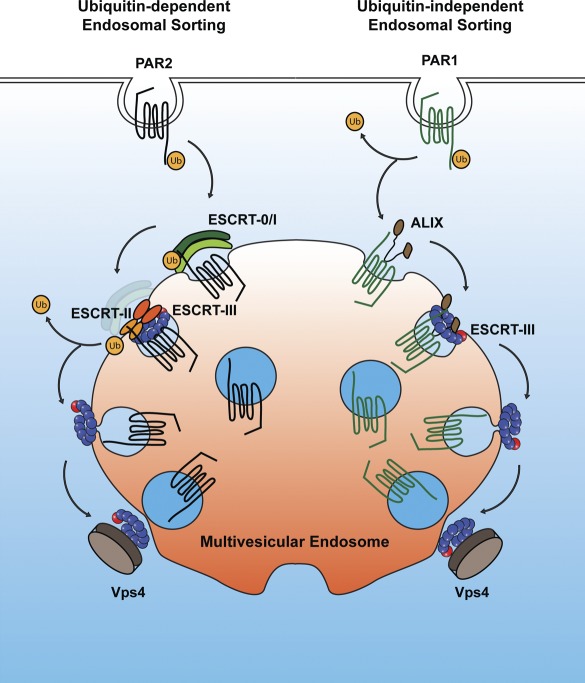
A novel MVB/lysosomal sorting pathway for signaling receptors bypasses the requirement for ubiquitination and ubiquitin-binding ESCRTs and may be broadly applicable to GPCRs containing YPXnL motifs.
Abstract
The sorting of signaling receptors to lysosomes is an essential regulatory process in mammalian cells. During degradation, receptors are modified with ubiquitin and sorted by endosomal sorting complex required for transport (ESCRT)–0, –I, –II, and –III complexes into intraluminal vesicles (ILVs) of multivesicular bodies (MVBs). However, it remains unclear whether a single universal mechanism mediates MVB sorting of all receptors. We previously showed that protease-activated receptor 1 (PAR1), a G protein–coupled receptor (GPCR) for thrombin, is internalized after activation and sorted to lysosomes independent of ubiquitination and the ubiquitin-binding ESCRT components hepatocyte growth factor–regulated tyrosine kinase substrate and Tsg101. In this paper, we report that PAR1 sorted to ILVs of MVBs through an ESCRT-III–dependent pathway independent of ubiquitination. We further demonstrate that ALIX, a charged MVB protein 4–ESCRT-III interacting protein, bound to a YPX3L motif of PAR1 via its central V domain to mediate lysosomal degradation. This study reveals a novel MVB/lysosomal sorting pathway for signaling receptors that bypasses the requirement for ubiquitination and ubiquitin-binding ESCRTs and may be applicable to a subset of GPCRs containing YPXnL motifs.
Introduction
G protein–coupled receptors (GPCRs) are the largest family of signaling receptors expressed in mammalian cells and mediate vast physiological responses. The temporal and spatial fidelity of GPCR signaling is critical for appropriate cellular responses. Moreover, dysregulated GPCR signaling has been implicated in numerous human diseases including neurodegeneration and cancer progression (Hanyaloglu and von Zastrow, 2008; Marchese et al., 2008). In addition to desensitization, GPCR trafficking is important for the precise regulation of signaling responses. This is particularly true for protease-activated receptor 1 (PAR1), a GPCR for thrombin (Coughlin, 2000; Arora et al., 2007). Thrombin cleaves the N terminus of PAR1, unmasking a new N-terminal domain, which functions as a tethered ligand that activates the receptor through intramolecular binding (Vu et al., 1991). Once activated, PAR1 is internalized and sorted directly to lysosomes and degraded, a process important for termination of G protein signaling (Trejo et al., 1998; Booden et al., 2004). The mechanism by which activated PAR1 is trafficked to lysosomes is not known.
The sorting of transmembrane proteins such as EGF receptor (EGFR) from the plasma membrane to lysosomes has been extensively studied and is mediated by the endosomal sorting complex required for transport (ESCRT). The ESCRT machinery is comprised of distinct complexes that function coordinately to sort ubiquitinated receptors to intraluminal vesicles (ILVs) of multivesicular bodies (MVBs; Hurley and Hanson, 2010). Hepatocyte growth factor–regulated tyrosine kinase substrate (HRS), a component of ESCRT-0, recruits ubiquitinated receptors and Tsg101, a ubiquitin-binding subunit of ESCRT-I (Lu et al., 2003). ESCRT-I and -II function in receptor sorting to ILVs and ILV formation (Wollert and Hurley, 2010). ESCRT-III polymerizes on endosomal membranes and is the main driver of ILV scission. The AAA-ATPase vacuolar protein sorting 4 (Vps4) disassembles and recycles ESCRT-III components and is essential for ESCRT function. In addition to receptor sorting at the MVB, ESCRT mediates viral budding and cytokinesis through processes that require ESCRT-I and -III and ALIX, an ESCRT-III–interacting protein, but not ESCRT-0 or -II (Strack et al., 2003; Carlton et al., 2008). Whether there are differences in ESCRT requirements for the sorting of signaling receptors at the MVB in mammalian cells remains unclear.
Most GPCRs require posttranslational modification with ubiquitin and ESCRTs for sorting from endosomes to lysosomes. The chemokine receptor CXCR4 is ubiquitinated after activation and sorted from endosomes to lysosomes through a pathway that requires HRS and Vps4 (Marchese et al., 2003). PAR2, a GPCR related to PAR1, also undergoes agonist-induced ubiquitination and is sorted to lysosomes through an HRS-dependent pathway (Hasdemir et al., 2007). However, not all GPCRs require direct ubiquitination for MVB sorting and lysosomal degradation, as exemplified by the δ-opioid receptor (DOR). A ubiquitination-deficient DOR mutant is efficiently sorted to ILVs of MVBs similar to wild-type (WT) receptor (Henry et al., 2011). However, degradation of DOR requires HRS and Vps4 but not Tsg101 (Hislop et al., 2004), indicating that receptor sorting can occur independent of ubiquitination and requires some but not all components of the ubiquitin-binding ESCRT machinery. Thus, it remains to be determined whether a signaling receptor can bypass the requirement for both ubiquitination and ubiquitin-binding components of the ESCRT machinery and sort to MVBs/lysosomes.
