SUMMARY
Determining the cellular source of new skeletal elements is critical for understanding appendage regeneration in amphibians and fish. Recent lineage-tracing studies indicated that zebrafish fin ray bone regenerates through the de-differentiation and proliferation of spared osteoblasts, with limited if any contribution from other cell types. Here, we examined the requirement for this mechanism by using genetic ablation techniques to destroy virtually all skeletal osteoblasts in adult zebrafish fins. Animals survived this injury and restored the osteoblast population within two weeks. Moreover, amputated fins depleted of osteoblasts regenerated new fin ray structures at rates indistinguishable from fins possessing a resident osteoblast population. Inducible genetic fate-mapping confirmed that new bone cells do not arise from de-differentiated osteoblasts under these conditions. Our findings demonstrate diversity in the cellular origins of appendage bone, and reveal that de novo osteoblasts can fully support the regeneration of amputated zebrafish fins.
INTRODUCTION
After amputation of an appendage in certain salamanders and fish, new cartilage or bone structures of correct size and pattern emerge from a mound of proliferative tissue called the blastema (Brockes and Kumar, 2005). A major objective in the field has been to define the cellular source(s) of regenerated skeletal elements. This includes identifying cell types within the appendage stump that normally give rise to regenerated cartilage or bone after amputation, as well as identifying cells that have the developmental capacity to create skeleton under additional conditions (Poss, 2010; Tanaka and Reddien, 2011). Proposed sources are the differentiated chondrocytes and osteoblasts themselves, or non-skeletal cells that undergo new differentiation or trans-differentiation events after amputation.
Grafting experiments in amphibians performed over the past century have attempted to resolve this issue. Surgical transplantation of dissected cartilage or bone indicated that skeletal tissues wholly or predominantly contribute like tissue, suggesting that lineage is restricted throughout blastema formation and patterning (Namenwirth, 1974; Steen, 1968; Steen, 1970). Yet, other experiments, including the transplantation of dye-labeled muscle cells to limb blastemas or non-skeletal tissue to irradiated limbs, indicated that additional cell types may act as progenitors for bone or cartilage (Lo et al., 1993; Morrison et al., 2006). A recent study of axolotl limb regeneration examined the contributions of tissues grafted from transgenic animals constitutively expressing a fluorescent reporter protein. These experiments generated the prevailing model for axolotl limb cartilage regeneration, which is that cartilage cells predominantly contribute like tissue, while one or more cell populations within the dermis also has the potential to form cartilage (Kragl et al., 2009).
Tissue grafts can be ineffective at resolving certain key questions of tissue origin, such as: 1) how host tissue naturally participates in regeneration; 2) the extent to which specific cell types contribute during regeneration; and 3) whether cells in the stump undergo developmental changes like de-differentiation in the process of creating new structures. Very recently, three studies examined similar questions during fin regeneration in zebrafish by genetic lineage-tracing of specific cell types. Adult zebrafish fins contain several cylinder-shaped, segmented bony fin rays that are lined by osteoblasts and encase fibroblasts, blood vessels, nerves, and pigment cells. By inducible fate-mapping of cells expressing the intermediate osteoblast marker osterix, Knopf and colleagues found that existing osteoblasts undergo partial de-differentiation, as defined by reduced expression of osteoblast markers, after which they proliferate and contribute solely to regenerated bone structures (Knopf et al., 2011). Tu and Johnson assessed the mosaicism of transgenes injected into embryos during rapid cell division, and found that transgenic clones containing labeled osteoblasts within regenerated fins do not possess other cell types (Tu and Johnson, 2011). Sousa and colleagues used live imaging of labeled osteocalcin-expressing cells to indicate contribution of differentiated osteoblasts to the regenerate (Sousa et al., 2011). Together, these studies supported a common conception that osteoblasts in the regenerate derive predominantly or wholly from the de-differentiation, proliferation, and migration of lineage-restricted stump osteoblasts.
Here, by creating a system to facilitate inducible ablation of resident osteoblasts in adult fins, we examined the extent to which zebrafish fin regeneration is dependent on these cells. We found by lineage-tracing of existing osteoblasts that they are restricted to contributing like cells during regeneration, in agreement with recent published work. Unexpectedly however, ablation of ostensibly all osteoblasts prior to amputation did not slow down the rate of zebrafish fin regeneration. Instead, new osteoblasts arose from cells that differentiated de novo after amputation, a result confirmed by genetic fate-mapping. Our findings indicate that stump osteoblasts are a dispensable source for regenerating appendage bone, and provide important new context for understanding mechanisms of robust skeletal regeneration.
RESULTS
Lineage-Restricted Contributions by Osteoblasts During Zebrafish Fin Regeneration
To identify contributions by osteoblasts after zebrafish fin amputation, we used an inducible genetic fate-mapping approach. We screened several candidate genes for bone-specific expression as a prerequisite for genetic fate-mapping. osterix (also known as sp7) is a zinc finger transcription factor whose expression is first seen during intermediate stages of osteoblast differentiation (Li et al., 2009; Renn and Winkler, 2009). osteocalcin (also known as bglap), expressed by mature osteoblasts, has been used as a marker of terminal osteogenesis (Inohaya et al., 2007). We generated transgenic reporter lines to visualize the activity of the teleost osterix and osteocalcin regulatory sequences. Tg(osterix:mCherry)pd43 (osx:mCherry) and Tg(osteocalcin:EGFP)pd44 (osc:EGFP) each showed osteoblast-specific fluorescence in uninjured adult zebrafish fins that was excluded from medially located fibroblasts and from epidermis. osx:mCherry visualized a larger pool of osteoblasts than osc:EGFP (Figure S1A–D). Regenerated osteoblasts labeled by osterix-driven mCherry expression could be detected as early as 2 days post-amputation (dpa), while osteocalcin-driven EGFP expression was not detectable until 7 dpa (Figure S1E–H).
