copies present in the pseudogene reference sequence, the 24-bp element was repeated 10 times in this sequence (data not shown). Furthermore, the sequence also contained the ancestral variant at ODS2. Interestingly, 10 copies of the 24-bp repeat element were also found in the pseudogene-specific haplotype amplified from nine individuals with the ODS2-specific primer together with the pseudogene-specific primer at PDS2. cDNA sequencing of the PMS2CL transcripts of five of these individuals showed that all carried the presumptive H1-reciprocal PMS2CL allele. Subsequent haplotype analysis between PDS2 in intron 12 and c.2253 in exon 13 showed in all five individuals the identical PMS2CL hybrid allele haplotype present also in individual Ci (data not shown). Taken together, these data strongly suggest that a H1-reciprocal PMS2CL allele exists. Furthermore, they show that H1 PMS2 and PMS2CL hybrid alleles can occur separately on different chromosomes, because none of the five individuals carried the H1 PMS2.
Finally, a phylogenetic network analysis of the H1 subhaplotypes is in agreement with the evolution of all observed H1 types from one common ancestor. Although the subhaplotype-discriminating SNPs in the exonic regions are deduced from the genotypes, all H1 alleles were haplotyped in intron 12. Taking into consideration only the 2.3-kb sequence properly haplotyped, the H1 subtypes fall into three major haplotypes (Supp. Table S3 and Table 1). The SNPs specific for H1-C, that is, c.2175–1203G>C (SNP2) and c.2175–691A>T (PDSxv), are listed in the SNP databases (www.ncbi.nlm.nih.gov/SNP/) as polymorphic in the gene and/or in the pseudogene. Hence, these two variants may have been introduced into a presumed ancestor H1 allele by gene conversion events (see also above). The tagging SNPs of the subhaplotype H1-B, c.2175–320C>T (SNP6) and c.2175–51G>C (SNP8), are not listed in the SNP databases and are likely to result from replication errors rather than from recombination events. Together with three other SNPs (c.2175–1017C>A 5 SNP3, c.2175–774A>G 5 SNP4, c.2175–186T>C = SNP7) they sum up to a minimum of five sites within the 2,277 analyzed basepairs that have been mutated since the formation of the anticipated ancestor H1 allele that originates in Africa before the dispersal of modern humans to Europe and Asia. Interestingly, no exclusively non-African hybrid allele has evolved afterward, indicating that this genomic region is rather stable as also supported by the stability of the repeat number of the 24-bp minisatellite on the hybrid allele.
The distribution of hybrid-allele haplotypes shows considerable differences among ethnic groups. H1 hybrid alleles were four to five times more frequent in African-Americans than in Caucasians and Hispanics, which accounted also for the overall higher frequency of hybrid alleles in African-Americans. On the other hand, Caucasians and Hispanics had a higher frequency of the H2 and H3 haplotypes. Particularly, H2 is three times more frequent in Caucasians than in African-Americans (0.18 vs. 0.06). Although H2 and H3 hybrid alleles appear by cDNA sequencing as two unique hybrid alleles, they may represent the outcome of recurrent gene conversion or crossover events between different PMS2 and PMS2CL alleles. Yet, it is striking that the H2 hybrid allele characterized by the presence of PSV c.*92dupA together with variant c.2466T>C was not found in any of the patients studied by Clendenning et al. [2006]. The most likely explanation for this observation would be allelic-drop out of a unique hybrid allele that carries pseudogene-derived sequence variants at one or both sites of the gene-specific long-range PCR primer used by Clendenning et al. [2006]. Allelic drop-out of this frequent variant would also explain the overall low hybrid-allele frequency in their patient cohort (i.e., 0.1 vs. 0.27 in Caucasians and 0.41 in African-Americans as determined in this study).
Taken together, we found that depending on the population at least 14–60% or, if we consider only exons 13 and 14, even up to nearly 100% of pseudogene-derived sequences found within the exonic regions of PMS2 alleles are of the H1 hybrid allele haplotype, and consequently, can be traced back to one ancient intrachromosomal crossover event exchanging the sequences of the paralogues.