We previously showed that activated PAR1 is efficiently sorted from endosomes to lysosomes and degraded independent of ubiquitination (Wolfe et al., 2007). In contrast to DOR, however, neither HRS nor Tsg101 is essential for lysosomal degradation of PAR1 (Gullapalli et al., 2006). Thus, it is not known whether activated PAR1 ultimately sorts to ILVs of MVBs and requires any components of the ESCRT machinery for lysosomal degradation. Here, we report that activated PAR1 sorts to ILVs of MVBs through a pathway that requires ALIX and ESCRT-III function but not receptor ubiquitination. Moreover, our findings indicate that ALIX binds to a YPX3L motif of PAR1 and recruits ESCRT-III to mediate MVB/lysosomal sorting.
Results
Activated PAR1 sorts to ILVs of MVBs independent of ubiquitination
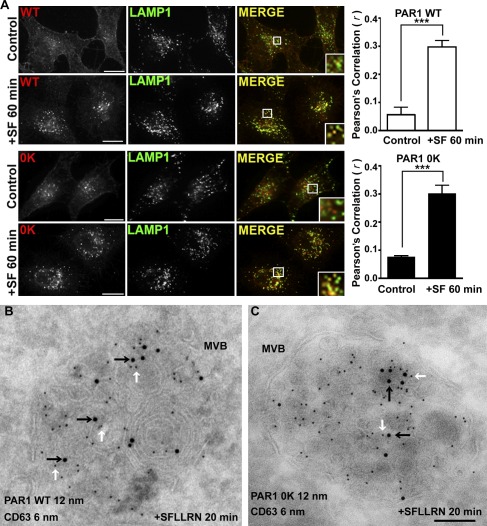
Activated PAR1 WT is internalized, sorted efficiently to lysosomes, and rapidly degraded with a half-life of 30 min (Trejo and Coughlin, 1999). In previous work, we showed that a PAR1 lysine-less 0K mutant defective in ubiquitination is degraded comparably with WT receptor after activation (Wolfe et al., 2007), suggesting that PAR1 degradation occurs independent of ubiquitination. To investigate the mechanism of ubiquitin-independent degradation of PAR1, we first determined whether agonist promoted lysosomal sorting of the PAR1 0K mutant by examining its colocalization with the lysosomal-associated membrane protein 1 (LAMP1) in HeLa cells using immunofluorescence confocal microscopy. In the absence of agonist stimulation, PAR1 WT and 0K mutant cycle constitutively between the plasma membrane and early endosomes (Paing et al., 2006; Wolfe et al., 2007). Consistent with these studies, we found that PAR1 WT and 0K mutant localized predominantly to the cell surface and early endosomes and showed no significant colocalization with LAMP1 under control conditions (Figs. 1 A and S1 A). However, after 60 min of agonist stimulation, PAR1 WT colocalized extensively with LAMP1-positive vesicles (Fig. 1 A), as previously reported (Trejo et al., 2000). Activation of PAR1 0K mutant with agonist for 60 min also resulted in substantial receptor colocalization with LAMP1 (Fig. 1 A). The colocalization of activated PAR1 WT and 0K mutant with LAMP1 was verified by determining Pearson’s correlation coefficient for PAR1 WT (r = 0.30 ± 0.05; n = 6) and 0K mutant (r = 0.30 ± 0.08; n = 6). Thus, after activation, PAR1 0K mutant is efficiently sorted to lysosomes like WT receptor, suggesting that ubiquitination is not essential for PAR1 lysosomal sorting.
Figure 1.
PAR1 is sorted to MVBs independent of ubiquitination. (A) HeLa cells expressing FLAG-PAR1 WT or 0K mutant were pretreated with 0.2 mM leupeptin. Cells were then incubated without (Control) or with 100 µM SFLLRN (SF) for 60 min at 37°C, fixed, permeabilized, and incubated with anti-FLAG and -LAMP1 antibodies. Immunolabeled cells were processed and imaged by confocal microscopy. The colocalization of PAR1 and LAMP1 is shown in yellow in the merged image and is representative of many cells examined in three independent experiments. Insets show magnifications of boxed areas. Bars, 10 µm. The data (mean ± SD) represent Pearson’s correlation coefficients that were calculated for PAR1 and LAMP1 colocalization for control versus agonist-stimulated conditions and were significant, as determined by Student’s t test (***, P < 0.001; n = 6). (B and C) HeLa cells expressing FLAG-PAR1 WT or 0K mutant were prelabeled with anti-FLAG antibody for 1 h at 4°C and then stimulated with 100 µM SFLLRN for 20 min at 37°C. Cells were fixed and processed for immuno-EM. Dual-labeled ultrathin sections revealed that PAR1 WT and 0K mutant detected with 12-nm gold-conjugated secondary antibody (black arrows) sorted to ILVs present in CD63-positive MVBs, which was detected with 6-nm gold-labeled secondary antibody (white arrows). Bar, 100 nm.
A critical step in lysosomal sorting and degradation of most proteins in mammalian cells requires incorporation into ILVs of the MVB. To determine whether PAR1 directly enters ILVs of MVBs independent of receptor ubiquitination, we performed immuno-EM on ultrathin cryosections of HeLa cells expressing PAR1 WT or ubiquitin-deficient 0K mutant. To examine endocytic sorting of the cell surface cohort of PAR1 WT and 0K mutant after agonist activation, receptors were prelabeled with anti-FLAG antibody at 4°C and then treated with agonist for 20 min. After agonist stimulation, activated PAR1 WT (12-nm gold particles) sorted to an endosomal compartment that colabeled with the late endosome/MVB marker CD63 (6-nm gold particles; Fig. 1 B), suggesting that activated PAR1 traffics to MVBs. Remarkably, the ubiquitination-deficient PAR1 0K mutant also localized to ILVs of CD63-positive MVBs after agonist activation (Fig. 1 C), indicating that PAR1 traffics to ILVs of MVBs independent of receptor ubiquitination. To ensure that antibody prelabeling did not alter the internalization and sorting of PAR1, we examined the colocalization of PAR1 WT and 0K mutant with LAMP1 by immunofluorescence confocal microscopy in parallel. As expected, PAR1 WT and 0K mutant localized to the cell surface and early endosomes and did not colocalize with LAMP1 after incubation for 20 min in the absence of agonist (Fig. S1, B and C). In contrast, however, stimulation of PAR1 WT and 0K mutant with agonist for 20 min caused significant colocalization with LAMP1 (Fig. S1, B and C), consistent with that observed in cells not prelabeled with antibody (Fig. 1 A). Together, these data demonstrate that ubiquitination of activated PAR1 is not required for entry into ILVs of MVBs and suggest that a novel pathway exists for the sorting of nonubiquitinated receptors to the lysosome.