To fate-map differentiated osteoblasts, we generated a transgenic line with osterix regulatory sequences driving a tamoxifen-inducible Cre recombinase-Estrogen receptor fusion protein Tg(osterix:mTagBFP-2A-CreER)pd45 (osx:CreER) (Figure S1I). An indicator line, Tg(bactin2-Lox-DsRed-STOP-Lox-EGFP)s928 (β-act2:RSG) permitted visualization of EGFP fluorescence after Cre-mediated excision of loxP-flanked stop sequences, and was expressed in adult osteoblasts, intraray fibroblasts, and epidermis (Figure S1J) (Kikuchi et al., 2010). To label osteoblasts, we incubated osx:CreER; β-act2:RSG animals with 4-hydroxytamoxifen (4-HT) or vehicle for one day (Figure 1A). Within 2 days, EGFP+ cells were visible lining the osteoblast compartment in fin rays of animals treated with 4-HT (Figure 1B–E). We did not observe EGFP+ cells in intraray fibroblasts, located medially to osteoblasts in longitudinal fin sections. These data indicate that osx:CreER inducibly and specifically labels osteoblasts.
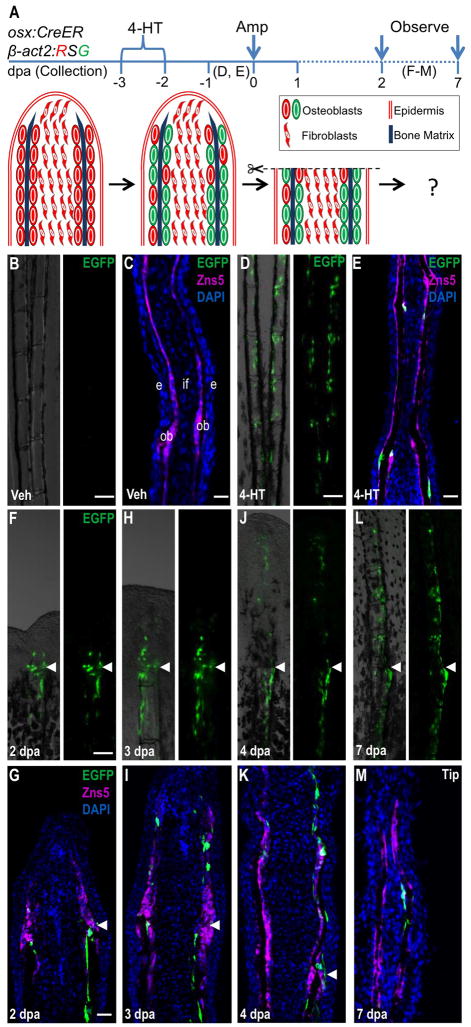
Figure 1. Resident Osteoblasts Contribute New Osteoblasts to Regenerating Fin Structures.
(A) Cartoon summarizing strategy for inducible, genetic fate-mapping of osteoblasts during zebrafish fin regeneration. 4-HT treatment labels osterix-expressing cells with EGFP prior to amputation.
(B, C) Uninjured osx:CreER; β-act2:RSG fins, shown as whole mount (B) and in a longitudinal section (C), display no labeling after vehicle treatment. Zns5 (magenta) is an uncharacterized antigen that helps identify osteoblasts lining hemiray bone (Johnson and Weston, 1995). This antibody stains cell membranes and visualizes as non-contiguous staining in sections. (C) The longitudinal fin section is labeled to show structures: intraray fibroblasts (if), osteoblasts (ob), and epidermis (e).
(D, E) 4-HT treatment labels many osteoblasts with EGFP in uninjured fins, shown as a whole-mount image (D) and a longitudinal section (E).
(F–M) EGFP+ osteoblasts labeled by 4-HT treatment prior to fin amputation contribute labeled progeny to the regenerate, visualized by whole-mount images and in sections at 2 (F, G), 3 (H, I), 4 (J, K), and 7 (L, M) dpa. EGFP fluorescence proximal and distal to the amputation plane is restricted to the osteoblast compartment and is not present in intraray fibroblasts or epidermis. Arrowheads indicate the plane of amputation. Scale bars = 100 μm.
To examine the contribution of osx-expressing cells to the regenerate, we amputated the caudal fins of zebrafish 2 days after 4-HT treatment (Figure 1A). EGFP+ cells were detected in regenerating fins from 2 dpa onwards, a result that indicated contribution from osteoblasts within the stump. As regeneration progressed to 3, 4 and 7 dpa, the domain of EGFP+ expression expanded distally within the regenerate. Confocal analysis of fin sections at 2, 3, 4, and 7 dpa revealed EGFP+ cells confined to regions lining bone matrix both below and above the amputation plane, indicating that a population of osteoblasts in the regenerate is derived from stump osteoblasts. No EGFP+ expression was observed in intraray fibroblasts or other cell types (Figure 1F–M). Thus, in agreement with similar experiments published recently (Knopf et al., 2011), our data support a mechanism in which contributions of osterix-expressing cells are restricted to the osteoblast lineage during zebrafish fin regeneration.
