Abstract
Histone deacetylase inhibitors (HDACis) have now emerged as a powerful new class of small-molecule therapeutics acting through the regulation of the acetylation states of histone proteins (a form of epigenetic modulation) and other non-histone protein targets. Over 490 clinical trials have been initiated in the last 10 years, culminating in the approval of two structurally distinct HDACis – SAHA (vorinostat, Zolinza™) and FK228 (romidepsin, Istodax™). However, the current HDACis have serious limitations, including ineffectively low concentrations in solid tumors and cardiac toxicity, which is hindering their progress in the clinic. Herein, we review the primary paradigms being pursued to overcome these hindrances, including HDAC isoform selectivity, localized administration, and targeting cap groups to achieve selective tissue and cell type distribution.
Balancing the epigenome
Cancer is vastly divergent, clever at avoiding therapeutic strategies, and lays a burden of pain, suffering and death on our society. Although billions of dollars, countless research institutions and the best scientific minds have all been engaged in attempting to eradicate this disease, there have been only flashes of success in a subset of cancers while a broad success across all cancer subtypes has so far remained elusive [1]. In that struggle, our knowledge of the complexities of cancer has grown rapidly, shedding light on the causes and character of neoplastic phenotypes. Mutagenesis, permanent alteration(s) to the genetic information within previously healthy cells, has long been the main suspect in cancer progression, but the improper regulation of non-mutated DNA is a major culprit as well [2].
Among abnormalities that lead to cancerous phenotypes, epigenetic misregulation is reversable by definition, unlike genetic mutations or deletions [3]. While our understanding of epigenetics is still burgeoning, a long list of regulatory mechanisms has been uncovered to date, including transcription factors [4,5], many types of noncoding RNA previously considered to be nonfunctional (including siRNA) [6–8], DNA methylation [9], histone modification [10,11] chromatin remodeling [12] and features of the nuclear architecture, including transcription factories [13] and chromosome terratories [14] (Figure 1). Much success in medicinal chemistry has been achieved in this area, targeting transcription factors (such as the estrogen and androgen receptors), utilization of RNA silencing, inhibiting DNA methyltransferases and histone modification enzymes, such as histone acetyl transferase (HAT) and histone deacetylase (HDAC) [15].
Figure 1.
Factors influencing epigenetic regulation of DNA information.
Since cancer is the result of the epigenetic differentiation program going in reverse [16], drugs aimed at pushing towards a terminal phenotype should lock it down, allowing the body to regain control and homeostasis [17]. As the mammoth information waves from proteomics, genomics and epigenomics converge, our biological understanding of the cellular world will pave the road to inumerable chemical interventions.
The focus of this review is HDAC inhibitors (HDACis), a particularly promising class of epigenetic drugs. We will discuss their successes and failures in the clinic, the possibility of various targeting approaches to address those failures and elaborate on the future prospect of a new paradigm in HDAC inhibition; namely, molecules with tissue-selective biodistribution profiles able to overcome systemic toxicity.
HDAC
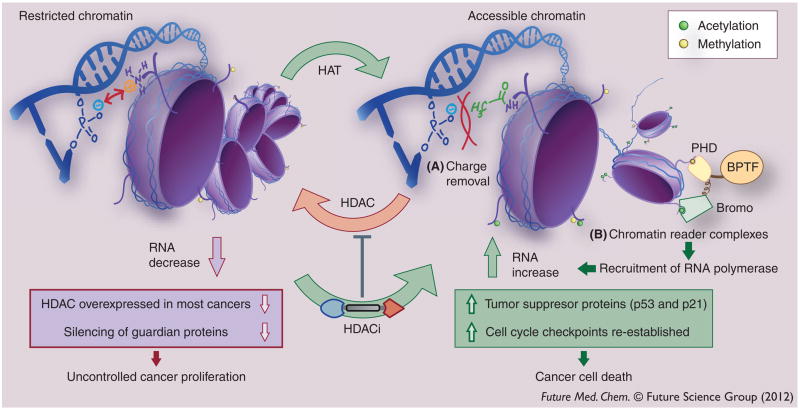
HDAC encourages silencing of genes by removing acetyl groups from lysine residues on the tails of histone proteins, which DNA wraps around (Figure 2). This creates a positive charge that causes the negatively charged phosphate backbone of DNA to tightly coil and restrict chromatin structures. In addition, HDAC-promoted deacetylation of acetylated lysine, a key epigenetic marker read by bromodomains within transcription factor complexes that recruit RNA polymerases, further dampens the transcriptional activity of hypoacetylated chromatin. This is contravened by HAT, which opens the structure by acetylating lysine residues on the histone, upregulating gene expression. Although the acetylation states of histone tails correlate well with chromatin accessability, HDACs have been found at sites of active transcription, suggesting they are used to reset chromatin acetylation after transcription [18]. For some complexes with HDACs present at sites of active transcription, they may also function to recognize acetylated lysine, rather than remove it [19].
Figure 2. The dynamic change in histone acetylation states and the accessability of the gene code is facilitated by the activities of two functionally opposed enzymes – histone acetyl transferase and histone deacetylase.
Acetylated lysines on core histone tails encourage gene expression via (A) reduction of electrostatic interaction between histone lysines and the DNA phosphate backbone, and also by (B) enabling binding of chromatin reader complexes, such as BPTF, equipped with an acetylated lysine reader (bromo, the bromodomain) and a methylated lysine reader (PHD) [145]. Inhibiting HDACs in the nucleus induces apoptosis via re-establishing expression of key tumor suppressor proteins, such as p53 and p21(Cip1/WAF1) [94].
BPTF: Bromodomain plant homeodomain finger transcription factor; HAT: Histone acetyl transferase; HDAC: Histone deacetlyase; HDACi: Histone deacetlyase inhibitor; PHD: Plant homeodomain.
HDAC activity plays a key role in cell differentiation [20], embryogenesis [21], cancers [22], neurodegenerative diseases [23], immunilogical responses [24], metabolic homeostasis [25] and many other biological phenomena. Small-molecule inhibitors of HDAC shift the equilibrium towards accessible chromatin, and restores expression of key genes [26].
While many disease states are characterized by epigenetic imbalance that could benefit from HDACis, much attention has been directed towards cancers. Silencing of tumor suppresor genes (such as p21) through hypoacetylation is a hallmark of many cancers, and turning these back on through HDAC inhibition has shown clinical benefit.
There are 18 known isoforms of HDAC (Table 1). The zinc-dependent metalloproteases are grouped into Class I, II and IV (based largely on cellular location and sequence homology) [27], with Class III being NAD+-dependent enzymes [28]. The zinc-dependent Class II is further divided into IIa (having both nuclear and cytoplasmic localization) and IIb (primarily cytoplasmic and the only class with two enzyme active sites). The structural differences among these isoforms is becoming clearer as more crystal structures of these enzymes complexed with inhibitors become available (Table 1, structures available as of 15 December 2011 from the Protein Data Bank) [29]. Nevertheless, gaps still exist in HDAC structural information, and these have to be filled in by homology models [30,31].
Table 1.
Various classes of zinc-dependent histone deacetylase isoforms.
| Class | I | IIa | IIb | IV | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Isoform | HDAC1 | HDAC2 | HDAC3 | HDAC8 | HDAC4 | HDAC5 | HDAC7 | HDAC9 | HDCA6 | HDAC10 | HDAC11 |
| Crystal structures | 0 | 1 | 0 | 21 | 8 | 0 | 3 | 1 | 0 | 0 | 0 |
| Cellular location | Nucleus primarily | Nucleus and cytoplasm | Cytoplasm primarily | Nucleus | |||||||
HDAC: Histone deacetylase.
HDAC inhibition successes
The US FDA has approved two HDACis, SAHA (Vorinostat, Zolinza™) [32] and FK228 (Romidepsin, Istodax™) [33], with many others at various stages of testing (Figure 3). These clinical validations have sustained a wave of research efforts aimed at:
Figure 3. Various classes of histone deacetylase inhibitors.
HDACi: Histone deacetylase inhibitor; SAHA: Suberoylanilide hydroxamic acid.
Synthesis of naturally occuring HDACis;
Synthesis of new synthetic HDACi compounds of a wide variety;
Solving of crystal structures for various isoforms of HDAC;
Determining structure–activity relationships (SAR) in terms of HDAC inhibition potency, isoform selectivity and/or anticancer activity;
Evaluating HDACis in the clinic both as stand alone and combination anticancer therapy.
HDACi pharmacophoric model
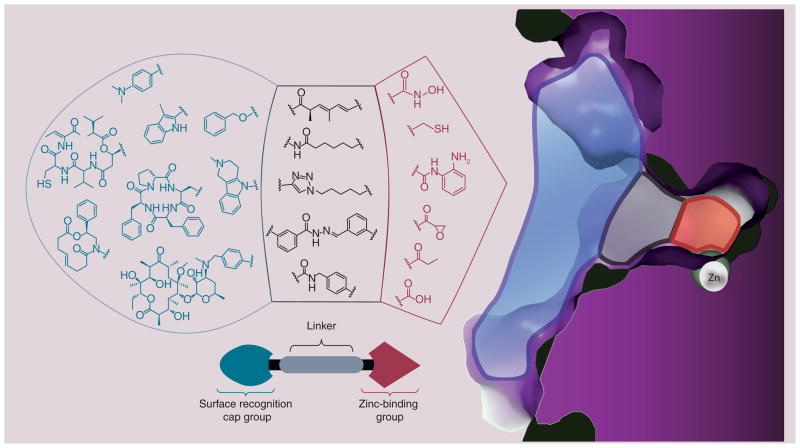
Mimicking the natural substrate (acetylated lysine residues), HDACi typically follow a structural motif (Figure 4) comprised of a surface recognition cap moiety that can tolerate extraordinary variability, a linker group that traverses the tunnel of the active site and a zinc-binding group (ZBG) that chelates active site zinc ion (Zn2+) [34,35]. Modulating these different pieces of the pharmacophore has been pursued in attempts to understand the structural basis for HDACi potency, isoform selectivity and efficacy against various diseases including cancers [36,37].