ESCRT-III and Vps4 mediate PAR1 lysosomal degradation
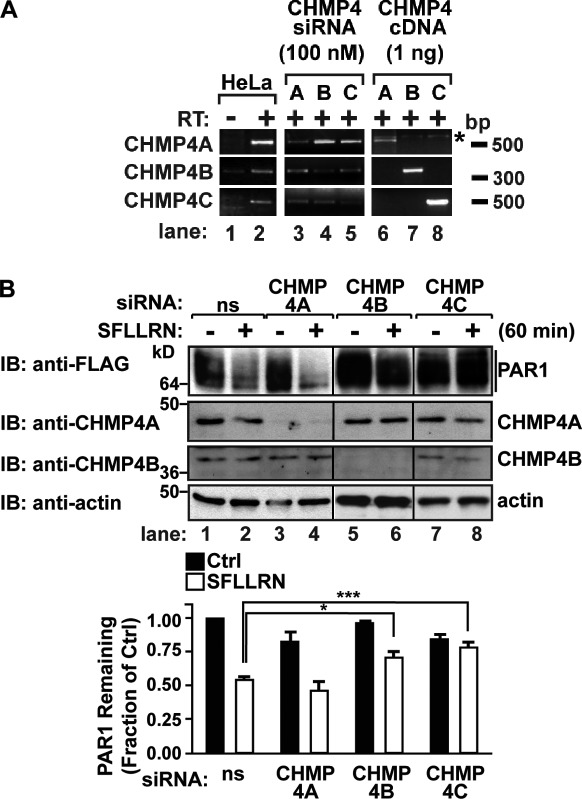
To define the ubiquitin-independent lysosomal degradation pathway of PAR1, we examined the function of ESCRT-III components, which do not contain any known ubiquitin-binding domains (Hurley and Hanson, 2010). Charged MVB protein 4 (CHMP4) is the most abundant subunit of ESCRT-III, which serves as the critical mediator of ILV scission, and exists as three isoforms: CHMP4A, B, and C (Wollert and Hurley, 2010). We first determined the expression of CHMP4 isoforms in HeLa cells using RT-PCR and CHMP4 isoform–specific primers (Fig. 2 A, lanes 6–8). RT-PCR assay of HeLa cell RNA revealed the presence of all three isoforms, including CHMP4A, B, and C (Fig. 2 A, lanes 1 and 2). To assess CHMP4 function in PAR1 degradation, siRNAs that specifically target each of the three CHMP4 isoforms expressed in HeLa cells were used. HeLa cells transfected with CHMP4 isoform–specific siRNA SMARTpools exhibited a substantial decrease in CHMP4A, B, and C mRNA transcripts, as revealed by RT-PCR (Fig. 2 A, lanes 3–5). Consistent with diminished mRNA transcripts, CHMP4-specific siRNAs also caused significant depletion of endogenous CHMP4A and CHMP4B protein, as detected by immunoblotting (Fig. 2 B). Because there are no anti-CHMP4C antibodies commercially available, we were unable to examine endogenous CHMP4C protein expression in siRNA-transfected cells.
Figure 2.
CHMP4 mediates PAR1 lysosomal degradation. (A) RT-PCR assays of HeLa RNA using CHMP4A, B, or C isoform–specific primers. No RT is indicated (−). RNA isolated from HeLa cells transfected with 100 nM CHMP4A-, B-, or C-specific siRNAs was subject to RT-PCR. As a control, 1 ng CHMP4A, B, and C cDNA plasmids was amplified by RT-PCR in parallel. The asterisk indicates a nonspecific band. (B) HeLa cells expressing FLAG-PAR1 were transfected with 100 nM of nonspecific (ns) or CHMP4 isoform–specific siRNAs and stimulated with 100 µM SFLLRN for 60 min at 37°C, and PAR1 degradation was assessed. Cell lysates were immunoblotted (IB) with anti-CHMP4A, -CHMP4B, and -actin antibodies. The data (mean ± SD) are expressed as the fraction of PAR1 remaining compared with untreated control (Ctrl). The amount of PAR1 detected in CHMP4B and CHMP4C siRNA–treated cells was significantly different from nonspecific siRNA-treated cells, determined by a two-way analysis of variance (*, P < 0.05; ***, P < 0.001; n = 3).
To assess the function of CHMP4 in PAR1 degradation, HeLa cells expressing FLAG-PAR1 were transfected with either nonspecific or CHMP4-specific siRNAs. Cells were then incubated with agonist for 60 min, and PAR1 degradation was assessed. In nonspecific siRNA-transfected cells, an ∼50% decrease in the amount of PAR1 protein was detected after 60 min of agonist incubation compared with unstimulated control cells (Fig. 2 B, lanes 1 and 2). The extent of PAR1 degradation was similar in nonspecific and CHMP4A siRNA–transfected cells after agonist stimulation, suggesting that CHMP4A is not essential for PAR1 degradation (Fig. 2 B, lanes 1–4). In contrast, agonist-induced degradation of PAR1 was significantly inhibited in cells transfected with either CHMP4B or CHMP4C siRNA SMARTpools (Fig. 2 B, lanes 5–8), indicating that both CHMP4B and C isoforms are necessary for PAR1 degradation. Cells transfected with single siRNAs targeting CHMP4B resulted in loss of endogenous CHMP4B expression and also blocked agonist-promoted PAR1 degradation (Fig. S2). These results suggest that CHMP4 mediates agonist-induced PAR1 lysosomal degradation.
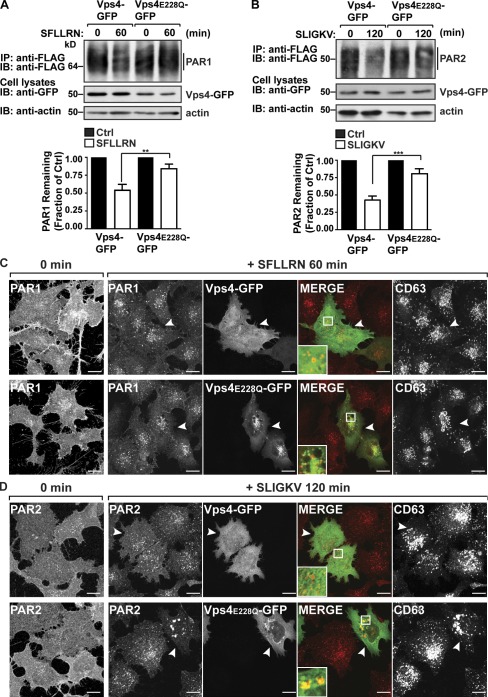
The AAA-ATPase Vps4 catalyzes ESCRT-III disassembly and recycling and is critical for ESCRT function (Hurley and Hanson, 2010). To examine the role of Vps4 in agonist-induced PAR1 degradation, we used a catalytically inactive Vps4 E228Q variant, which functions as a dominant-negative mutant (Babst et al., 1998; Bishop and Woodman, 2000). HeLa cells coexpressing PAR1 and either Vps4 WT or E228Q mutant fused to GFP were incubated with agonist, and receptor degradation was assessed. Interestingly, agonist-promoted PAR1 degradation was significantly inhibited in cells coexpressing Vps4 E228Q mutant compared with Vps4 WT–expressing cells (Fig. 3 A), indicating that Vps4 mediates MVB sorting of PAR1. Agonist-promoted PAR2 degradation was similarly inhibited in Vps4 E228Q mutant–expressing cells compared with control cells expressing WT Vps4 (Fig. 3 B). PAR1 and PAR2 degradation induced by agonist was also inhibited in cells transfected with siRNAs targeting both Vps4A and B isoforms (Fig. S3, A and B). These results suggest that Vps4 is required for both ubiquitin- and nonubiquitin-mediated lysosomal sorting of GPCRs.