Genetic Ablation and Recovery of Zebrafish Osteoblasts
Although these data indicated that spared osteoblasts contribute new bone during regeneration, we and others (Knopf et al., 2011; Sousa et al., 2011) could not assess the relative level of contribution versus other potential sources due to incomplete labeling efficiency. In addition and perhaps more importantly, these findings did not address the extent to which fin regeneration is dependent on cellular contributions by resident osteoblasts. To probe the regenerative capacity of zebrafish fins after massive osteoblast loss, we generated a transgenic line containing a mCherry-tagged, human codon-optimized version of the Escherichia coli enzyme Nitroreductase (NTR) downstream of osterix regulatory sequences, Tg(osterix:mCherry-NTRo)pd46 (osx:NTR) (Grohmann et al., 2009). NTR reduces exogenously added metronidazole (Mtz) pro-drug to form a cytotoxic product with negligible bystander effects, and has been employed successfully to ablate specific cell types in zebrafish larvae (Curado et al., 2007; Pisharath et al., 2007). Treatment of osx:NTR; osc:EGFP fish for 24 hours with 10 mM Mtz caused a dramatic loss of osx- and osc-driven fluorescence throughout the fish by 4 days post-treatment (dpt) (Figure 2A). We did not observe impaired movement or behavior in these animals. TUNEL staining of caudal fins indicated extensive induction of apoptosis in osteoblasts lining the fin bone of osx:NTR fish that had been treated one day prior with Mtz (Figure 2B; Figure S2A).
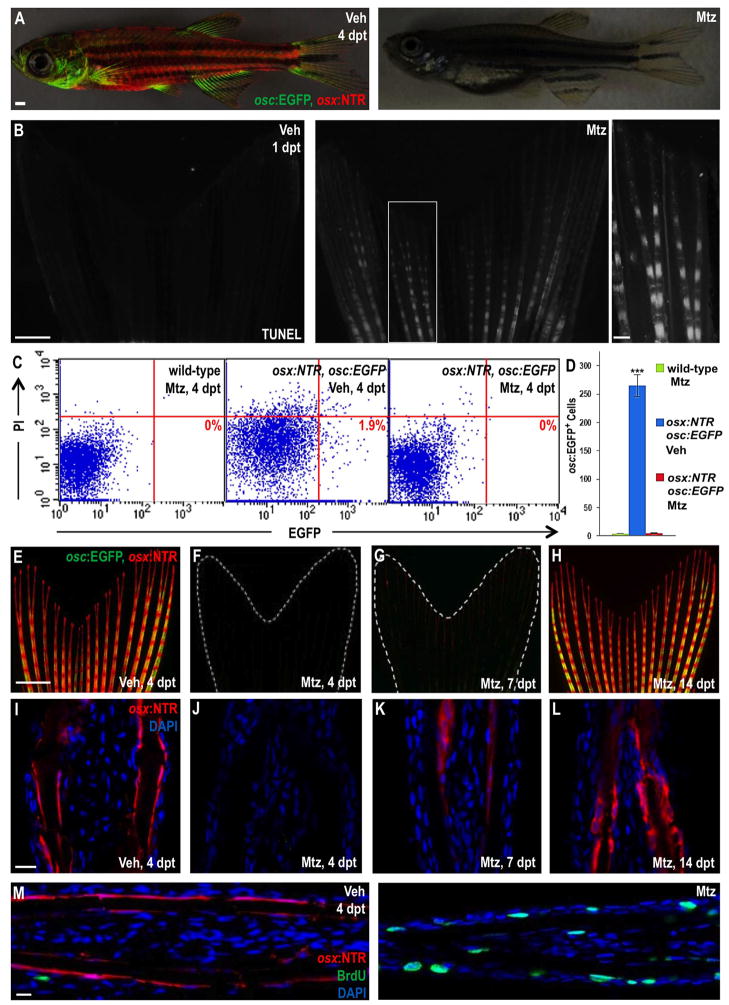
Figure 2. Inducible Ablation and Recovery of Adult Zebrafish Osteoblasts.
(A) Juvenile osx:NTR; osc:EGFP fish treated with vehicle (left) or Mtz (right) and assessed for fluorescence 4 days later. There was no detectable marker expression in Mtz-treated animals. Scale bar = 1 mm.
(B) TUNEL staining of osx:NTR fins 24 hours after vehicle (left) or Mtz (right) treatment, indicating profound, osteoblast-specific apoptosis (white) in Mtz-treated fish. Higher magnification images are shown in Figure S2. Scale bar = 100 μm.
(C) Flow cytometric analysis of caudal fin cells from wild-type (non-transgenic) and osx:NTR; osc:EGFP fish treated with vehicle or Mtz. Single cell suspensions were stained with propidium iodide (PI) and analyzed for EGFP. Representative plots are shown in (C); numbers in the lower right box indicate relative percentages of osc:EGFP+ cells.
(D) Absolute osc:EGFP cell counts (per 10,000 cells) from data in (C). Data are mean ± SEM from 9 animals each. ***p < 0.001, Student’s t-test. Wild-type and Mtz-treated osx:NTR; osc:EGFP samples show no significant difference in osc:EGFP+ cells, indicative of complete osteoblast loss.