Figure 4. Histone deacetylase 3 pharmacophoric model for Zn2+-chelating inhibitors.
The crystal structure shown highlights the surface (blue), hydrophobic tunnel (gray) and zinc-sequestering active site (red). Each histone deacetylase inhibitor pharmacophore is color-coded to reflect its binding within the histone deacetylase enzyme active site.
Hydroxamate (e.g., trichostatin A [TSA] and suberoylanilide hydroxamic acid [SAHA]) is the most common ZBG by far, owing it’s success to the fact that most of the binding energy associated with the strength of inhibition is derived from the bidentate chelation of this popular functional group (found in naturally occuring HDACis). Second to that is the naturally occuring prodrugs, the depsipeptides (largazole, FK-228 and the spiruchostatins), which have a latent alkyl-thiol that is unmasked in vivo to achieve excellent HDAC inhibition potency in an isoform-selective manner. A third common ZBG in the benzamide moiety (MS-275), which trades off potency for Class I isoform selectivity. The diversity among the linkers has not been systematically explored, but nonetheless they exhibit limited chemical diversity surrounding chain-like alkyl linkers with various degrees of saturation and often include substituted aryl groups, dictated by the diameter and hydrophobicity of the tunnel region. The surface-recognition cap groups enjoy the widest range of chemotype tolerance, and have been the topic of extensive study in attempts to toggle potency [36,38,39], biodistribution [40], isoform selectivity [41], cardiotoxicity [42] and, more recently, tissue targeting [43].
HDACis in the clinic
The interest in the clinical application of HDACis has exploded over the last few years, with over 490 clinical trials, excluding diseases other than cancer, of which there are a few examples [44]. The weakly HDAC-inhibiting phenyl butyrate was the first to enter clinical trials for cancer in the mid-1990s [45], followed by FK-228 [46] and a rush of hydroxamic-based HDACi in the last decade (Figure 5). As stated earlier, the FDA approved SAHA (Vorinostat) in 2006 and, later in 2009, FK-228 (Romidepsin) joined it in the medicine cabinet, both for treating cutaneous T-cell lymphoma (CTCL) [47,48].
Figure 5. Clinical timeline: the explosion of histone deacetylase inhibitor cancer clinical trials, over 490 to date.
Count was compiled from the clinicaltrials.gov databank. Numbers for 2010 and 2011 are incomplete given the time lag between start date and appearance in the databank, and do not reflect a decrease in medical excitement surrounding histone deacetylase inhibitors.
SAHA: Suberoylanilide hydroxamic acid.
Suberoylanilide hydroxamic acid (SAHA; vorinostat)
The approval of SAHA was the consequence of a Phase II multicenter trial in patients with refractory CTCL [48]. Of the 74 patients who received 400 mg of vorinostat orally daily, 29.7% had an objective response with a median duration of response ≥185 days and median time to progression of ≥299 days [49]. Additionally, 65 patients in this trial have pruritis, a symptom often associated with CTCL [50]. Of these patients presented with pruritis, 32% experienced relief of symptoms, which was independent of the response to the treatment. In another Phase II trial of oral Vorinostat for refractory CTCL where various dosing regimen and schedule were used, 45% patients with pruritis were relieved and attenuation of this condition was higher in patients with severe pruritis before the treatment. The most common side effects noticed during these trials were constitutional and gastrointestinal effects, including nausea, diarrhea, dysgeusia, and hematologic effects such as thrombocytopenia. Serious dose-dependent side effects such as anemia, infection, dehydration, sepsis, hypotension and pulmonary embolism were also observed [51].
FK228 (romidepsin)
In a study that evaluated Romidepsin as a monotherapy for the treatment of CTCL, 68 patients with refractory or relapsed CTCL were administered Romidepsin intravenously at 14 mg/m2 on days 1, 8 and 15 during a 28-day cycle. The observed treatment response was 34% with median duration of response of 13.7 months. Three patients with Sézary syndrome had complete remission and one patient continued to be in remission at 63 months. Constitutional and gastrointestinal adverse effects were fatigue, nausea and vomiting. Hematologic toxicities, such as leucopenia, lymphopenia, thrombocytopenia and anemia, were also observed. Asymptomatic ECG changes were present in 71% of patients [35,52]. Similar results were also reported by another Phase II clinical trial, establishing the efficacy of Romidepsin for the treatment of refractory CTCL [35,53].
Lack of efficacy against solid tumors
Despite promising results in the treatment of CTCL, these two HDACis have not been effective in clinical trials involving solid tumors. Many clinical trials have assessed the efficacy of Vorinostat against different solid tumors, including refractory breast, colorectal, non-small cell lung and thyroid cancers. Disappointingly, none of the patients in these trials showed partial or complete response to treatment, but the prevalence of drug-induced side effects was very high: constitutive (fatigue 62%), gastrointestinal (anorexia 81% and diarrhea 56%) and hematologic (thrombocytopenia 50%). A total of 63% also experienced QT interval prolongation less or equal to 30 ms and one patient had QT interval prolongation between 30 and 60 ms [54,55]. The only silver lining in these studies is that approximately 50–56% of patients experienced stabilization of their diseases. This leaves open a narrow window of opportunity for the use of vorinostat and similar HDACis in solid tumor therapy, most likely in combination with other chemotherapeutic agents.
Romidepsin has also been evaluated as a monotherapy against solid tumors. Similarly to vorinostat, romidepsin has also been ineffective against solid tumors. Stadler et al. reported that the treatment of patients with refractory metastatic renal cell cancer with Romidepsin resulted in only 7% objective response with one patient achieving and remaining in complete remission for 14 months. In addition to hematologic (anemia, neutropenia and thrombocytopenia), gastrointestinal (nausea, vomiting and diarrhea) and constitutional adverse effects, serious cardiotoxicity was also observed. Prolonged QT interval was detected in two patients, one patient developed atrial fibrilation, another had tachycardia and there was an occurrence of sudden death [56]. Romidepsin was also ineffective against metastatic colorectal cancer. In a 25-patient trial, no objective responses were seen, and only four patients had stable disease states for a period of time ranging from 44 to 161 days. Treatment was stopped in six patients due to the prevalence of serious side effects, such as thrombocytopenia, dehydration and QT interval prolongation [57]. Although these patients received similar dose of Romidepsin at the same rate and during the same 28-day cycle as patients with refractory CTCL, patients with CTCL had significantly better outcomes compared to those with solid tumors. In cancers of the blood, such as CTCL and multiple myeloma, the metabolic instability of these HDACi compounds may not preclude their effectiveness, compared with less permeable malignancies [58].
In addition to Romidepsin and Vorinostat, QT interval prolongation has been associated with other hydroxamate-based HDACis such as LBH589 and LAQ-824 [59]. The progress of HDACis through clinical trials has been the subject of recent review articles; we have restricted the focus of this review to the clinical trials of SAHA and Romidepsin [60–62]. In the sections below, we will use the information gleaned from these trials to discuss ways forward for HDACi as chemotherapy agents.
Cardiotoxicity: a hurdle for HDACis in the treatment of solid tumors?
HDACis such as romidepsin and SAHA have been associated with serious cardiotoxicity. Such cardiotoxicity include T-wave flattening, ST segment depression and QT interval prolongation [52]. QT interval prolongation has been to date the most severe cardiac event in patients treated with HDACi due to their ability to lead to potentially fatal ventricular arrhythmia, known as torsades de pointes [63]. Prior to its approval by the FDA, there have been six cases of unexpected deaths in patients treated with Romidepsin. Pulmonary embolus was believed to be responsible for one death, while the other five cases were attributed to sudden cardiac arrest [64,65]. Addressing this cardiotoxicity becomes crucial as various HDACis are being studied in clinical trials against solid tumors.
Although not completely understood, the mechanism of QT interval prolongation has been explained by aberrant cellular trafficking and/or functioning of the human ether-ago-go (hERG) K+ channel [66]. The latter being the most accepted mechanism for the HDACi induced QT interval prolongation [59]. The activation of the hERG K+ channel leads to ventricular repolarization, hence blocking of this channel may result in QT interval prolongation [66]. HDACi are not the only class of drugs that can interact with the hERG K+ channel; other drug classes also have that capacity due to the large size of the channel’s inner cavity and the presence of aromatic residues favoring hydrophobic interactions with lipophilic molecules inside [67].
In addition to the aforementioned mechanisms, drug-induced QT prolongation may be caused by increased turnover rate of mature hERG channels from the plasma membrane [68]. Though most drug-induced QT prolongations have been associated with the hERG channels, other ions channels such Na+ channel may be involved as well [69]. Lacerda and coworkers reported that Alfuzosin, an α1-adrenergic receptor antagonist with clinical evidence of QT prolongation, did not bind to hERG K+ channel. Instead, Alfuzosin mechanism of QT prolongation resides in its ability to enhance Na+ current [70]. Furthermore, the proper functioning of hERG in vivo required the coexpression of many other proteins such as MinK and MinK-related peptide 1 (MiRP1) [71,72]. Mutations or lack of these peptides have been linked to QT prolongation [72,73]. For drugs known to modulate gene expression, such as HDACi, altering the expression of hERG and any of these genes may lead to QT prolongation even in the absence of a direct interaction with the hERG channels at therapeutic doses. In fact, emerging evidence in the literature suggests that the QT prolongation associated with HDACis may be the consequence of such altered gene expression and possibly the inhibition of specific HDAC isoforms [74,75]. Therefore, changes in hERG expression or that of the coregulators of hERG activity may represent yet another mechanism of QT prolongation. This and other alternative mechanisms of QT prolongation discussed therein, may explain the findings that SAHA did not affect hERG K+ channels up to 300 μm [76] and that SB939, another hydroxamate-based HDACi, did not bind to hERG channels up to 10 μm but showed evidence of QT prolongation during Phase I clinical trials [77,78]. A study looking at the impact of HDACis on the expression of hERG and of its coregulators is needed to elucidate other potential mechanisms of drug-induced QT prolongation.