Figure 3.
PAR1 does not accumulate in Vps4 E228Q–positive endosomes. (A and B) HeLa cells expressing FLAG-PAR1 or FLAG-PAR2 were transfected with either Vps4-GFP or Vps4 E228Q–GFP. Cells were stimulated with 100 µM SFLLRN or 100 µM SLIGKV, as indicated, and lysed, and equivalent amounts of lysates were immunoprecipitated (IP), and PAR degradation was assessed by immunoblotting (IB). Cell lysates were immunoblotted with anti-GFP and -actin antibodies. The data (mean ± SD) are expressed as the fraction of PAR1 remaining compared with untreated control (Ctrl) and were analyzed by a two-way analysis of variance (**, P < 0.01; ***, P < 0.001; n = 3). (C and D) HeLa cells expressing FLAG-PAR1 or FLAG-PAR2 and either GFP-tagged Vps4 or E228Q mutant were prelabeled with anti-FLAG antibody for 1 h at 4°C, washed, and stimulated with 100 µM SFLLRN or 100 µM SLIGKV at 37°C, as indicated. Cells were fixed, permeabilized, and immunostained for PAR1 and CD63. Cells coexpressing PAR1 or PAR2 and either Vps4 WT or E228Q mutant are indicated by the arrowheads. The insets are magnifications of the boxed areas. Bars, 10 µm.
Next, we evaluated Vps4 function in MVB/lysosomal sorting of activated PAR1 and PAR2 by immunofluorescence confocal microscopy. Cells coexpressing FLAG-PAR1 or FLAG-PAR2 and either Vps4 WT or E228Q mutant fused to GFP were incubated with anti-FLAG antibody at 4°C to ensure only the cell surface receptors would be labeled and then stimulated with agonist. In the absence of agonist, PAR1 and PAR2 localized predominantly to the cell surface, whereas agonist-stimulated PAR1 and PAR2 redistributed from the cell surface to endosomes (Fig. 3, C and D). As expected, coexpression of Vps4 E228Q mutant resulted in accumulation of internalized PAR2 in enlarged vesicles, defined as a class E phenotype that results from disruption of ESCRT-III recycling (Fig. 3 D; Babst et al., 1998; Bishop and Woodman, 2000). In contrast, PAR1-containing endosomes were unperturbed in Vps4 E228Q mutant–expressing cells and appeared similar to those in adjacent untransfected cells and cells expressing Vps4 WT (Fig. 3 C). Aberrant localization of CD63 observed in cells coexpressing PAR1 and Vps4 E228Q mutant indicates disruption of the MVB compartment (Fig. 3 C). Thus, in contrast to ubiquitinated cargo, PAR1 fails to accumulate at the MVB when the pathway is disrupted, suggesting that MVB sorting of PAR1 is distinctly regulated.
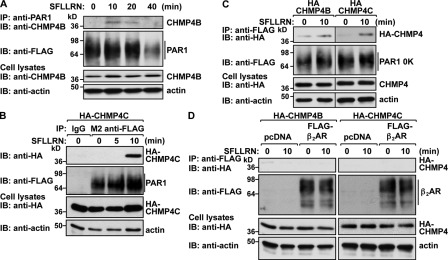
To confirm ESCRT-III function in PAR1 lysosomal degradation, we examined whether activated PAR1 forms a complex with CHMP4. We first assessed the coassociation of endogenous CHMP4B with activated PAR1 in HeLa cells by coimmunoprecipitation. HeLa cells expressing FLAG-PAR1 WT were incubated with agonist for various times, PAR1 was immunoprecipitated, and association of CHMP4B was examined. In the absence of agonist, PAR1 and CHMP4B interaction was undetectable (Fig. 4 A). However, an interaction between PAR1 and endogenous CHMP4B was detected after 10 min of agonist stimulation, and the proteins remained associated for ∼20 min (Fig. 4 A). These findings coincide with the time course for agonist-induced PAR1 localization to ILVs of MVBs and suggest that CHMP4B is a component of the ESCRT-III complex that binds to activated PAR1. Next, we examined PAR1 interaction with CHMP4C in HeLa cells transfected with an HA-tagged CHMP4C, as CHMP4C antibodies are not available. HeLa cells coexpressing FLAG-PAR1 and HA-CHMP4C were incubated with agonist for various times, PAR1 was immunoprecipitated, and CHMP4C association was assessed. In unstimulated control cells, CHMP4C failed to interact with PAR1 (Fig. 4 B). However, after activation of PAR1 for 10 min, a marked increase in CHMP4C association with PAR1 was detected (Fig. 4 B). Next, we examined whether the ubiquitin-deficient PAR1 0K mutant interacted with CHMP4 proteins. Similar to WT PAR1, both CHMP4B and CHMP4C coimmunoprecipitated with activated PAR1 0K mutant after 10 min of agonist stimulation (Fig. 4 C), indicating that CHMP4B and CHMP4C are components of the PAR1 ubiquitin–independent sorting complex. In contrast, FLAG-tagged β2-adrenergic receptor (β2AR), a classical GPCR, failed to coimmunoprecipitate with HA-CHMP4B or HA-CHMP4C in PAR1-expressing cells stimulated with the PAR1-specific agonist for 10 min (Fig. 4 D). Together, these findings demonstrate that ESCRT-III binds specifically to activated PAR1 and regulates MVB/lysosomal degradation.
Figure 4.