(E–H) Caudal fins of osx:NTR; osc:EGFP fish lose osteoblast fluorescence within 4 days of Mtz treatment (E, F). Expression of osx:NTR can be detected beginning at 7 days post-treatment (dpt) (G), more easily in tissue sections than whole-mount images. Expression recovers completely by 14 dpt (H). Scale bar = 1 mm.
(I–L) Longitudinal sections of osx:NTR fins at timepoints indicated in (E–H). osx:NTR fluorescence disappears by 4 dpt and recovers in the osteoblast compartment by 14 dpt. Scale bar = 100 μm.
(M) BrdU immunofluorescence (green) analysis of vehicle- (left) or Mtz-treated (right) osx:NTR animals 4 days post-treatment, indicating enhanced cellular DNA synthesis in Mtz-treated samples. Scale bar = 100 μm.
While we could not detect osx- or osc-driven fluorescence in Mtz-treated fin tissue, we cannot exclude the possibility that a small number of fin osteoblasts were spared. To confirm depletion of fin osteoblasts, we performed flow cytometry on caudal fin tissues of osx:NTR; osc:EGFP fish that had been treated with vehicle or Mtz (Figure 2C, D). Mtz treatment decimated the osc:EGFP+ cell population, yielding fins with no significant difference in osc:EGFP+ events from non-transgenic animals (Figure 2D). Thus, we created a system that permitted massive depletion of virtually all adult zebrafish fin osteoblasts.
To determine the longer term consequences of this procedure, we followed osx-driven mCherry-NTR fusion protein and osc:EGFP fluorescence after Mtz treatment. osx:NTR; osc:EGFP caudal fins began to recover osteoblast marker expression by 7 dpt, with virtually complete restoration by two weeks (Figure 2E–L). Similar loss of marker expression was observed in cranial structures and hypurals at 4 dpt, with significant recovery by 7 and 14 dpt (Figure S2B, C). Fin osteoblast loss and recovery was accompanied by cell proliferation, indicative of an injury response. We detected increased nuclear Bromodeoxyuridine (BrdU) incorporation, a marker of DNA synthesis, mainly in epidermal cells at 4 dpt (Figure 2M; Figure S2D). We also observed BrdU-positive cells between hemirays; these cells did not express detectable osterix:NTR fluorescence or periostin, prrx1a, or prrx1b, orthologues of genes that mark mammalian periosteal cells (data not shown). Our results indicate that zebrafish regenerate their fin osteoblast compartment within two weeks of its genetic depletion.
Osteoblast-Depleted Fins Regenerate Normally After Amputation
To examine whether regeneration of amputated fins can initiate and progress without a notable source of bone, we treated osx:NTR fish with Mtz or vehicle for 24 hours, returned animals to aquarium water for 2 days, visually confirmed loss of transgene fluorescence, and amputated fins (Figure 3A). We then imaged and measured fin regenerates every two days after amputation over a course of 14 days. As negative controls, we measured the rate of fin regeneration of wild-type fish treated with Mtz 2 days prior to amputation. Unexpectedly, we observed no significant difference in the rates of regeneration among these three groups at any timepoints (Figure 3B). By contrast, a post-amputation Mtz treatment (4 dpa) slowed regenerative events significantly, indicating that depletion of osteoblasts participating in regeneration impedes the process (Figure S3A–C). The lengths of regenerated fins at 30 dpa were similar after each of these treatments (Figure S3D). Importantly, our data indicate that, although osteoblasts in the appendage stump make contributions to regenerated bone, they are dispensable for regeneration of bony fin rays.
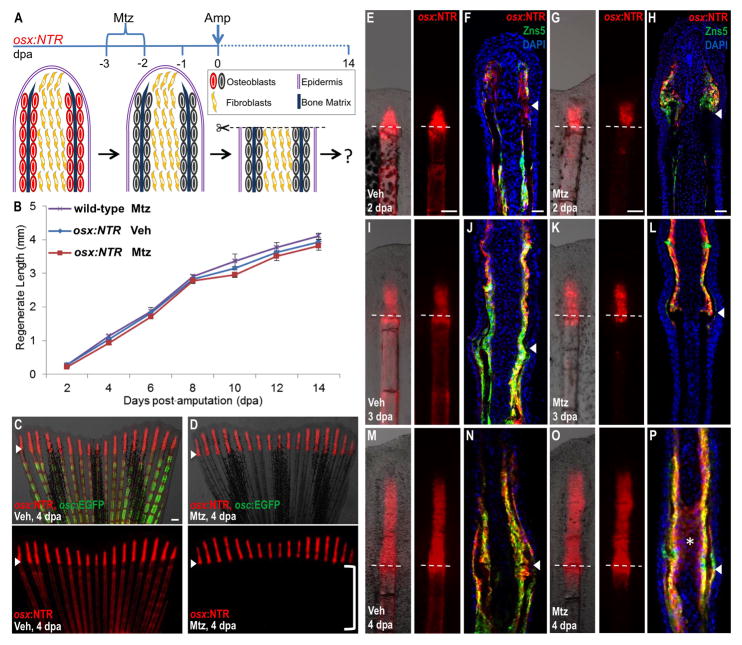
Figure 3. Osteoblast-Depleted Fins Regenerate Normally After Amputation.
(A) Cartoon summarizing strategy to assess regeneration of amputated fins after genetic ablation of osteoblasts.