Although it has been seen in different clinical trials that HDAC inhibition can lead to QT interval prolongation; there is, however, an increased risk in patients with certain predisposing factors such as diabetes mellitus, obesity, hypothyroidism and congenital long QT syndrome [79]. Other risk factors include gender, advanced age, previous cardiovascular and cerebrovascular diseases [66,80]. In a study by Barbey et al., baseline ECG in cancer patients prior to treatment revealed cardiac abnormalities, such as sinus tachycardia, atrial fibrillation and previous myocardial infarction in 36% of patients [81]. This study, as well as others, highlighted the importance of detecting and treating pre-existing cardiovascular diseases in cancer patients as these can be underestimated [81,82]. Predisposing factors to QT interval prolongation can be iatrogenic, following administration of various drugs, such as antipsychotics, and serotonin agonists and antagonists. In the UK and Italy, 2–3% of all drugs prescribed may provoke QT interval prolongation [83]. De Ponti et al. have compiled a more comprehensive list of drugs with QT interval prolongation potential [84]. Cancer patients, due to concomitant use of antiemetics, antibiotics and antifungal for the treatment of chemotherapy induced side effects, may be at an increased risk of QT interval prolongation, as these drugs may increase the QT interval [79,84]. Antidepressants, which may be used to treat symptomatic depression present in 24% of cancer patients, can also prolong the QT interval [79,85]. Metabolic disturbances are other QT prolongation predisposing factors. Electrolyte imbalance, such as hypokalemia, hypomagnesemia and hypocalcemia, which can be consequences of the chemotherapy-induced anorexia or vomiting, may also lead to QT prolongation [59,86].
Approaches to overcoming roadblocks against HDACi in the clinic
Delivering increased potency at the site of action, while eliminating the toxicities that result from off target effects of chemotherapies, is the hope of up-and-coming cancer treatments of all kinds [87]. Targeting in cancer therapy can mean:
Target preference: designing and developing drugs with extremely high potency and selectivity for a unique molecular entity and not others;
Selective delivery: directing the medicine to the organ, tissue, cell or subcellular location of interest.
Approaches being explored to overcome the problems seen with first-generation HDACis in the clinic include either or both of these targeting paradigms. We will explore two approaches from the ‘target preference’ paradigm; namely, isoform selectivity and hERG binding reduction; and two examples from the ‘selective delivery’ paradigm; namely, localized administration and targeting cap groups (Figure 6).
Figure 6. Overcoming clinical roadblocks to histone deacetylase inhibitor cancer therapy.
HDACi: Histone deacetylase inhibitor.
Isoform selectivity
It stands to reason that if the isoforms of HDAC have various locations, expression levels and functions, then an understanding of those differences, combined with an arsenal of isoform selective or isoform-specific HDACis could yield tremendous clinical benefit. However, it is not yet clear if hitting one HDAC isoform and not others will translate into clinical benefit. Here, we take a brief look at some of the most promising molecules that will help set the future direction of isoform selectivity. For more detailed reviews on isoform selectivity, we direct the reader to previous reviews [23,41,88,89].
Pan-HDAC inhibitors
The first-in-class drugs approved to date (as well as many candidates in clinical trials) act broadly on all isoforms of the zinc-dependent classes with little discrimination and are regarded as pan-HDAC inhibitors (pan-HDACis). While there are countless examples, three pre-eminent ones include the synthetic analogue SAHA, the naturally occuring TSA and the Novartis discovered LAQ-824, all of which show activity against all isoforms (Figure 7A).
Figure 7. Pan histone deacetylase inhibitors.
(A) Traditional, non-selective inhibitors (SAHA and TSA data averaged from four independent groups) [43,88,99,109]. (B) Pandacostat, profiled against values calculated from Ki reported by the traditional inhibitors SAHA, TSA and LAQ-824 (IC50 Bradner et al. using the Cheng–Prusoff equation) [19].
HDAC: Histone deacetylase; SAHA: Suberoylanilide hydroxamic acid; TSA: Trichostatin A.
Recently, the activity of these compounds against Class IIa HDACs has been brought into question primarily by the results from assay development and screening efforts of James Bradner and Ralph Mazitschek [19]. A novel, more sensitive Class IIa enzyme substrate was utilized, allowing for improved catalytic turnover and lower enzyme concentrations. With these tools in hand, hydroxamic acids such as SAHA were shown to have a surprisingly attenuated Class IIa inhibition activity (Figure 7B), and a true pan-HDACi was discovered, Pandacostat [19]. Class IIa HDACs were suggested as readers of acetylation marks on chromatin rather than erasers, raising important questions as to interplay between Class IIa inhibition and cancer progression. It is instructive to state here that assays probing for Class IIa specific HDACis have been demonstrated to be frequently contaminated with more active HDAC isoforms, an additional factor that may skew isoform selectivity data [90].
The cause(s) of ineffectiveness for these first-in-class HDACis against solid tumors at doses that have proven effective in CTCL are not well understood. It is conceivable that doses needed to see clinical benefit may be achievable if isoform selectivity reduces or prevents dose-limiting side effects. Thus, effort to develop inhibitors selective for isoforms has been thought to be a significant step towards successful HDACi therapy.
Inhibitors selective for HDAC1, 2 & 3
Within Class I, there are four isoforms (Table 1), with HDAC1, 2 and 3 sharing the most sequence homology; they therefore are usually hit with similar strength for any given inhibitor. HDAC1, 2 and 3 are located in the nucleus (almost entirely) and are found in all healthy cell types [91]. However, in certain cancers overexpression of these HDACs has correlated with poor survival rates [91–93]. Highest levels of Class I HDAC have been found especially in late stage, aggressive malignancies [91] and inhibiting these nuclear HDACs induces apoptosis by re-establishing expression of key on cosuppressor proteins, such as p21(Cip1/WAF1) [94].
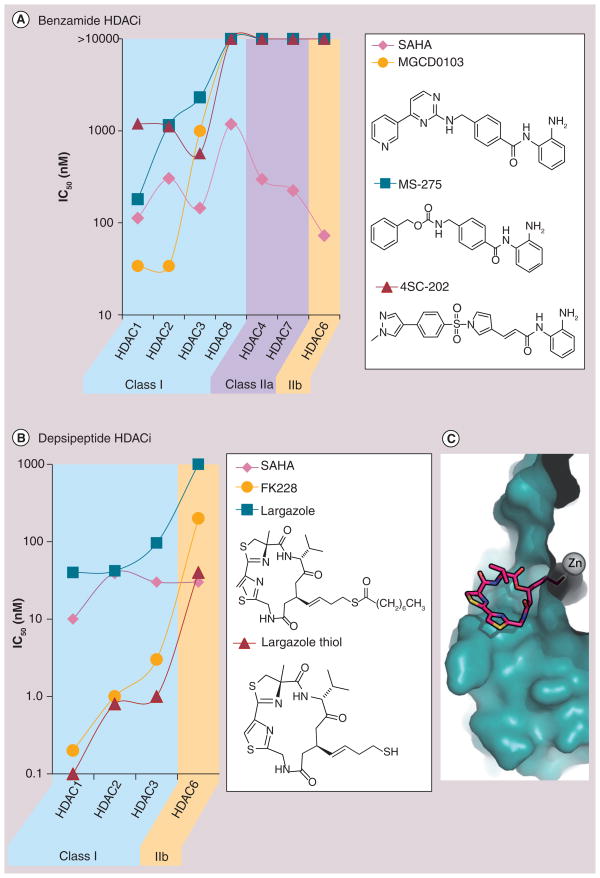
Summarized in Figure 8 are inhibition data for the clinically relevant benzamides and the natural product depsipeptides HDACi, which have varying degrees of selectivity for HDAC1, 2 and 3.
Figure 8. Histone deacetylase isoform selectivity of clinically relevant benzamides and depsipeptide histone deacetylase inhibitors relative to suberoylanilide hydroxamic acid.
(A) Clinically relevant benzamide HDACi are selective for HDAC 1,2 and 3 of Class 1, but not HDAC8. (B) Depsipeptide HDACi are selective for Class 1 HDAC and more potent than the benzamides and SAHA. (C) Crystal structure of largazolethiol, the product of in vivo hydrolysis of the thioester bond, bound to HDAC8 shows extensive interaction of the macrocycle with HDAC outer surface rims which may explain the enhanced potency of depsipeptides relative to other HDACis [104].
HDAC: Histone deacetlyase; HDACi; Histone deacetylase inhibitor.
The first major Class I selective HDACi with high hopes was benzamide MS-275, due to the lack of cardiotoxicity. The isoform selectivity of MS-275, MGCD0103 (Mocetinostat) [95] and more recently 4SC-202 [96] are typical of the benzamide class of HDACis [88]. While they are extremely selective (Figure 8A), their half-maximal inhibitory concentration lies in the micromolar regime, much higher than the low nanomolar activity of most hydroxamic acid-based HDACis, a concern that may be responsible for the poor performance of MS-275 in the clinic. In various Phase I clinical trials involving MS-275 in patients with refractory solid and hematologic malignancies, no cardiotoxicity attributed to MS-275 was detected [97–99]. There were also no deaths related to MS 275 administration [100]. Although Phase I studies showed promising results, MS-275 as a monotherapy had little efficacy in patients with refractory leukemia and metastatic melanoma [100,101]. In a latter study, no objective response was observed; however, disease stabilization was seen in 25% of the patients, with time to progression ranging from 5 to 385 days and median survival of 8.84 months [101]. Similar toxicity profile and efficacy were also reported for MGCD0103 [102,103]. Despite the limitations seen with Class I selective benzamies so far, 4SC-202 [96] is still charging full steam ahead, although results showing improved clinical benefit have yet to be released.