During this study we have developed a simple gDNA-based PCR assay that tests for the presence of H1 hybrid alleles with a sensitivity of 100% and a specificity of 99%. The missense variant p.N775S present on the H1 hybrid allele is of so far unknown biological significance, and could play a role as a cancer susceptibility allele. We used our assay to assess the frequency of H1 hybrids in colorectal cancer patients and a control cohort, but without significant differences between the cohorts. However, these results have to be considered with caution. Due to the limited number of cases, only major differences among the groups would be uncovered, whereas small differences as expected for a low-penetrance allele may remain undetected. Further, our assay can currently not distinguish between homozygous or heterozygous carriers, and there are three missense variants, p.A423T, p.T511A, and p.L571I in exon 11 that were allocated to some of the H1 subhaplotypes (Supp. Table S3). With the current multiplex assay we do not test for the presence of these variants that may modulate the functionality of the resulting gene product independently or in conjunction with p.N775S.
Whether our multiplex PCR assay could be useful in diagnostic settings where RNA-based mutation analysis cannot be applied remains to be tested. In principle, it could render information whether or not allelic drop-out is to be expected when using gene-specific primers for exons 13 and 14. Currently, however, it does not assay the presence of the reciprocal PMS2CL hybrid allele that presumably leads to coamplification of the pseudogene. Also, it may render false negative results for presumably rare alleles that might carry isolated PSVs at the intronic PDS used for gene-specific primers. Therefore, we believe it is currently still advisable to use RNA-based PMS2 testing [Etzler et al., 2008] wherever possible in clinical and research settings.
Supplementary Material
Acknowledgments
This work was supported by the Austrian “Fonds zur Förderung der wissenschaftlichen Forschung” (FWF), grant no P21172-B12.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Auclair J, Leroux D, Desseigne F, Lasset C, Saurin JC, Joly MO, Pinson S, Xu XL, Montmain G, Ruano E, Navarro C, Puisieux A, Wang Q. Novel biallelic mutations in MSH6 and PMS2 genes: gene conversion as a likely cause of PMS2 gene inactivation. Hum Mutat. 2007;28:1084–1890. doi: 10.1002/humu.20569. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- Clendenning M, Hampel H, LaJeunesse J, Lindblom A, Lockman J, Nilbert M, Senter L, Sotamaa K, de la Chapelle A. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–495. doi: 10.1002/humu.20318. [DOI] [PubMed] [Google Scholar]
- De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzler J, Peyrl A, Zatkova A, Schildhaus HU, Ficek A, Merkelbach-Bruse S, Kratz CP, Attarbaschi A, Hainfellner JA, Yao S, Messiaen L, Slavc I, Wimmer K. RNA-based mutation analysis identifies an unusual MSH6 splicing defect and circumvents PMS2 pseudogene interference. Hum Mutat. 2008;29:299–305. doi: 10.1002/humu.20657. [DOI] [PubMed] [Google Scholar]
- Feuk L, MacDonald JR, Tang T, Carson AR, Li M, Rao G, Khaja R, Scherer SW. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 2005;1:e56. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward BE, De Vos M, Valleley EM, Charlton RS, Taylor GR, Sheridan E, Bonthron DT. Extensive gene conversion at the PMS2 DNA mismatch repair locus. Hum Mutat. 2007;28:424–430. doi: 10.1002/humu.20457. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Hotspots of homologous recombination in the human genome: not all homologous sequences are equal. Genome Biol. 2004;5:242. doi: 10.1186/gb-2004-5-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senter L, Clendenning M, Sotamaa K, Hampel H, Green J, Potter JD, Lindblom A, Lagerstedt K, Thibodeau SN, Lindor NM, Young J, Winship I, Dowty JG, White DM, Hopper JL, Baglietto L, Jenkins MA, de la Chapelle A. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamalek JM, Cooper DN, Schempp W, Minich P, Kohn M, Hoegel J, Goidts V, Hameister H, Kehrer-Sawatzki H. Polymorphic micro-inversions contribute to the genomic variability of humans and chimpanzees. Hum Genet. 2006;119:103–112. doi: 10.1007/s00439-005-0117-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, Bannwart F, Yurtsever H, Neuweiler J, Riehle HM, Cattaruzza MS, Heinimann K, Schar P, Jiricny J, Marra G. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–1171. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg? Hum Genet. 2008;124:105–122. doi: 10.1007/s00439-008-0542-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.