Activated PAR1 coassociates with CHMP4. (A) HeLa cells expressing FLAG-PAR1 were stimulated with 100 µM SFLLRN for various times at 37°C. Equivalent amounts of cell lysates were immunoprecipitated (IP) with anti-PAR1 antibody, and the presence of endogenous CHMP4B was determined by immunoblotting (IB). Cell lysates were immunoblotted with anti-CHMP4B and -actin antibodies. (B and C) HeLa cells expressing FLAG-PAR1 (B) or FLAG-PAR1 0K (C) and coexpressing HA-CHMP4C or HA-CHMP4B were stimulated with 100 µM SFLLRN at 37°C, as indicated. Equivalent amounts of cell lysates were immunoprecipitated with M2 anti-FLAG antibody or IgG control, and the presence of FLAG-PAR1 or PAR1 0K and associated HA-CHMP4 was determined by immunoblotting. Cell lysates were immunoblotted with anti-HA and -actin antibodies. (D) PAR1-expressing HeLa cells were cotransfected with FLAG-tagged β2-adrenergic receptor (β2AR) and either HA-CHMP4B or HA-CHMP4C and stimulated with 100 µM SFLLRN at 37°C for 10 min. FLAG-β2AR was immunoprecipitated with anti-FLAG antibody, and immunoprecipitates were analyzed by immunoblotting using anti-HA antibodies. Cell lysates were immunoblotted with anti-HA and -actin antibodies as controls.
The ESCRT-III–associated protein ALIX is required for PAR1 degradation
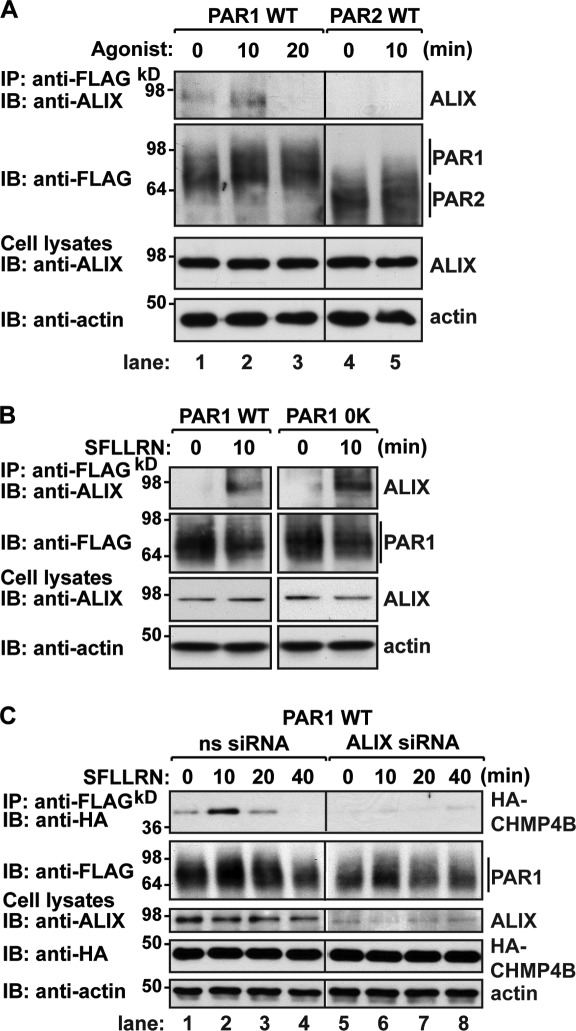
To further define the PAR1 ubiquitin–independent lysosomal sorting pathway, we investigated the function of ALIX, a CHMP4-interacting protein that recruits ESCRT-III to membranes (Fisher et al., 2007). We first examined whether PAR1 interacted with endogenous ALIX in HeLa cells. Cells expressing FLAG-PAR1 were stimulated with agonist for various times, PAR1 was immunoprecipitated, and association of endogenous ALIX was assessed. In cells treated with agonist for 10 min, PAR1 association with endogenous ALIX was markedly increased compared with untreated control cells (Fig. 5 A, lanes 1 and 2), whereas PAR1 and ALIX interaction was diminished after 20 min of agonist exposure (Fig. 5 A, lanes 1–3). In contrast, PAR2 and ALIX interaction was negligible in control and agonist-treated cells (Fig. 5 A, lanes 4 and 5). PAR1 also associated with HA-ALIX in an agonist-dependent manner (Fig. S4 A), whereas HA-ALIX failed to bind to β2AR in cells treated with PAR1-specific agonist (Fig. S4 B) or to activated PAR2 (Fig. S4 C). Next, we examined the association of endogenous ALIX with ubiquitination-deficient PAR1 in HeLa cells. In contrast to control cells, activation of the PAR1 0K mutant for 10 min resulted in endogenous ALIX association (Fig. 5 B), suggesting that ubiquitination is not required for ALIX interaction with activated PAR1.
Figure 5.
ALIX associates with PAR1 and mediates interaction with CHMP4. (A and B) HeLa cells expressing either FLAG-tagged PAR1 WT, 0K mutant, or PAR2 were stimulated with 100 µM SFLLRN or 100 µM SLIGKV for various times at 37°C. Equivalent amounts of cell lysates were immunoprecipitated (IP) and examined as described in Fig. 4 B. IB, immunoblotting. (C) HeLa cells expressing FLAG-PAR1 were cotransfected with HA-CHMP4B and either 100 nM nonspecific (ns) or ALIX-specific siRNAs. Equivalent amounts of cell lysates were immunoprecipitated and analyzed as described in Fig. 4 B.
ALIX has been shown to link certain viral proteins to the ESCRT-III complex, a process important for viral budding (Usami et al., 2007). Therefore, we examined whether ALIX was required for agonist-induced PAR1 interaction with CHMP4. HeLa cells coexpressing FLAG-PAR1 and HA-CHMP4B transfected with nonspecific or ALIX-specific siRNAs were treated with agonist, PAR1 was immunoprecipitated, and CHMP4 association was assessed. In nonspecific siRNA control cells, a 10-min incubation with agonist resulted in robust PAR1 association with CHMP4B (Fig. 5 C, lanes 1–4). In striking contrast, activated PAR1 interaction with CHMP4B was virtually ablated in ALIX-deficient cells (Fig. 5 C, lanes 5–8). These findings suggest that ALIX links PAR1 to the ESCRT-III machinery to facilitate lysosomal sorting.