The naturally occurring depsipeptides FK-228 (Romidepsin, Figure 3) and largazole are HDAC1, 2 and 3 selective owing to the unique ability to recognize amino acid side chains and amide backbones on the enzyme outer rim (the most structurally divergent location on all HDAC enzymes), via a multitude of binding interactions from their complex macrocyclic ring structures (Figure 8C) [104]. These molecules require in vivo unmasking of their alkyl-thiol ZBG, but once revealed the strength chelation leads to low nanomolar inhibition of HDAC1, 2 and 3 (Figure 8B). This increased potency, in combination with its isoform selectivity, are likely the attributes that carried FK-228 through the clinic culminating in approval for CTCL, a blood cancer that may not be subject to drug-penetration issues typical of many solid tumors.
Inhibitors selective for HDAC6
It can be misleading to discuss HDAC6 in regards to ‘epigenetic’ cancer therapy. It is not truly a HDAC, as its primary cellular localization is in the cytoplasm where it regulates acetylation states (and thereby the functionality) of tubulin, HSP90 and other extra-nuclear proteins [105]. The cell motility and metastatic potential result from the influence of HDAC6 on microtubule formation [106]. HDAC6 allows progression and growth of malignancies by enabling them to survive even in the absence of adequate anchoring to the extracellular matrix [106]. It is also needed for the development malignancy through the RAS/MAPK signaling pathway, and plays many other roles that make it an intriguing therapeutic target [107].
One of the first major breakthroughs in isoform selectivity was in the discovery and use of Tubacin. This aided in elucidating the distinct activity of HDAC6 on tubulin, but demonstrated poor drug properties (low water solubility and synthetically challenging) [108]. Recently, a major success in HDAC6 selectivity was achieved by Alan Kozikowski’s group, guided by homology modeling in absence of HDAC6 crystal structures bound to inhibitors [7,31]. The resulting lead, Tubastatin A (Figure 9), exhibits an excess of 1000-fold selectivity for HDAC6 over HDAC1, 57-fold over HDAC8 and at least 2000-fold over every other isoform. This was achieved without compromising activity, and in-fact Tubastatin A is more potent than SAHA at inhibiting HDAC6. The structural basis for the selectivity is due to the widening of the outer rim that connects to the Zn2+-containing active site of HDAC6 (17 Å compared with 12 Å for HDAC1), a difference thoroughly investigated by Kozikowski’s group through designing of steric bulk into the inhibitor’s cap group. This is a key observation that may explain the strong selectivity for HDAC6 found in the synthetic macrocyclic hydroxymate compounds designed recently by Auzzas et al., of which (R)-9 is a lead example (Figure 9) [109]. Efforts in the Pflum lab to modify the C-3 position on SAHA with short alkanes showed HDAC6 preference; albeit with 1000-fold loss in activity [110]. The HDAC6 selective inhibitor ACY-1215, in combination studies with clinically approved proteasome inhibitor bortezomib, is being investigated for treatment of multiple myeloma [111]. These selective inhibitors have shown promise, as HDAC6 is known to be overexpressed in various cancers and its complete knockdown does not impair normal functions, predicting a lack of major clinical side effects [106].
Figure 9.
Histone deacetylase 6-selective inhibitors.
Inhibitors selective for HDAC8
HDAC8 has an increased expression profile in smooth muscle tissue and has been proposed to regulate the ability of smooth muscle cells to perform contractions [112]. HDAC8 is differentially expressed and associated with various cancers. Noteably, HDAC8 is the only HDAC (so far) relevant in neuroblastoma [113], making its selective inhibition of high interest in the etiology and treatment of this form of cancer. Early reports of inhibitors selective for HDAC8 included short [114] and linkerless hydroxamates [115]. Highlighted in Figure 10 are HDAC inhibition profiles of two classes of exciting HDAC 8-selective molecules that were reported within the year.
Figure 10. Histone deacetylase 8-selective inhibitors.
(A) Hydrazide aryl hydroximatic acids and (B) (R)-α-amino-ketones.
SAHA: Suberoylanilide hydroxamic acid; TSA: Trichostatin A.
HDAC8 is most often the least inhibited isoform within Class I. It is especially unresponsive to HDACi derived from the most common ZBG, the hydroxamate. The highthroughput screening efforts by James Bradner and Stuart Schrieber have produced libraries of small-molecule HDACis [19], which recently furnished a new linker motif that exhibits selectivity for HDAC8 (Figure 10A) [116]. Novartis reported two lead HDACi that have an (R)-α-amino-ketone moiety as a unique ZBG. These compounds show selectivity for HDAC8 principally through interaction with the acetate exit tunnel of HDAC8. The spatial arrangement of the functional groups in these novel HDACis do not fit the traditional ‘cap-linker-ZBG’ pharmacophoric model (Figure 10B) [117]. It will be exciting to see pharmacological testing of these compounds, promised by the authors as forthcoming in a future report.
The clinical benefits of HDAC8 isoform selectivity may be useful though limited, as it has been shown that selective inhibition of HDAC8 induces apoptosis in T-cell cancers, such as leukemia, but has little antiproliferative activity against cells derived from solid tumors. This observation suggests an important connection between isoform selectivity and cancer-type HDACi selectivity [114], which had been suggested for acute myeloid leukemia [115]. Nevertheless, the biochemical understanding of HDAC8 isoform is much deeper than most, having the advantages of robust collection of very selective compounds and by far the most structural information.
The pursuit of isoform specific/selective HDACis is of tremendous importance, particularly for unique HDAC isoforms such as HDAC6 and HDAC8; it may, however, not be sufficient to address all the problems that have beleaguered HDACis in the clinic. Additionally, the functional redundancy of closely related isoforms, such as HDAC1, HDAC2 and HDAC3, may offset any benefit derived from selective inhibition of a member of such related HDAC isoforms [118]. While selecting for one or several HDAC isoform targets will likely play an important role in the road to reducing off target toxicity, systemic inhibition of any single isoform is still a potential health hazard, leaving a need for selective delivery to the desired location.
hERG binding reduction
Cardiac toxicity is one of the major side effects/ concerns preventing progress of HDACi in the clinics. Understanding the molecular entities that are being hit by HDACis to produce this off-target effect is an alternative approach to increase the safety for this class of drugs. Recently, Novartis has performed a study to design non-cardiotoxic hydroxamate-based HDACis [42]. Starting from LAQ824 (Figure 7), one of the most potent HDACis in vitro [119], a SAR was performed with the objective retaining potency while decreasing its hERG affinity. Using the in vitro cardiac safety index (iCSI) – the ratio of the hERG IC50 to cellular IC50 – researchers were able to determine the potential cardiotoxicity of several derivatives of LAQ824 early in the SAR study. The incorporation of this index early in their design and in vitro characterization enabled the synthesis of two compounds that achieve single-digit nanomolar IC50 HDACi activity and low hERG affinity with iCSI values greater than 6000, providing a safety margin for in vivo and clinical studies [42]. Using similar in vivo testing Shultz et al. have reported the synthesis of isoindoline-based HDACis [120]. The use of the iCSI as a parameter for HDACi candidate selection may decrease the number of clinical trials being terminated for cardiotoxicity.
Interaction of HDACis with the hERG K+ channel, which is currently viewed as a downside of HDACi therapy, may paradoxically be an advantage, as hERG K+ channels are involved in proliferation of various malignant cell lines [121]. Besides their epigenetic mechanism of action, HDACis, which are able to block the hERG K+ channels, may also induce apoptosis through an additional pathway. To fully benefit from such dual activity, selective distribution of those HDACis will have to be achieved, inflicting potent cytotoxicity onto cancer cells while minimizing delivery to the heart to avoid QT prolongation. Such an approach may enable the full anticancer potential of HDACis to be harnessed.
Localized administration
A target-independent methodology that has a great potential in overcoming many of the systemic toxicity issues associated with HDACi usage is to locally administer compounds into the tumor tissue. Localized drug administration has been achieved through intratumoral injection [122], topical application [123], and surgically placed biodegradable polymers [124]. to mention but a few. Three different HDACis in topical formulations are currently in early stages of clinical trials.
In a Phase I trial, Kong et al. studied the safety of topical FK-228 in patients with CTCL [201]. Through direct application of FK-228 to the skin lesions, selective delivery can be achieved minimizing systemic side effects. This led to a patent application for the topical formulation of FK-228 [125] for CTCL and other skin diseases. Following the same paradigm, another clinical trial began in 2008 for topically administered pan-inhibitor DAC060 from Genextra [123]. Exciting initial results from the Phase II trial have been reported, showing complete or near complete remission in 16 of 22 patients with non-melanoma skin cancer, and partial regression from all others, with only mild inflammatory side effects [123]. Although the structure of DAC060 has not been disclosed, Genextra recently published studies on N-hydroxyphenylacrylamide and spiro[benzofuran-2,4′-piperidine] hydroxamic acid HDACi [126,127]. The latest clinical trial, started in 2011, by Shape Pharmaceutical Inc., is evaluating the safety, pharmacokinetics and pharmacodynamics of topical formulation of SHP141, a novel HDACi [202]. This clinical trial was initiated after encouraging results in a mouse model of CTCL [128].
All the clinical trials aforementioned are examples of selective delivery through direct physical application of the HDACi to the malignant tissues. This method is not applicable to most malignancies as they may involve parenchyma of organs and may also have metastatic lesions. However, they do illustrate the potential of selective delivery as a powerful means of utilizing HDACis without inducing dangerous side effects.
Targeting cap groups for tissue–cell-selective drug accumulation
Equipping HDACis with a surface recognition cap group capable of binding unique biological targets, such as over expressed or uniquely expressed receptors, could confer interesting and desirable tissue-selective accumulation properties on HDACis. Because the HDAC enzyme outer surface rims are highly tolerant of variations on HDACi surface-recognition cap group, some of these tissue-selective compounds could be incorporated into the design of next-generation drugs. Such HDACis will retain or even have enhanced HDAC inhibition and possess targeted anticancer activity due to the selective tissue distribution conferred by the appended targeting moiety. Additionally, the increased potency afforded by drug accumulation at the site of disease will likely translate to lower therapeutic doses, thereby minimizing detrimental off target effects, which are often presented at high drug doses. We highlight here two examples of such molecules that have the potential to shape the future of HDACi therapy.