(B) Lengths of fin regenerates after osteoblast ablation and amputation. As a negative control, wild-type animals were treated with Mtz 2 days before amputation and 4 days after amputation (wild-type, Mtz). osx:NTR animals treated with vehicle (osx:NTR, Veh) or Mtz (osx:NTR, Mtz) prior to amputation regenerated fins with similar efficacy. Data are mean ± SEM from 15 animals each.
(C, D) osx:NTR; osc:EGFP animals had indistinguishable regenerative lengths at 4 dpa whether or not osteoblasts were present prior to amputation, and indistinguishable osterix-driven expression in the regenerate. Osteoblast depletion proximal to the amputation plane is evident in Mtz-treated animals by the absence of marker expression (D; bracket). Bottom images show osx:NTR fluorescence only. Arrowheads indicate the plane of amputation. Scale bar = 100 μm.
(E–P) Whole-mount views and longitudinal sections of fins at 2 (E–H), 3 (I–L), and 4 (M–P) dpa, highlighting osterix-driven NTR fluorescence. osx:NTR is undetectable below the amputation plane of fins from Mtz-treated animals at 3 and 4 dpa, except for a trail of fluorescent cells at the amputation site. Tissue sections indicate expression of osx:NTR in osteoblasts at each of the 3 time points, and ectopic osx:NTR fluorescence in intraray fibroblasts at 4 dpa in the Mtz treated group (asterisk in (P)). Dotted lines and arrowheads indicate the plane of amputation. Scale bar = 100 μm.
To identify unique developmental responses of osteoblast-depleted fins during regeneration, we analyzed marker expression in osx:NTR; osc:EGFP animals at various time points post-amputation. Fins from animals treated with Mtz prior to amputation lacked detectable osx:NTR fluorescence proximal to the amputation plane at 2–4 dpa, except for a small trail of osx-expressing cells at the injury site in 3 and 4 dpa fins. In situ hybridization for endogenous osx and runx2a, a transcription factor that marks early osteoblasts (Li et al., 2009), gave similar results. However, they displayed prominent osx:NTR fluorescence and endogenous osx and runx2a expression distal to the amputation plane, indicating that the differentiation kinetics of osteoblasts in regenerating structures were distinct from those in existing fin tissue (Figure 3C–E, G, I, K, M, O; Figure S3E, F). Analysis of sections revealed that recovered osx:NTR fluorescence was restricted to the typical osteoblast compartment in the regenerate at 2 dpa (Figure 3F, H). However, by 4 dpa, and occasionally detectable at 3 dpa, osx:NTR fluorescence was present in a portion of medially located intraray fibroblasts near the amputation plane in Mtz-treated animals (Figure 3J, L, N, P). We occasionally detected osx mRNA in a similar region of intraray fibroblasts at 3 dpa in Mtz-treated animals (Figure S3F). By 7–8 dpa, this ectopic expression was no longer detectable (data not shown), and osx:NTR; osc:EGFP fluorescence began to recover throughout the proximodistal axis of the fins (Figure S3B). These observations of: 1) normal regeneration after massive osteoblast ablation; 2) temporally isolated activation of the osteoblast regulatory program in regenerating tissue versus uninjured areas; and 3) osterix–driven expression in medial fibroblast areas, were consistent with existence of an alternative regenerative source to stump osteoblasts.
Osteoblasts Arise De Novo in Fin Regenerates after Ablation of the Resident Population
While these findings implicated a non-osteoblast source, it remained formally possible that a portion of existing osteoblasts mimicked ablation by downregulating osteoblast markers upon Mtz treatment, and then recovered to contribute a new pool of osteoblasts to the regenerate. To address this mechanism, we first bathed uninjured osx:CreER; β-act2:RSG; osx:NTR animals in 4-HT for 24 hours to tag osteoblasts with an irreversible β-actin2-driven label (Figure 4A, B). Two days after genetic labeling, we treated these fish with Mtz for 24 hours, depleting all detectable EGFP fluorescence within 2 days (Figure 4C, H). We then amputated these fins and examined them at 4 dpa for reemergence of β-actin2-driven EGFP fluorescence in the regenerate. No EGFP fluorescence was detectable after this protocol (Figure 4D). To confirm these results, we assessed fin samples by cell dissociation and flow cytometry for β-actin2-driven EGFP. While uninjured fins from 4-HT-treated animals had many EGFP+ events, Mtz treatment depleted these events to background (EtOH-treated) levels (Figure S4A). EGFP+ events in Mtz-treated fish remained at background levels after amputation and 4 days of regeneration (Figure S4B). These data indicate that no new osteoblasts were contributed to the regenerate from resident osterix-expressing cells that had escaped ablation.