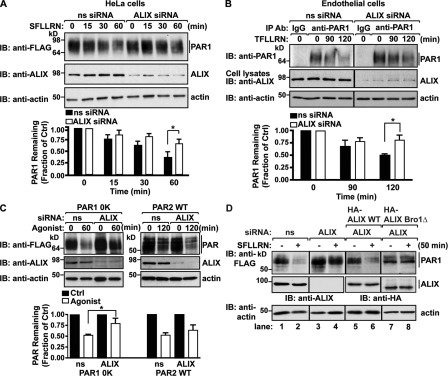
Next, we assessed ALIX function in agonist-induced PAR1 lysosomal degradation. HeLa cells expressing FLAG-PAR1 were transfected with ALIX-specific siRNAs and stimulated with agonist for various times, and PAR1 degradation was examined. In ALIX-deficient cells, agonist-promoted degradation of PAR1 was significantly inhibited compared with the extent of PAR1 degradation observed after 60 min of agonist stimulation in siRNA-treated control cells (Fig. 6 A). However, internalization of activated PAR1 occurred normally in ALIX-deficient cells (Fig. S4 D), indicating that ALIX mediates PAR1 sorting to a lysosomal degradative pathway after internalization. Depletion of endogenous ALIX expression also caused a significant defect in agonist-promoted degradation of endogenous PAR1-expressed human endothelial cells (Fig. 6 B). Moreover, agonist-induced degradation of ubiquitination-deficient PAR1 0K mutant was also impaired in ALIX-deficient cells, whereas activated PAR2 degradation remained intact (Fig. 6 C). Together, these findings support a function for ALIX in ubiquitin-independent MVB sorting of activated PAR1.
Figure 6.
ALIX is required for agonist-induced PAR1 degradation. (A and C) HeLa cells expressing FLAG-tagged PAR1 (A), PAR1 0K mutant, or PAR2 (C) were transfected with 100 nM nonspecific (ns) or ALIX-specific siRNAs and stimulated with 100 µM SFLLRN or 100 µM SLIGKV at 37°C, as indicated. PAR degradation was assessed by immunoblotting (IB). The data (mean ± SD) are expressed as the fraction of PAR1 remaining compared with untreated control (Ctrl) and were analyzed by a two-way analysis of variance (*, P < 0.05; n = 3). (B) Human endothelial cells transfected with 100 nM of nonspecific or ALIX-specific siRNAs stimulated with 100 µM TFLLRNPNDK at 37°C, as indicated, and immunoprecipitated (IP) with anti-PAR1 or IgG antibodies (Ab). Endogenous PAR1 was detected by immunoblotting. Cell lysates were immunoblotted with anti-ALIX and -actin antibodies. The data (mean ± SD) are expressed as a fraction of untreated control and were analyzed by a two-way analysis of variance (*, P < 0.05; n = 3). (D) HEK293 cells cotransfected with either 100 nM of nonspecific or two ALIX-specific siRNAs together with FLAG-PAR1 and either siRNA-resistant HA-ALIX WT or HA-ALIX Bro1Δ were incubated with or without 100 µM SFLLRN for 50 min at 37°C. Equivalent amounts of lysates were analyzed for PAR1, endogenous ALIX (lanes 1–4), HA-ALIX WT (lanes 5 and 6), or HA-ALIX Bro1Δ (lanes 7 and 8) expression by immunoblotting.
To confirm that ALIX is required for PAR1 degradation, we performed knockdown rescue experiments in cells cotransfected with two siRNAs targeting endogenous ALIX together with FLAG-tagged PAR1 and an siRNA-resistant HA-ALIX WT variant. In cells depleted of endogenous ALIX, agonist-induced degradation of PAR1 was significantly impaired compared with siRNA-treated control cells (Fig. 6 D, lanes 1–4). However, coexpression of siRNA-resistant HA-ALIX WT restored activated PAR1 degradation in cells lacking endogenous ALIX (Fig. 6 D, lanes 5 and 6). In contrast, agonist failed to induce PAR1 degradation in cells depleted of endogenous ALIX and expressing an siRNA-resistant HA-ALIX Bro domain deletion mutant (Fig. 6 D, lanes 7 and 8). The N-terminal Bro domain of ALIX mediates interaction with CHMP4 (Kim et al., 2005). These findings suggest that ALIX-mediated MVB/lysosomal sorting of PAR1 requires interaction with CHMP4 and is a distinct pathway from that used by ubiquitinated GPCRs.
ALIX binds to a highly conserved YPXnL motif in PAR1 via its central V domain
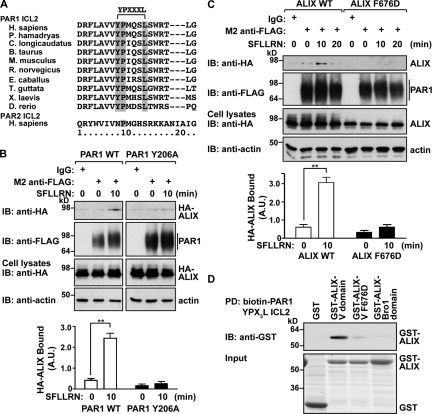
ALIX contains an N-terminal Bro1 domain, a central V domain, and C-terminal proline-rich region. ALIX binds to YPXnL motifs present in the late domain of viral Gag proteins via its central V domain (Fisher et al., 2007; Zhai et al., 2008) and is critical for ESCRT-III recruitment and viral budding (Usami et al., 2007). A search of the PAR1 cytoplasmic domains for such a sequence revealed a highly conserved motif within the second intracellular loop of the receptor (Fig. 7 A). The PAR1 Y206PMQSL211 motif conforms to the ALIX-binding motif YPXnL (with n = 3) and is not present in PAR2 cytoplasmic domains (Fig. 7 A). A search of several hundred class A GPCRs for such a motif revealed the presence of a highly conserved sequence that conforms to the YPXnL motif residing in the second intracellular loop of seven other mammalian GPCRs (Table 1). Only one class A GPCR harbors a conserved YPXnL motif within the cytoplasmic tail domain (Table 1). To examine the importance of the YPX3L motif in PAR1 degradation, we mutated the critical Y206 to alanine, as analogous Y mutations in YPXnL motifs of viral Gag proteins abrogated ALIX interaction and function (Strack et al., 2003). The PAR1 Y206A mutant expressed at the surface and internalized after activation comparable with WT PAR1 (Fig. S5, A and B), suggesting that the PAR1 Y206A mutant traffics to and from the cell surface normally.
Figure 7.