CHR-2845 is a hydroxamic-based HDACi endowed with an ester linkage which can be hydrolyzed by human carboxylesterase-1 (hCE-1), an enzyme present mainly in macrophages, monocytes and kupffer cells. Hydrolysis of CHR-2845 yields CHR-2847 (the active metabolite) which accumulates in cells expressing hCE-1 [129]. This accumulation results in a 20–100-fold increase in potency against monocytes derived malignancies relative to non-monocytic malignancies [130]. In a Phase I multicenter trial of CHR-2845 in patients with advanced hematological malignancies, no dose-limiting toxicities were detected. In terms of efficacy, one patient with chronic myelomonocytic leukemia achieved bone marrow response and symptoms relief after completion of nine cycles of CHR-2845 [130].
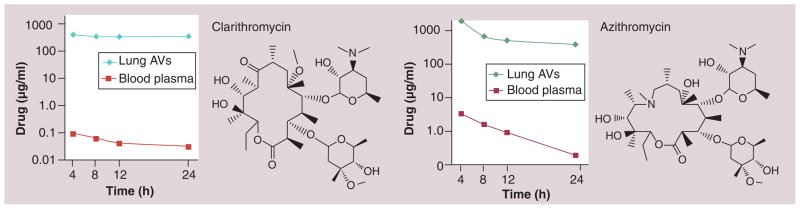
Our laboratory has been developing HDACis incorporating various nonpeptide macrocyclic ring systems known to selectively accumulate in the lungs. The macrocyclic templates we have chosen were derived from the medically successful antibiotics azithromycin (AZ) and clarithromycin (CL) (Figure 11), as well as a triketolide (TE-802) that has demonstrated superior efficacy in mice model of respiratory tract infection [35,131,132]. Our choice of these macrocyclic compounds was informed by their extraordinary tissue distribution profiles in data presented to the FDA and subsequently confirmed by various independent laboratories [133,134]. The lung tissue selective accumulation of AZ [135] and CL [136] (Figure 11) is a major determinant of their effectiveness against various respiratory tract infections [135]. For the 15-membered AZ, targeting to the lung tissue occurs via rapid uptake into monocytes, phagocytes, alveolar macrophages, fibroblasts and lymphocytes, which themselves have a selective distribution to lungs especially in response to diseased states such as infection and inflammation [131].
Figure 11. Lung selective distribution profile of clarithromycin and azithromycin in human patients [135].
Lung AV: Lung aveolar macrophage.
Using a combination of the tools of synthetic organic chemistry, computational chemistry and cell based assays, we have identified a series of macrocyclic HDACis derived from AZ, CL and TE-802. These elicit selective and potent anti-proliferative activity against human lungs, prostate and breast cancer cell lines [43,137]., Overall, these compounds have improved enzyme inhibition potency and isoform-selectivity (subclass isoform preference for HDAC1 and 2, over HDAC8). They possess both linker-length and macrolide-type dependent HDAC inhibition activities (Figure 12). The alkyl linker length is optimized at seven carbons (n = 2) across all macrocyclic cap groups (Figure 12). The presence or absence of the cladinose sugar on AZ and CL derivatives (cladinose-containing AO-AZ versus AO-AZH, and AO-CL versus AO-CLH, Figure 12) has little effect on the HDAC inhibition profile. Computational analyses enabled an understanding of the linker length preference and the roles of the interaction between the HDAC enzymes outer rim and the inhibitors’ macrocyclic templates that are responsible for the enhanced affinity and isozyme selectivity [43,137]. Ongoing efforts in our lab have revealed interesting patterns of tissue-selective accumulation in a subset of these macrolide-derived HDACis. The prospect of tissue-specific HDACi delivery is a particularly enticing alternative to isoform-selective HDACis [137].
Figure 12. Targeted histone deacetylase inhibitor with non-peptide macrocyclic cap groups.
(A) Targeting with the cap group from the traditional HDACi pharmacophoric model. (B) Linker length dependance on the activity of non-pepide macrolide HDACi. AO-AZ#: Azithromycin-triazole-HDACi of chain length #n; AO-AZH#: Azithromycin-triazole-HDACi of chain length #n with H replacing the cladinose sugar; AO-CL#: Clarithromycin-triazole-HDACi of chain length #n; AO-CLH#: Clarithromycin-triazole-HDACi of chain length #n with H replacing the cladinose sugar; AO-TK#: Triketolide-triazole-HDACi of chain length #n; AZH: Azithromycin; HDACi: Histone deacetylase inhibitor.
Future perspective
The approval of SAHA and FK-228 has firmly laid the foundation for the exploration of HDAC inhibition as a therapeutic approach for other cancer subtypes and related diseases. The next 5–10 years will see new and unprecedented therapeutic opportunities based on HDACi regiments, although not without challenges.
We anticipate more isoform-selective HDACis, specifically for Class IIa HDAC4 and HDAC7, since their crystal structures are now available [138,139]. Based on the current trends, we expect more FDA approvals will arrive in the next decade, either for new compounds based on: the desirable targeting characteristics outlined in this review, new combination therapies; and/or new indications for other cancer types other than CTCL [140]. With the aspiration of finding real cures for cancer and other difficult-to-treat diseases for which HDACi could be beneficial, we make a bold claim here that the paradigm of tissue- and cell-targeted delivery will gain prominence in the design of new generation of HDACis. This approach will be a natural complement to investigations centered on identifying isoform selective HDACis.
In order to fashion HDACis that preferentially accumulate in certain tissues, many more small molecules that have inherent tissue-selective distribution profiles and are compatible with HDAC inhibition must be identified. This endeavor may be complicated by the fact that drug tissue distribution profiles are not one of the routine pharmacokinetic properties (adsorption, distribution, metabolism and excretion) investigated due to the relative difficulties of obtaining tissue samples [141]. As methods for analysis of biodistribution improve, more and more chemical entities will be unveiled to aid this approach. Meanwhile, a treasure trove of information that is accessible to researchers who maintain interest in tissue-selective drug accumulation, are supporting documents for several drugs currently approved by the FDA.
As nanotechnology comes of age, we speculate that targeted nanoparticle formulations of HDACis will answer some of the delivery problems associated with treating solid malignancies [142]. The technological innovations driving decreased expense will spur a dramatic increase in genetic and epigenetic screening, allowing more in-depth, routine and comprehensive correlations to be made in order to map the epigenetic landscape. HDACis will be key players in this arena, not only as personalized, targeted therapeutic agents, but also as tools to parse out an understanding of epigenetic states [143]. Many difficulties that accompany such a massive endeavor will be unburdened through advanced and globally integrated computing technologies for storing, accessing, automatically updating, and utilizing the seemingly intractable amount of genetic, epigenetic, proteomic and clinical information.
The gains so far recorded in HDACi therapy could not have come at a better time. The information gleaned from these advances will extend the reach of HDAC inhibition to other diseases likely in combination with other epigenetic modifiers, such as siRNA and inhibitors of DNA methylation, allowing for more precise control over the epigenetic program [144]. The future is bright for HDAC inhibition.
Executive summary.
Histone deacetylase inhibitors (HDACis) are an exciting new class of medicines, with broad applications, currently most notable in cancer.
Serious dose-limiting (and, therefore, efficacy-limiting) side effects need to be overcome, especially cardiac toxicity, although at high systemic concentrations other serious effects are expected.
-
Approaches for overcoming systemic toxicities and increasing potency against solid tumors include:
-
Target preference methodologies
Isoform selectivity, whereby newly designed or discovered HDACis are able to hit only one or a few of the 11 known HDACs.
Weakening hERG binding, whereby the cardiac toxicity may be limited by reducing efficacy for hERG without comprimising HDACi potency.
-
Selective delivery methodologies
Localized administration, whereby much higher concentrations of the drug are achieved at the site of action by topical drug application, intratumoral injection or by other means.
Targeted cap groups, whereby the structural and chemical flexibility of the HDACi surface recognition cap group is exploited to introduce ligands known to selectively accumulate within certain organs, tissue, cells or subcellular compartments.