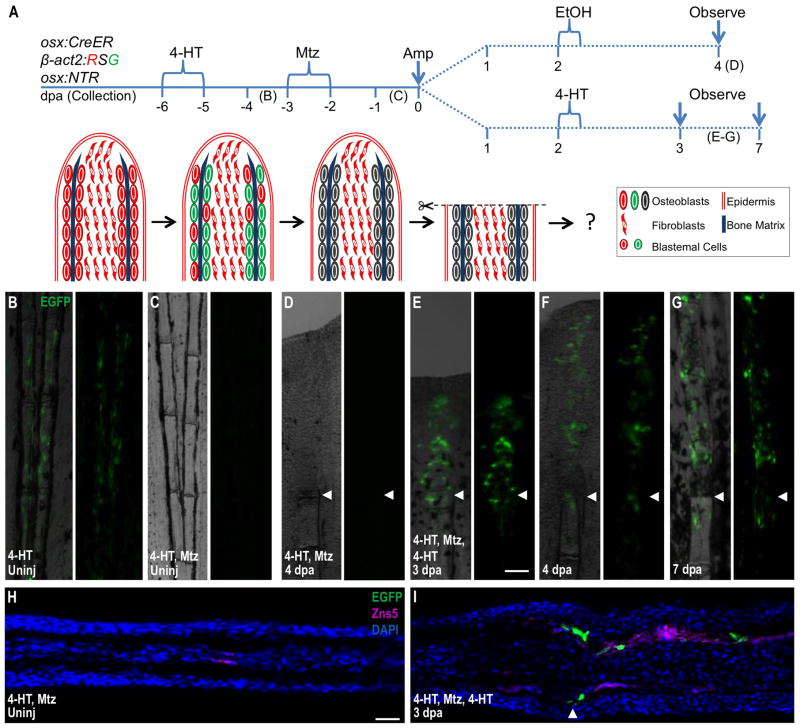
Figure 4. New Osteoblasts Arise in the Regenerates of Osteoblast-Depleted Fins Through De Novo Differentiation.
(A) Cartoon depicting strategy that combines inducible lineage-tracing and cell ablation in the osx:CreER; β-act2:RSG; osx:NTR background. 4-HT treatment imparts a β-actin2-driven EGFP label in osteoblasts.
(B, C) 4-HT labels osteoblasts of uninjured fins with EGFP fluorescence (B). This label was undetectable after Mtz treatment (C), indicating efficient osteoblast ablation.
(D) After amputation of fins of 4-HT-labeled and Mtz-treated animals, EGFP label was not detectable in 4 dpa regenerates or portions of the fins proximal to the injury site. Because the label was driven by the β-actin2 promoter, this result indicates that EGFP loss in (C) was due to cell ablation and not downregulation of an osteoblast marker.
(E–G) A second 4-HT treatment at 2 dpa generated EGFP+ cells in 3, 4, and 7 dpa regenerates, but not in portions of the fins proximal to the injury site. This result indicates that, although β-actin2 expressing osteoblasts are not contributed by uninjured fin regions, the regenerated osteoblasts can still be labeled by EGFP via their expression of osx:CreER and β-act2:RSG after amputation.
(H) Longitudinal section of uninjured fin corresponding to (C), indicating lack of EGFP fluorescence after 4-HT labeling and subsequent Mtz treatment.
(I) Longitudinal section of 3 dpa regenerate corresponding to (E), indicating that EGFP fluorescence induced by a postamputation 4-HT label is present in the osteoblast compartment of the regenerated portion only. Scale bar = 100 μm.
To confirm that the β-actin2 promoter remained active in new osteoblasts within these structures that had regenerated from osteoblast-depleted fins, we gave these animals an additional treatment with 4-HT for 12 hours at 2 dpa, and analyzed fins at 3, 4 and 7 dpa. Two days post-amputation represents the earliest time point at which we could detect recovered osx:NTR (and presumably osx:CreER expression) in the regenerate. This protocol labeled many cells in regenerating structures, and the proximodistal domain of EGFP+ cells increased with the length of the regenerate. EGFP was restricted to cells lining bone, and was not detected proximal to the amputation plane except for a small trail of cells at the injury site (Figure 4E–G, I). These findings indicated that existing osteoblasts had been effectively removed by genetic ablation and were not a source of regenerated bone. We conclude that in these experiments, regenerating bone has a lineage distinct from mature osteoblasts in the appendage stump.
DISCUSSION
Appendage regeneration has been studied for nearly 250 years, but only recently have technologies become available to resolve the cellular basis of these events. Here, we applied a combination of genetic cell ablation and fate-mapping approaches to define the importance of existing osteoblasts in regeneration of the skeletal bone of zebrafish fins. Our study adds key context to the results of recent lineage-tracing experiments in regenerating zebrafish fins, which highlighted a primary cellular mechanism for bone regeneration in which existing osteoblasts undergo de-differentiation, proliferate, and contribute new osteoblasts (Knopf et al., 2011; Sousa et al., 2011; Tu and Johnson, 2011). Although we also observed this mechanism, our data indicate that such events are dispensable, and that osteoblasts can regenerate readily after amputation through de novo differentiation. Thus, there are multiple cellular sources with the potential to contribute substantially to bone regeneration.
The combination of technologies we employed builds upon strategies that have been reported over the past century and have suggested the occurrence of trans-differentiation during skeletal regeneration. In salamanders, new bone can develop in the regenerate after removal of the skeletal elements and subsequent amputation through the affected area (Thornton, 1938). Additionally, although irradiated limbs fail to regenerate after amputation, transplantation of non-skeletal tissues can rescue this capacity (Dunis and Namenwirth, 1977; Namenwirth, 1974). Similar experiments have been performed in teleost fins in which entire fin rays were extirpated before amputation, a manipulation that has multiple interpretations and has yielded mixed results (Goss and Stagg, 1957; Nabrit, 1929, 1931; Turner, 1941). Our approach using genetic tools clearly indicates that non-osteoblast cells can be a primary or exclusive source of new, patterned appendage skeleton.