A YPX3L motif of PAR1 mediates binding to ALIX. (A) Alignment of PAR1 and PAR2 ICL2 sequences from various species. The conserved residues of the YPX3L motif are shaded in gray. (B) HeLa cells coexpressing HA-ALIX WT and either FLAG-PAR1 WT or Y206A mutant were stimulated with 100 µM SFLLRN at 37°C, as indicated. Cell lysates were immunoprecipitated and examined as described in Fig. 4 B. IB, immunoblotting. The data (mean ± SD) represent the amount of immunoprecipitated ALIX normalized to the amount of immunoprecipitated PAR1 and were significant, as determined by Student’s t test (**, P < 0.01; n = 3). A.U., arbitrary unit. (C) HeLa cells transfected with FLAG-PAR1 and either HA-ALIX WT or F676D mutant were stimulated with agonist and processed as previously described. Cell lysates were immunoblotted with anti-HA and -actin antibodies. The data (mean ± SD) were calculated, as previously described, and significant, as determined by Student’s t test (**, P < 0.01; n = 3). (D) Biotinylated PAR1 ICL2 peptide containing the YPX3L motif was immobilized on streptavidin beads and incubated with GST, GST–ALIX V domain, GST–ALIX V F676D, or the GST–ALIX Bro1 domain. Pulldowns (PD) were analyzed for the presence of bound protein by immunoblotting. Input was analyzed by Coomassie staining.
Table 1.
Human GPCRs with conserved YPXnL motifs
| Family | IUPHAR nomenclature | NCBI Protein database accession no. | YPXnL location | Sequence |
| Adrenoceptors | α1B Adrenoceptor | P35368 | ICL2 | 141-DRYIGRYSLQYPTLVT-158 |
| Angiotensin | AT2 | P50052 | ICL2 | 140-DRYQSVIYPFLSQRRN-157 |
| Galanin | GAL2 | O43603 | ICL2 | 121-DRYLAIRYPLHSRELRT-139 |
| Histamine | H2 | P25021 | ICL2 | 114-DRYCAVMDPLRYPVLVT-132 |
| Neuropeptide FF | NPFF2 | A0PJM9 | ICL2 | 141-DRFQCVVYPFKPKLTIK-159 |
| Neuropeptide S | NPS receptor | Q6W5P4 | ICL2 | 144-DRYHAIVYPMKFLQGE-161 |
| Protease activated | PAR1 | P25116 | ICL2 | 198-DRFLAVVYPMQSLSWRT-216 |
| P2Y | P2Y1 | P47900 | ICL2 | 147-HRYSGVVYPLKSLGRLK-165 |
| Melanocortin | MC4 | P32245 | Cytoplasmic tail | 318-YPLGGLCDLSSRY-stop |
A search of 397 class A GPCR cytoplasmic sequences for YPXnL motifs was completed by sequence alignment using the GPCR database (Vroling et al., 2011). Of these, several GPCRs contained sequences conforming to the YPXnL motif in the ICL2 (bold) and were aligned starting with the DRY/F motif. The list includes the GPCR family name, International Union of Basic and Clinical Pharmacology (IUPHAR) nomenclature, and NCBI accession numbers.
Next, we examined whether the PAR1 YPX3L motif was necessary for interaction with ALIX. HeLa cells coexpressing FLAG-PAR1 WT or Y206A mutant together with HA-ALIX were stimulated with agonist, and coassociation was examined. PAR1 WT robustly associated with ALIX after 10 min of agonist stimulation, whereas activated PAR1 Y206A mutant interaction with ALIX was negligible (Fig. 7 B). PAR2, which lacks a YPXnL motif, also failed to bind to HA-ALIX (Fig. S4 C), suggesting that the YPXnL motif is an important determinant for ALIX interaction. We also examined whether the ALIX V domain is required for association with PAR1. The ALIX central V domain binds directly to YPXnL sequences of viral Gag proteins, and mutation of the critical phenylalanine to aspartic acid (F676D) was shown to disrupt this interaction (Zhai et al., 2008). In contrast to PAR1 interaction with WT ALIX, the HA-ALIX F676D mutant failed to bind to PAR1 after 10 min of agonist incubation (Fig. 7 C). To confirm direct interaction between the PAR1 YPX3L motif and the ALIX V domain, we performed in vitro binding assays using a biotinylated PAR1 ICL2 peptide containing the YPX3L motif in pull-down experiments with purified GST-tagged ALIX. Strikingly, the ALIX V domain showed robust interaction with the PAR1 YPX3L containing ICL2 peptide (Fig. 7 D), whereas neither the ALIX V domain harboring the F676D mutant nor the ALIX Bro1 domain bound to the PAR1 ICL2 peptide (Fig. 7 D). Together, these findings strongly suggest that ALIX can bind directly to the YPX3L motif of PAR1 via its central V domain.
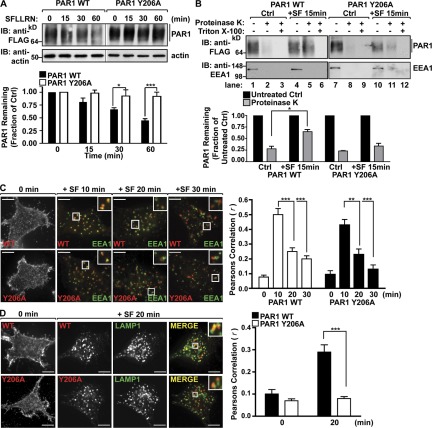
Our experiments indicate that the PAR1 YPX3L motif is critical for binding to ALIX. Therefore, we tested whether the PAR1 YPX3L motif was necessary for agonist-promoted PAR1 degradation. Incubation with agonist for 60 min caused an ∼50% loss of PAR1 protein (Fig. 8 A). In contrast to WT PAR1, agonist-stimulated degradation of the PAR1 Y206A mutant was significantly inhibited (Fig. 8 A). Next, we examined whether PAR1 Y206A mutant remained at the limiting membrane of MVBs or sorted to an internal compartment by evaluating its sensitivity to proteinase K digestion after activation and internalization. Proteinase K protection assays have been used previously to assess EGFR sorting to ILVs of MVBs (Malerød et al., 2007). HeLa cells expressing PAR1 WT or Y206A mutant were incubated with agonist for 15 min at 37°C, washed, and gently permeabilized with digitonin. Membranes were isolated, divided into aliquots, and either left untreated, treated with proteinase K, or treated with proteinase K supplemented with 0.1% Triton X-100. Activation of PAR1 WT resulted in a greater accumulation of receptor in proteinase K–treated samples compared with unstimulated control samples (Fig. 8 B, lanes 2 and 5), indicating that activated PAR1 WT is sorted into protective endosomal compartments. In contrast, PAR1 Y206A mutant failed to accumulate in protective endosomal compartments compared with unstimulated control (Fig. 8 B, lanes 8 and 11), suggesting the PAR1 Y206A mutant is defective in MVB sorting. However, both PAR1 WT and Y206A mutant were efficiently degraded after exposure to proteinase K in samples supplemented by Triton X-100, which allows proteinase K access to internal membranous compartments (Fig. 8 B, lanes 3, 6, 9, and 12). The early endosomal antigen protein 1 (EEA1) was also efficiently degraded in all proteinase K–treated samples, demonstrating effective plasma membrane permeabilization and proteinase K activity (Fig. 8 B). In addition, the PAR1 Y206A mutant internalized to EEA1-positive endosomes like WT receptor in cells treated with agonist for 10 min (Fig. 8 C), and both sorted from the EEA1-containing endosomes after 20 min (Fig. 8 C). The extent of colocalization with EEA1 at 10 min was quantified by determining the Pearson’s correlation coefficient for PAR1 WT (r = 0.50 ± 0.1; n = 6) and Y206A (r = 0.43 ± 0.09; n = 6). Interestingly, however, PAR1 Y206A mutant showed minimal colocalization with LAMP1 after 20 min of agonist stimulation compared with WT PAR1 (Fig. 8 D), which was verified by Pearson’s correlation coefficient for PAR1 WT (r = 0.28 ± 0.08; n = 6) and Y206A (r = 0.08 ± 0.02; n = 6). Thus, the YPX3L motif is a critical determinant for PAR1 ubiquitin–independent MVB/lysosomal sorting (Fig. 9).