-
Key Terms
- Chromatin
DNA coiled around histone proteins and compacted into highly ordered structures in the nucleus
- Histone acetyl transferase
Class of enzymes that add an acetyl group onto the tails of histone protieins
- Histone deacetylase
Class of enzymes that remove acetyl groups from the tails of histone proteins (and also other, non-histone protiens)
- Isoforms
Also known as or isozymes, are different forms of a protein or enzyme that all have a similar function, but may differ in subcellular location, subtrates, sequence, size, and shape
- Cutaneous T-cell lymphoma
Immune system malignancy involving, but not limited to, skin lesions
- hERG K+ channel
Ion channel involved in the electrical repolarization of the heart
- QT prolongation
Potentially fatal increased time interval of ventricular depolarization of the heart
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
The authors acknowledge support from the Georgia Institute of Technology, the Blanchard fellowship, and NIH Grant R01CA131217 (AK Oyelere). BE Gryder, QH Sodji are recipients of the GAANN predoctoral fellowship from the Georgia Tech Center for Drug Design, Development and Delivery. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
References
- 1.Mariotto AB, Robin Yabroff K, Shao Y, Feuer EJ, Brown ML. Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst. 2011;103(2):117–128. doi: 10.1093/jnci/djq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadikovic B, Al-Romaih K, Squire JA, Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics. 2008;9(6):394–408. doi: 10.2174/138920208785699580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang YW, Kuo CT, Stoner K, Huang THY, Wang LS. An overview of epigenetics and chemoprevention. FEBS Lett. 2011;585(13):2129–2136. doi: 10.1016/j.febslet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz WA, Hoffmann MJ. Transcription factor networks in embryonic stem cells and testicular cancer and the definition of epigenetics. Epigenetics. 2007;2(1):37–42. doi: 10.4161/epi.2.1.4067. [DOI] [PubMed] [Google Scholar]
- 5.Jagannathan V, Robinson-Rechavi M. The challenge of modeling nuclear receptor regulatory networks in mammalian cells. Mol Cell Endocrinol. 2011;334(1–2):91–97. doi: 10.1016/j.mce.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4(5):296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willingham AT, Gingeras TR. TUF love for ‘junk’ DNA. Cell. 2006;125(7):1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Fire AZ. Gene silencing by double-stranded RNA (Nobel lecture) Angew Chem Int Ed Engl. 2007;46(37):6967–6984. doi: 10.1002/anie.200701979. [DOI] [PubMed] [Google Scholar]
- 9.Blackledge NP, Klose RJ. CpG island chromatin A platform for gene regulation. Epigenetics. 2011;6(2):147–152. doi: 10.4161/epi.6.2.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frederiks F, Stulemeijer IJE, Ovaa H, Van Leeuwen F. A modified epigenetics toolbox to study histone modifications on the nucleosome core. ChemBioChem. 2011;12(2):308–313. doi: 10.1002/cbic.201000617. [DOI] [PubMed] [Google Scholar]
- 11.Gunjan A, Singh RK. Epigenetic therapy: targeting histones and their modifications in human disease. Future Med Chem. 2010;2(4):543–548. doi: 10.4155/fmc.10.18. [DOI] [PubMed] [Google Scholar]
- 12.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 13.Dorier J, Stasiak A. The role of transcription factories-mediated interchromosomal contacts in the organization of nuclear architecture. Nucleic Acids Res. 2010;38(21):7410–7421. doi: 10.1093/nar/gkq666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8(2):104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 15.Chapman-Rothe N, Brown R. Approaches to target the genome and its epigenome in cancer. Future Med Chem. 2009;1(8):1481–1495. doi: 10.4155/fmc.09.105. [DOI] [PubMed] [Google Scholar]
- 16.Garber K. HDAC inhibitors overcome first hurdle. Nat Biotechnol. 2007;25(1):17–19. doi: 10.1038/nbt0107-17. [DOI] [PubMed] [Google Scholar]
- 17.Nalabothula N, Carrier F. Cancer cells’ epigenetic composition and predisposition to histone deacetylase inhibitor sensitization. Epigenomics. 2011;3(2):145–155. doi: 10.2217/epi.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradner JE, West N, Grachan ML, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6(3):238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Hart SRL, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38(1):32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 21.Brunmeir R, Lagger S, Seiser C. Histone deacetylase 1 and 2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53(2–3):275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 23.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7(10):854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 24.Georgopoulos K. From immunity to tolerance through HDAC. Nat Immunol. 2009;10(1):13–14. doi: 10.1038/ni0109-13. [DOI] [PubMed] [Google Scholar]
- 25.Karpac J, Jasper H. Metabolic homeostasis: HDACs take center stage. Cell. 2011;145(4):497–499. doi: 10.1016/j.cell.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 27.Verdin E, Dequiedt F, Kasler HG. Class 2 histone deacetylases: versatile regulators. Trends Genet. 2003;19(5):286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 28.Bieliauskas AV, Pflum MKH. Isoform-selective histone deacetylase inhibitors. Chem Soc Rev. 2008;37(7):1402–1413. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burton SG, Dorrington RA. Hydantoin-hydrolysing enzymes for the enantioselective production of amino acids: new insights and applications. Tetrahedron Asymmetry. 2004;15(18):2737–2741. [Google Scholar]
- 30.Wang DF, Helquist P, Wiech NL, Wiest O. Toward selective histone deacetylase inhibitor design: homology modeling, docking studies, and molecular dynamics simulations of human Class 1 histone deacetylases. J Med Chem. 2005;48(22):6936–6947. doi: 10.1021/jm0505011. [DOI] [PubMed] [Google Scholar]
- 31.Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132(31):10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26(9):1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 33.Grant C, Rahman F, Piekarz R, et al. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther. 2010;10(7):997–1008. doi: 10.1586/era.10.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller TA, Witter DJ, Belvedere S. Histone deacetylase inhibitors. J Med Chem. 2003;46(24):5097–5116. doi: 10.1021/jm0303094. [DOI] [PubMed] [Google Scholar]
- 35.Mwakwari SC, Patil V, Guerrant W, Oyelere AK. Macrocyclic histone deacetylase inhibitors. Curr Top Med Chem. 2010;10(14):1423–1440. doi: 10.2174/156802610792232079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patil V, Guerrant W, Chen PC, et al. Anti-malarial and antileishmanial activities of histone deacetylase inhibitors with triazole-linked cap group. Bioorg Med Chem. 2010;18(1):415–425. doi: 10.1016/j.bmc.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marson CM. Histone deacetylase inhibitors: design, structure-activity relationships and therapeutic implications for cancer. Anticancer Agents Med Chem. 2009;9(6):661–692. doi: 10.2174/187152009788679976. [DOI] [PubMed] [Google Scholar]
- 38.Rajak H, Agarawal A, Parmar P, et al. 2,5-Disubstituted-1,3,4-oxadiazoles/thiadiazole as surface recognition moiety: design and synthesis of novel hydroxamic acid based histone deacetylase inhibitors. Bioorg Med Chem Lett. 2011;21(19):5735–5738. doi: 10.1016/j.bmcl.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Zhang YJ, Feng JH, Liu CX, et al. Design, synthesis and preliminary activity assay of 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid derivatives as novel Histone deacetylases (HDACs) inhibitors. Bioorg Med Chem Lett. 2010;18(5):1761–1772. doi: 10.1016/j.bmc.2010.01.060. [DOI] [PubMed] [Google Scholar]
- 40.Canzoneri JC, Chen PC, Oyelere AK. Design and synthesis of novel histone deacetylase inhibitor derived from nuclear localization signal peptide. Bioorg Med Chem Lett. 2009;19(23):6588–6590. doi: 10.1016/j.bmcl.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Design. 2008;14(6):505–528. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- 42.Shultz MD, Cao XY, Chen CH, et al. Optimization of the in vitro cardiac safety of hydroxamate-based histone deacetylase inhibitors. J Med Chem. 2011;54(13):4752–4772. doi: 10.1021/jm200388e. [DOI] [PubMed] [Google Scholar]
- 43.Mwakwari SC, Guerrant W, Patil V, et al. Non-peptide macrocyclic histone deacetylase inhibitors derived from tricyclic ketolide skeleton. J Med Chem. 2010;53(16):6100–6111. doi: 10.1021/jm100507q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rotili D, Simonetti G, Savarino A, Palamara AT, Migliaccio AR, Mai A. Non-cancer uses of histone deacetylase inhibitors: effects on infectious diseases and β-hemoglobinopathies. Curr Top Med Chem. 2009;9(3):272–291. doi: 10.2174/156802609788085296. [DOI] [PubMed] [Google Scholar]
- 45.Gilbert J, Baker SD, Bowling MK, et al. A Phase 1 dose escalation and bioavailability study of oral sodium phenylbutyrate in patients with refractory solid tumor malignancies. Clin Cancer Res. 2001;7(8):2292–2300. [PubMed] [Google Scholar]
- 46.Sandor V, Bakke S, Robey RW, et al. Phase 1 trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res. 2002;8(3):718–728. [PubMed] [Google Scholar]
- 47.Porcu P, Wong HK. We should have a dream: unlocking the workings of the genome in cutaneous T-cell lymphomas. Clin Lymphoma Myeloma. 2009;9(6):409–411. doi: 10.3816/CLM.2009.n.081. [DOI] [PubMed] [Google Scholar]
- 48.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12(10):1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 49.Olsen EA, Kim YH, Kuzel TM, et al. Phase 2B multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25(21):3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 50.Meyer N, Paul C, Misery L. Pruritus in cutaneous T-cell lymphomas: frequent, often severe and difficult to treat. Acta Dermatol Venereologica. 2010;90(1):12–17. doi: 10.2340/00015555-0789. [DOI] [PubMed] [Google Scholar]