Which cells regenerate bone in the absence of contributions by skeletal osteoblasts? Intraray fibroblasts are the predominant cell type in fins along with epidermis and share with osteoblasts the expression of markers like msxb, msxc, sox9a, and Col2a1 (Akimenko et al., 1995; Smith et al., 2006), genes known in mammals to instruct and/or indicate osteoblast fate decisions (Karsenty, 2008). Thus, they represent primary candidates. Indeed, the ectopic induction of osterix-driven fluorescence that we observed in medially located fibroblasts after osteoblast ablation and amputation may be a signature of their trans-differentiation. If the fibroblasts are a bone source in zebrafish, their contributions are analogous to the dermal contributions to cartilage indicated in a recent study of axolotl limb regeneration (Kragl et al., 2009), and suggest an evolutionarily shared regenerative strategy. The use of inducible, Cre-based lineage-tracing experiments is recent to the zebrafish model system, and to our knowledge there is no marker or regulatory sequence with demonstrated specificity to zebrafish fin fibroblasts. Thus, the establishment of new reagents for specifically fate-mapping fibroblasts, as well as other important fin cell types, will advance the findings we report here.
Recent clonal analyses suggested that osteoblasts are not clonal partners with intraray fibroblasts or other recognized fin cell types during ontogeny or regeneration (Tu and Johnson, 2011). It is possible that a modified clonal analysis approach, employing stable transgenic lines and irreversible labeling (e.g. Cre recombinase technology), would provide new opportunity to represent all fin cells and recognize heterogeneous clone partners (Tanaka and Reddien, 2011). On the other hand, it is possible that osteoblast depletion triggers a novel source that does not normally participate in regeneration of amputated fin rays. There are precedents for dormant regenerative mechanisms that emerge predominantly in special contexts. For instance, pancreatic β-cells regenerate after resection injury by self-replication (Dor et al., 2004), yet can be replenished by duct cell or α-cell transdifferentiation after injuries of ischemia or extreme β-cell loss, respectively (Thorel et al., 2010; Xu et al., 2008). Thus, it will be important to determine the extent to which non-osteoblasts contribute bone in the presence of a full complement of osteoblasts, and to identify signals that recognize source availability and regulate output from diverse sources. Determining the breadth and plasticity of cellular sources in spectacular examples of bone reconstitution like zebrafish fin regeneration stands to illuminate potential therapies of major bone injury or loss in humans.
EXPERIMENTAL PROCEDURES
Zebrafish
Wild-type or transgenic zebrafish of the outbred Ekkwill (EK) or a hybrid EK/AB strain of 4–6 months of age were used for all experiments. Caudal fin amputations were performed with a razor blade on fish anesthetized with tricaine, and removed one-half of fins. The transgenic β-act2:RSG line has been described previously (Kikuchi et al., 2010). osx:mCherry and osc:EGFP constructs were generated by subcloning mCherry and EGFP cassettes downstream of published promoter sequences of medaka osterix and osteocalcin genes (Inohaya et al., 2007; Renn and Winkler, 2009). For osx:NTR, we subcloned mCherry, fused to a human codon-optimized version of the Escherichia coli enzyme Nitroreductase, downstream of the osterix regulatory fragment (Grohmann et al., 2009). To generate osx:CreER, a bicistronic construct containing the coding sequence for mTagBFP (Evrogen) (Subach et al., 2008) and sequences encoding a tamoxifen-inducible Cre recombinase-estrogen receptor fusion protein, separated by a 2A viral linker sequence (Provost et al., 2007; Trichas et al., 2008), were subcloned downstream of the osterix promoter. mTagBFP aided visualization of CreER expression in embryos, useful for identifying and maintaining the transgenic line. Plasmid constructs were co-injected with I-SceI into one-cell zebrafish embryos for linearization, and all transgenic strains were analyzed as hemizygotes.
For 4-HT labeling, adult zebrafish were incubated with aquarium water containing 5 μM 4-HT, made from a 1 mM stock solution in 100% ethanol. Fish were maintained in 4-HT in the dark for the indicated periods of time, and then were rinsed and returned to recirculating aquarium water. For osteoblast ablation, fish were incubated with 10 mM Mtz (Sigma, M1547) dissolved in aquarium water, and maintained for 24 hours in the dark before they were rinsed and returned to recirculating aquarium water.
Fin Length Measurement and Analysis
Leica Application Suite software was used to measure fin regenerates from images of live, anaesthetized fish. The distances from the amputation plane to the distal tips of the 2nd and 3rd lateral-most rays on the dorsal lobe was measured. These lengths were averaged to give one value per animal. Unpaired Student’s t-tests were performed to determine p-values.
Histological Methods
TUNEL staining on whole-mount fins was performed using a previously described protocol (Wills et al., 2008). For BrdU-labeling experiments, animals were injected intraperitoneally with ~0.05 ml of a 2.5 mg/ml solution of BrdU dissolved in water. BrdU was injected 5 hours before fin collection, and immunodetection of BrdU was performed as described (Lee et al., 2005). In situ hybridization on cryosections was performed using digoxygenin-labeled runx2a (Li et al., 2009) or osterix RNA probes as described previously (Poss et al., 2002). The monoclonal Zns5 (1:50 dilution; Zebrafish International Resource Center), polyclonal DsRed (1:500 dilution; Clontech) and polyclonal p63 (1:200 dilution; Abcam) antibodies were used for immunofluorescence analysis.