- 51.Duvic M, Talpur R, Ni X, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109(1):31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piekarz RL, Frye R, Turner M, et al. Phase 2 multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27(32):5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whittaker SJ, Demierre MF, Kim EJ, et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol. 2010;28(29):4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 54.Vansteenkiste J, Van Cutsem E, Dumez H, et al. Early Phase 2 trial of oral vorinostat in relapsed or refractory breast, colorectal, or non-small cell lung cancer. Invest N Drugs. 2008;26(5):483–488. doi: 10.1007/s10637-008-9131-6. [DOI] [PubMed] [Google Scholar]
- 55.Woyach JA, Kloos RT, Ringel MD, et al. Lack of therapeutic effect of the histone deacetylase inhibitor vorinostat in patients with metastatic radioiodine-refractory thyroid carcinoma. J Clin Endocrinol Metab. 2009;94(1):164–170. doi: 10.1210/jc.2008-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stadler WM, Margolin K, Ferber S, Mcculloch W, Thompson JA. A Phase 2 study of depsipeptide in refractory metastatic renal cell cancer. Clin Genitourin Cancer. 2006;5(1):57–60. doi: 10.3816/CGC.2006.n.018. [DOI] [PubMed] [Google Scholar]
- 57.Whitehead R, Rankin C, Hoff P, et al. Phase 2 trial of romidepsin (NSC-630176) in previously treated colorectal cancer patients with advanced disease: a Southwest oncology group study (S0336) Invest New Drugs. 2009;27(5):469–475. doi: 10.1007/s10637-008-9190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinet N, Bertrand P. Interpreting clinical assays for histone deacetylase inhibitors. Cancer Manage Res. 2011;3(1):117–141. doi: 10.2147/CMR.S9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strevel EL, Ing DJ, Siu LL. Molecularly targeted oncology therapeutics and prolongation of the QT interval. J Clin Oncol. 2007;25(22):3362–3371. doi: 10.1200/JCO.2006.09.6925. [DOI] [PubMed] [Google Scholar]
- 60.Rasheed WK, Johnstone RW, Prince HM. Histone deacetylase inhibitors in cancer therapy. Expert Opin Invest Drugs. 2007;16(5):659–678. doi: 10.1517/13543784.16.5.659. [DOI] [PubMed] [Google Scholar]
- 61.Tan J, Cang S, Ma Y, Petrillo R, Liu D. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J Hematol Oncol. 2010;3(1):5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Federico M, Bagella L. Histone deacetylase inhibitors in the treatment of hematological malignancies and solid tumors. J Biomed Biotechnol. 2011 doi: 10.1155/2011/475641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolbrette D. Drugs that cause torsades de pointes and increase the risk of sudden cardiac death. Curr Cardiol Rep. 2004;6(5):379–384. doi: 10.1007/s11886-004-0041-8. [DOI] [PubMed] [Google Scholar]
- 64.Bates SE, Rosing DR, Fojo T, Piekarz RL. Challenges of evaluating the cardiac effects of anticancer agents. Clin Cancer Res. 2006;12(13):3871–3874. doi: 10.1158/1078-0432.CCR-06-1017. [DOI] [PubMed] [Google Scholar]
- 65.Shah MH, Binkley P, Chan K, et al. Cardiotoxicity of histone deacetylase inhibitor depsipeptide in patients with metastatic neuroendocrine tumors. Clin Cancer Res. 2006;12(13):3997–4003. doi: 10.1158/1078-0432.CCR-05-2689. [DOI] [PubMed] [Google Scholar]
- 66.Ponte ML, Keller GA, Girolamo GD. Mechanisms of drug induced QT interval prolongation. Curr Drug Saf. 2010;5(1):44–53. doi: 10.2174/157488610789869247. [DOI] [PubMed] [Google Scholar]
- 67.Hoffmann P, Warner B. Are hERG channel inhibition and QT interval prolongation all there is in drug-induced torsadogenesis? A review of emerging trends. J Pharmacol Toxicol Methods. 2006;53(2):87–105. doi: 10.1016/j.vascn.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Guo J, Li X, Shallow H, et al. Involvement of caveolin in probucol-induced reduction in hERG plasma-membrane expression. Mol Pharmacol. 2011;79(5):806–813. doi: 10.1124/mol.110.069419. [DOI] [PubMed] [Google Scholar]
- 69.Perrin MJ, Subbiah RN, Vandenberg JI, Hill AP. Human ether-a-go-go related gene (hERG) K+ channels: function and dysfunction. Prog Biophys Mol Biol. 2008;98(2–3):137–148. doi: 10.1016/j.pbiomolbio.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 70.Lacerda AE, Kuryshev YA, Chen Y, et al. Alfuzosin delays cardiac repolarization by a novel mechanism. J Pharmacol Exp Ther. 2008;324(2):427–433. doi: 10.1124/jpet.107.128405. [DOI] [PubMed] [Google Scholar]
- 71.Kupershmidt S, Yang T, Anderson ME, et al. Replacement by homologous recombination of the minK gene with lacZ reveals restriction of minK expression to the mouse cardiac conduction system. Circulation Res. 1999;84(2):146–152. doi: 10.1161/01.res.84.2.146. [DOI] [PubMed] [Google Scholar]
- 72.Abbott GW, Sesti F, Splawski I, et al. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97(2):175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 73.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress lKs function. Nat Genet. 1997;17(3):338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 74.Munster PN, Rubin EH, Van Belle S, et al. A single supratherapeutic dose of vorinostat does not prolong the QTc interval in patients with advanced cancer. Clin Cancer Res. 2009;15(22):7077–7084. doi: 10.1158/1078-0432.CCR-09-1214. [DOI] [PubMed] [Google Scholar]
- 75.Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kerr JS, Galloway S, Lagrutta A, et al. Nonclinical safety assessment of the histone deacetylase inhibitor vorinostat. Int J Toxicol. 2010;29(1):3–19. doi: 10.1177/1091581809352111. [DOI] [PubMed] [Google Scholar]
- 77.Razak ARA, Hotte SJ, Siu LL, et al. Phase 1 clinical, pharmacokinetic and pharmacodynamic study of SB939, an oral histone deacetylase (HDAC) inhibitor, in patients with advanced solid tumours. Br J Cancer. 2011;104(5):756–762. doi: 10.1038/bjc.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, Yu N, Chen D, et al. Discovery of (2E)-3-{2-Butyl-1-[2-(diethylamino)ethyl]-1H-benzimidazol-5-yl}-N-hydroxyacrylamide (SB939), an orally active histone deacetylase inhibitor with a superior preclinical profile. J Med Chem. 2011;54(13):4694–4720. doi: 10.1021/jm2003552. [DOI] [PubMed] [Google Scholar]
- 79.Brana I, Tabernero J. Cardiotoxicity. Annu Oncol. 2010;21(Suppl 7):vii173–vii179. doi: 10.1093/annonc/mdq295. [DOI] [PubMed] [Google Scholar]
- 80.Roden DM. Drug-induced prolongation of the QT interval. N Engl J Med. 2004;350(10):1013–1022. doi: 10.1056/NEJMra032426. [DOI] [PubMed] [Google Scholar]
- 81.Barbey JT, Pezzullo JC, Soignet SL. Effect of arsenic trioxide on QT interval in patients with advanced malignancies. J Clin Oncol. 2003;21(19):3609–3615. doi: 10.1200/JCO.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 82.Ederhy S, Cohen A, Dufaitre G, et al. QT interval prolongation among patients treated with angiogenesis inhibitors. Targeted Oncol. 2009;4(2):89–97. doi: 10.1007/s11523-009-0111-3. [DOI] [PubMed] [Google Scholar]
- 83.De Ponti F, Poluzzi E, Montanaro N, Ferguson J. QTc and psychotropic drugs. Lancet. 2000;356(9223):75–76. doi: 10.1016/S0140-6736(05)73412-7. [DOI] [PubMed] [Google Scholar]
- 84.De Ponti F, Poluzzi E, Montanaro N. Organising evidence on QT prolongation and occurrence of Torsades de Pointes with non-antiarrhythmic drugs: a call for consensus. Eur J Clin Pharmacol. 2001;57(3):185–209. doi: 10.1007/s002280100290. [DOI] [PubMed] [Google Scholar]
- 85.Hann D, Winter K, Jacobsen P. Measurement of depressive symptoms in cancer patients: Evaluation of the center for epidemiological studies depression scale (Ces-d) J Psychosomatic Res. 1999;46(5):437–443. doi: 10.1016/s0022-3999(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 86.Ijaz AK. Clinical and therapeutic aspects of congenital and acquired long QT syndrome. Am J Med. 2002;112(1):58–66. doi: 10.1016/s0002-9343(01)01011-7. [DOI] [PubMed] [Google Scholar]
- 87.Sawyers C. Targeted cancer therapy. Nature. 2004;432(7015):294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 88.Khan N, Jeffers M, Kumar S, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409(2):581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 89.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009;280(2):211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 90.Jones P, Altamura S, De Francesco A, et al. Probing the elusive catalytic activity of vertebrate Class 2a histone deacetylases. Bioorg Med Chem Lett. 2008;18(6):1814–1819. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 91.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;280(2):168–176. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 92.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDACI in hormone refractory prostate cancer. Prostate. 2004;59(2):177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 93.Barlesi F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non-small-cell lung cancer. J Clin Oncol. 2007;25(28):4358–4364. doi: 10.1200/JCO.2007.11.2599. [DOI] [PubMed] [Google Scholar]
- 94.Huang BH, Laban M, Leung CHW, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21(Cip1/WAF1) expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12(4):395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 95.Fournel M, Bonfils C, Hou Y, et al. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol Cancer Ther. 2008;7(4):759–768. doi: 10.1158/1535-7163.MCT-07-2026. [DOI] [PubMed] [Google Scholar]
- 96.Henning SW, Doblhofer R, Kohlhof H, et al. Preclinical characterization of 4SC-202, a novel isotype specific HDAC inhibitor. Eur J Cancer Suppl. 2010;8(7):61. [Google Scholar]
- 97.Ryan QC, Headlee D, Acharya M, et al. Phase 1 and pharmacokinetic study of MS-275, a histone deacetylase inhibitor, in patients with advanced and refractory solid tumors or lymphoma. J Clin Oncol. 2005;23(17):3912–3922. doi: 10.1200/JCO.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 98.Kummar S, Gutierrez M, Gardner ER, et al. Phase 1 trial of MS-275, a histone deacetylase inhibitor, administered weekly in refractory solid tumors and lymphoid malignancies. Clin Cancer Res. 2007;13(18):5411–5417. doi: 10.1158/1078-0432.CCR-07-0791. [DOI] [PubMed] [Google Scholar]
- 99.Gore L, Rothenberg ML, O’Bryant CL, et al. A Phase 1 and pharmacokinetic study of the oral histone deacetylase inhibitor, MS-275, in patients with refractory solid tumors and lymphomas. Clin Cancer Res. 2008;14(14):4517–4525. doi: 10.1158/1078-0432.CCR-07-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gojo I, Jiemjit A, Trepel JB, et al. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood. 2007;109(7):2781–2790. doi: 10.1182/blood-2006-05-021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hauschild A, Trefzer U, Garbe C, et al. Multicenter Phase 2 trial of the histone deacetylase inhibitor pyridylmethyl-N-{4-[(2-aminophenyl)-carbamoyl]-benzyl}-carbamate in pretreated metastatic melanoma. Melanoma Res. 2008;18(4):274–278. doi: 10.1097/CMR.0b013e328307c248. [DOI] [PubMed] [Google Scholar]
- 102.Blum KA, Advani A, Fernandez L, et al. Phase 2 study of the histone deacetylase inhibitor MGCD0103 in patients with previously treated chronic lymphocytic leukaemia. Br J Haematol. 2009;147(4):507–514. doi: 10.1111/j.1365-2141.2009.07881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siu LL, Pili R, Duran I, et al. Phase 1 study of MGCD0103 given as a three-times-per-week oral dose in patients with advanced solid tumors. J Clin Oncol. 2008;26(12):1940–1947. doi: 10.1200/JCO.2007.14.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cole KE, Dowling DP, Boone MA, Phillips AJ, Christianson DW. Structural basis of the antiproliferative activity of largazole, a depsipeptide inhibitor of the histone deacetylases. J Am Chem Soc. 2011;133(32):12474–12477. doi: 10.1021/ja205972n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene. 2007;26(37):5468–5476. doi: 10.1038/sj.onc.1210614. [DOI] [PubMed] [Google Scholar]
- 106.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277(1):8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 107.Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J Biomed Biotechnol. 2011 doi: 10.1155/2011/875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100(8):4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Auzzas L, Larsson A, Matera R, et al. Non-natural macrocyclic inhibitors of histone deacetylases: design, synthesis, and activity. J Med Chem. 2010;53(23):8387–8399. doi: 10.1021/jm101092u. [DOI] [PubMed] [Google Scholar]
- 110.Choi SE, Weerasinghe SVW, Pflum MKH. The structural requirements of histone deacetylase inhibitors: suberoylanilide hydroxamic acid analogs modified at the C3 position display isoform selectivity. Bioorg Med Chem Lett. 2011;21(20):6139–6142. doi: 10.1016/j.bmcl.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Santo L, Hideshima T, Kung AL, et al. Selective inhibition of HDAC6 with a new prototype inhibitor (ACY-1215) overcomes bortezomib resistance in multiple myeloma (MM) ASH Annu Meet Abstracts. 2010;116(21):2997. [Google Scholar]
- 112.Waltregny D, Glenisson W, Tran SL, et al. Histone deacetylase HDAC8 associates with smooth muscle α-actin and is essential for smooth muscle cell contractility. FASEB J. 2005;19(8):966–968. doi: 10.1096/fj.04-2303fje. [DOI] [PubMed] [Google Scholar]
- 113.Oehme I, Deubzer HE, Wegener D, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15(1):91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 114.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22(5):1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 115.Krennhrubec K, Marshall BL, Hedglin M, Verdin E, Ulrich SM. Design and evaluation of ‘Linkerless’ hydroxamic acids as selective HDAC8 inhibitors. Bioorg Med Chem Lett. 2007;17(10):2874–2878. doi: 10.1016/j.bmcl.2007.02.064. [DOI] [PubMed] [Google Scholar]
- 116.Tang WP, Luo TP, Greenberg EF, Bradner JE, Schreiber SL. Discovery of histone deacetylase 8 selective inhibitors. Bioorg Med Chem Lett. 2011;21(9):2601–2605. doi: 10.1016/j.bmcl.2011.01.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Whitehead L, Dobler MR, Radetich B, et al. Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorg Med Chem. 2011;19(15):4626–4634. doi: 10.1016/j.bmc.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 118.Haberland M, Johnson A, Mokalled MH, Montgomery RL, Olson EN. Genetic dissection of histone deacetylase requirement in tumor cells. Proc Natl Acad Sci USA. 2009;106(19):7751–7755. doi: 10.1073/pnas.0903139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Khan N, Jeffers M, Kumar S, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409(2):581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 120.Shultz M, Fan J, Chen C, et al. The design, synthesis and structure–activity relationships of novel isoindoline-based histone deacetylase inhibitors. Bioorg Med Chem Lett. 2011;21(16):4909–4912. doi: 10.1016/j.bmcl.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 121.Jehle J, Schweizer PA, Katus HA, Thomas D. Novel roles for hERG K(+) channels in cell proliferation and apoptosis. Cell Death Dis. 2011;2:e193. doi: 10.1038/cddis.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mahvi DM, Henry MB, Albertini MR, et al. Intratumoral injection of IL-12 plasmid DNA – results of a Phase 1/1B clinical trial. Cancer Gene Ther. 2007;14(8):717–723. doi: 10.1038/sj.cgt.7701064. [DOI] [PubMed] [Google Scholar]
- 123.Botti E, Mercurio C, Spallone G, et al. Histone deacetylases as new therapeutical targets for the treatment of non-melanoma skin cancer: results of Phase 1/2a trial with topical DAC060. J Invest Dermatol. 2011;131:S46–S46. [Google Scholar]
- 124.De Souza R, Zahedi P, Allen CJ, Piquette-Miller M. Polymeric drug delivery systems for localized cancer chemotherapy. Drug Deliv. 2010;17(6):365–375. doi: 10.3109/10717541003762854. [DOI] [PubMed] [Google Scholar]
- 125.Government of the United States of America, represented by the Secretary, Department of Health and Human Services, topical formulations of histone deacetylase inhibitors and methods using the same. 20080292616A1. US. 2008
- 126.Virasi M, Thaler F, Abate A, et al. Discovery, synthesis, and pharmacological evaluation of spiropiperidine hydroxamic acid based derivatives as structurally novel histone deacetylase (HDAC) inhibitors. J Med Chem. 2011;54(8):3051–3064. doi: 10.1021/jm200146u. [DOI] [PubMed] [Google Scholar]
- 127.Thaler F, Colombo A, Mai A, et al. Synthesis and biological evaluation of N-hydroxyphenylacrylamides and N-hydroxypyridin-2-ylacrylamides as novel histone deacetylase inhibitors. J Med Chem. 2009;53(2):822–839. doi: 10.1021/jm901502p. [DOI] [PubMed] [Google Scholar]
- 128.Bradner J, Greenberg E, Ponduru S, et al. A soft-drug histone deacetylase inhibitor for cutaneous T-cell lymphoma. ASH Annu Meet Abstracts. 2007;110(11):800. [Google Scholar]
- 129.Tong W, Stevenson W, Cortes J, et al. In vitro and in vivo anti-leukemia activity of CHR-2845, a cell-targeted HDAC inhibitor for use in monocytic leukemia. ASCO Meet Abstracts. 2009;27(15S):e14579. [Google Scholar]
- 130.Ossenkoppele G, Lowenberg B, Zachee P, et al. CHR-2845, a monocyte/macrophage targeted histone deacetylase inhibitor in a first in man clinical trial in subjects with advanced haematological malignancies. ASH Annu Meet Abstracts. 2010;116(21):3279. [Google Scholar]
- 131.Hoepelman IM, Schneider MME. Azithromycin – the first of the tissue-selective azalides. Int J Antimicrob Agents. 1995;5(3):145–167. doi: 10.1016/0924-8579(95)00009-w. [DOI] [PubMed] [Google Scholar]
- 132.Ono T, Kashimura M, Suzuki K, et al. In vitro and in vivo antibacterial activities of the tricyclic ketolide TE-802 and its analogs. J Antibiot. 2004;57(8):518–527. doi: 10.7164/antibiotics.57.518. [DOI] [PubMed] [Google Scholar]
- 133.Togami K, Chono S, Seki T, Morimoto K. Distribution characteristics of telithromycin, a novel ketolide antimicrobial agent applied for treatment of respiratory infection, in lung epithelial lining fluid and alveolar macrophages. Drug Metab Pharmacokinet. 2009;24(5):411–417. doi: 10.2133/dmpk.24.411. [DOI] [PubMed] [Google Scholar]
- 134.Di Paolo A, Barbara C, Chella A, Angeletti CA, Del Tacca M. Pharmacokinetics of azithromycin in lung tissue, bronchial washing, and plasma in patients given multiple oral doses of 500 and 1000 mg daily. Pharmacol Res. 2002;46(6):545–550. doi: 10.1016/s1043661802002384. [DOI] [PubMed] [Google Scholar]
- 135.Patel KB, Xuan DW, Tessier PR, Russomanno JH, Quintiliani R, Nightingale CH. Comparison of bronchopulmonary pharmacokinetics of clarithromycin and azithromycin. Antimicrob Agents Chemother. 1996;40(10):2375–2379. doi: 10.1128/aac.40.10.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rodvold KA. Clinical pharmacokinetics of clarithromycin. Clin Pharmacokinet. 1999;37(5):385–398. doi: 10.2165/00003088-199937050-00003. [DOI] [PubMed] [Google Scholar]
- 137.Oyelere AK, Chen PC, Guerrant W, et al. Non-peptide macrocyclic histone deacetylase inhibitors. J Med Chem. 2009;52(2):456–468. doi: 10.1021/jm801128g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bottomley MJ, Lo Surdo P, Di Giovine P, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008;283(39):26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schuetz A, Min J, Allali-Hassani A, et al. Human HDAC7 harbors a Class 2a histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J Biol Chem. 2008;283(17):11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang HS, Dymock BW. New patented histone deacetylase inhibitors. Expert Opin Ther Patents. 2009;19(12):1727–1757. doi: 10.1517/13543770903393789. [DOI] [PubMed] [Google Scholar]
- 141.Lin JH. Tissue distribution and pharmacodynamics: a complicated relationship. Curr Drug Metab. 2006;7(1):39–65. doi: 10.2174/138920006774832578. [DOI] [PubMed] [Google Scholar]
- 142.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7(9):771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 143.Schilsky RL. Personalized medicine in oncology: the future is now. Nat Rev Drug Discov. 2010;9(5):363–366. doi: 10.1038/nrd3181. [DOI] [PubMed] [Google Scholar]
- 144.Vucic EA, Brown CJ, Lam WL. Epigenetics of cancer progression. Pharmacogenomics. 2008;9(2):215–234. doi: 10.2217/14622416.9.2.215. [DOI] [PubMed] [Google Scholar]
- 145.Ruthenburg AJ, Li HT, Milne TA, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145(5):692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 201.National Cancer Institute. [Accessed 11 October 2011];Topical romidepsin in treating patients with Stage 1 or Stage 2 cutaneous T-cell non-Hodgkin’s lymphoma. http://clinicaltrials.gov/show/NCT00477698 NLM Identifier: NCT00477698.
- 202.Shape Pharmaceuticals Inc. [Accessed 11 October 2011];Safety, Pharmacodynamics (PD), Pharmacokinetics (PK) Study of SHP141 in 1A, 1B, or 2A Cutaneous T-Cell Lymphoma (CTCL) http://clinicaltrials.gov/show/NCT01433731 NLM Identifier: NCT01433731.