Flow Cytometry
Adult zebrafish caudal fins were amputated and dissociated by vigorous shaking for 20 minutes at room temperature in a solution of Liberase DH Research Grade (Roche) reconstituted in Hank’s Buffered Salt Solution (HBSS) buffer. The cells were briefly spun down and resuspended in HBSS, and then passed through a 40-μm filter. Propidium iodide (Sigma) was added to a concentratin of 1 μg/ml. Flow cytometry analysis was performed using a BD FACSCanto II (BD Biosciences), using forward and side scatter parameters to exclude cell debris.
Supplementary Material
HIGHLIGHTS.
Resident osteoblasts provide new bone cells to regenerating fins after amputation
A new transgenic model for inducible osteoblast ablation and recovery in zebrafish
Amputated fins regenerate normally after genetic depletion of osteoblasts
De novo osteoblasts can fully support regeneration of patterned skeletal bone
Acknowledgments
We thank B. Mayer for DNA injections, R. Anderson and D. Stainier for transgenic animals, J. Burris, A. Eastes, P. Williams, and N. Blake for zebrafish care, A. Nechiporuk and Poss lab members for comments on the manuscript, A. Kudo and A. Kawakami for helpful suggestions, D. Walther, C. Winkler, S. Srinivas, S. Leach, and H. Roehl for plasmids, and M. Cook for help with FACS analysis. K.D.P. is an Early Career Scientist of the Howard Hughes Medical Institute. This work was supported by NIH grant GM074057 to K.D.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimenko MA, Johnson SL, Westerfield M, Ekker M. Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development. 1995;121:347–357. doi: 10.1242/dev.121.2.347. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Dunis DA, Namenwirth M. The role of grafted skin in the regeneration of X-irradiated axolotl limbs. Developmental Biology. 1977;56:97–109. doi: 10.1016/0012-1606(77)90157-9. [DOI] [PubMed] [Google Scholar]
- Goss RJ, Stagg MW. The regeneration of fins and fin rays in Fundulus heteroclitus. The Journal of Experimental Zoology. 1957;136:487–507. doi: 10.1002/jez.1401360306. [DOI] [PubMed] [Google Scholar]
- Grohmann M, Paulmann N, Fleischhauer S, Vowinckel J, Priller J, Walther D. A mammalianized synthetic nitroreductase gene for high-level expression. BMC Cancer. 2009;9:301. doi: 10.1186/1471-2407-9-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohaya K, Takano Y, Kudo A. The teleost intervertebral region acts as a growth center of the centrum: In vivo visualization of osteoblasts and their progenitors in transgenic fish. Developmental Dynamics. 2007;236:3031–3046. doi: 10.1002/dvdy.21329. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G. Transcriptional Control of Skeletogenesis. Annual Review of Genomics and Human Genetics. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, et al. Bone Regenerates via Dedifferentiation of Osteoblasts in the Zebrafish Fin. Developmental Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Li N, Felber K, Elks P, Croucher P, Roehl HH. Tracking gene expression during zebrafish osteoblast differentiation. Developmental Dynamics. 2009;238:459–466. doi: 10.1002/dvdy.21838. [DOI] [PubMed] [Google Scholar]
- Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci U S A. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JI, Loof S, He P, Simon A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J Cell Biol. 2006;172:433–440. doi: 10.1083/jcb.200509011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabrit SM. The role of fin rays in the regenreation in the tail-fins of fishes. Biol Bull. 1929;56:235–266. [Google Scholar]
- Nabrit SM. The role of the basal plate in regeneration in the tail-fins of fishes. Biol Bull. 1931:60–63. [Google Scholar]
- Namenwirth M. The inheritance of cell differentiation during limb regeneration in the axolotl. Developmental Biology. 1974;41:42–56. doi: 10.1016/0012-1606(74)90281-4. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mechanisms of development. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- Renn J, Winkler C. Osterix-mCherry transgenic medaka for in vivo imaging of bone formation. Developmental Dynamics. 2009;238:241–248. doi: 10.1002/dvdy.21836. [DOI] [PubMed] [Google Scholar]
- Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblast differentiation and function. Developmental Biology. 2006;299:438–454. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simões M, Leon J, Roehl H, Cancela ML, Jacinto A. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- Steen TP. Stability of chondrocyte differentiation and contribution of muscle to cartilage during limb regeneration in the axolotl (Siredon mexicanum) The Journal of Experimental Zoology. 1968;167:49–78. doi: 10.1002/jez.1401670105. [DOI] [PubMed] [Google Scholar]
- Steen TP. Origin and Differentiative Capacities of Cells in the Blastema of the Regenerating Salamander Limb. American Zoologist. 1970;10:119–132. doi: 10.1093/icb/10.2.119. [DOI] [PubMed] [Google Scholar]
- Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Grüenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Conversion of Red Fluorescent Protein into a Bright Blue Probe. Chemistry & Biology. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Developmental Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel F, Népote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton CS. The histogenesis of the regenerating fore limb of larval Amblystoma after exarticulation of the humerus. Journal of Morphology. 1938;62:219–241. [Google Scholar]
- Trichas G, Begbie J, Srinivas S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biology. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Developmental Cell. 2011;20:725–732. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CJ. Regeneration of the gonopodium of Gambusia during morphogenesis. J Exp Zool. 1941;87:181–210. doi: 10.1002/jez.1401060202. [DOI] [PubMed] [Google Scholar]
- Wills AA, Kidd AR, 3rd, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, D’Hoker J, Stange G, Bonne S, De Leu N, Xiao X, Van de Casteele M, Mellitzer G, Ling Z, Pipeleers D, et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.