Figure 8.
The YPX3L motif of PAR1 is essential for MVB/lysosomal sorting. (A) HeLa cells expressing FLAG-PAR1 WT or Y206A mutant were incubated with 100 µM SFLLRN at 37°C, as indicated. PAR1 degradation was analyzed as described in Fig. 2 B. IB, immunoblotting. The data (mean ± SD) are expressed as a fraction of untreated control (Ctrl) and were analyzed by a two-way analysis of variance (*, P < 0.05; ***, P < 0.001; n = 3). (B) HeLa cells expressing FLAG-PAR1 WT or Y206A were untreated (control) or treated with 100 µM SFLLRN (SF) for 15 min at 37°C. Membranes were isolated and treated with proteinase K or proteinase K with 0.1% Triton X-100, and PAR1 or EEA1 was detected by immunoblotting. The data (mean ± SD) are expressed as the fraction of PAR1 remaining after proteinase K treatment compared with untreated control (−) from either the control or SFLLRN (+SF)-treated samples and were analyzed by Student’s t test (*, P < 0.05; n = 3). (C and D) HeLa cells expressing either FLAG-PAR1 WT or Y206A mutant were prelabeled with anti-FLAG antibody at 4°C and incubated with 100 µM SFLLRN at 37°C, as indicated. Cells were immunostained for either EEA1 or LAMP1 and analyzed, as described in Fig. 1 A. Insets show magnifications of boxed areas. Bars, 10 µm. The data (mean ± SD) represent Pearson’s correlation coefficients that were calculated, as described in Fig. 1 A, and were analyzed by Student’s t test (**, P < 0.01; ***, P < 0.001; n = 6).
Figure 9.
GPCR ubiquitin-dependent and -independent MVB sorting. The ubiquitin-mediated MVB sorting pathway of PAR2 requires the ESCRT-0, -I, and -II complexes before ESCRT-III–mediated incorporation into ILVs. In contrast, PAR1 MVB sorting is mediated by ALIX recruitment of the ESCRT-III complex, independent of ubiquitination (Ub).
Discussion
In this study, we define a new ubiquitin-independent MVB/lysosomal sorting pathway for signaling receptors that requires ALIX and ESCRT-III function in mammalian cells. We demonstrate that activated PAR1 is sorted to ILVs of MVBs independent of receptor ubiquitination but requires ESCRT-III function. Our data further indicate that PAR1 degradation is regulated by ALIX, which binds directly to a YPX3L motif localized within the PAR1’s intracellular loop via its central V domain and mediates CHMP4/ESCRT-III recruitment to the receptor. Thus, PAR1 bypasses the requirement for ubiquitination and ubiquitin-binding ESCRTs but requires ALIX and ESCRT-III for sorting to ILVs of MVBs (Fig. 9).
We previously showed that lysosomal sorting of PAR1 occurs independent of ubiquitination and the ubiquitin-binding ESCRT components HRS and Tsg101 (Gullapalli et al., 2006; Wolfe et al., 2007), but whether PAR1 sorts to ILVs of MVBs remained unknown. Here, we show that a ubiquitination-deficient PAR1 mutant sorted to ILVs similar to WT receptor, indicating that ubiquitination is not essential for ILV sorting of PAR1. Our experiments further indicate that CHMP4, the major subunit of ESCRT-III and driver of ILV scission, is essential for agonist-induced PAR1 degradation. In contrast to other ESCRT complexes, ESCRT-III does not contain any known ubiquitin-binding subunits, suggesting that ubiquitin binding is not essential for ESCRT-III–mediated PAR1 degradation. Moreover, ubiquitination is not a prerequisite for cargo passage into ILVs, as ESCRT-III subunits recruit enzymes that appear to deubiquitinate cargo before ILV entry (Kyuuma et al., 2007). Thus, unlike most cargo sorted to ILVs of MVBs via the canonical ESCRT pathway, our findings indicate that PAR1 is uniquely sorted by ESCRT-III independent of ubiquitination. This pathway is also distinct from the entirely ESCRT-independent ILV sorting pathway previously described for the melanosome constituent Pmel and the proteolipid protein (Theos et al., 2006; Trajkovic et al., 2008; van Niel et al., 2011). Other cargo, such as the yeast Sna3 and mammalian DOR, are also not directly ubiquitinated, but certain ubiquitin-binding ESCRT components facilitate MVB/lysosomal sorting (Hislop et al., 2004; McNatt et al., 2007; Oestreich et al., 2007; Watson and Bonifacino, 2007).
Our findings also reveal a novel function for ALIX, a CHMP4–ESCRT-III interacting protein, in mediating MVB/lysosomal sorting of PAR1. ALIX directly binds to and recruits CHMP4–ESCRT-III to membranes via interaction with its Bro domain and is important for abscission during cytokinesis and budding of certain viruses (Strack et al., 2003; Morita et al., 2007). However, ALIX function in cargo sorting at MVBs in mammalian cells is not known. Bro1, the yeast homolog of ALIX, facilitates the sorting of ubiquitinated cargo at MVBs (Odorizzi et al., 2003). In yeast, Bro1 recruits the deubiquitinating enzyme Doa4 to the late endosome, which is essential for cargo deubiquitination and sorting at the MVB. However, in mammalian cells, ALIX does not appear to function in lysosomal sorting of ubiquitinated cargo, as depletion of ALIX by siRNA failed to affect degradation of EGFR (Cabezas et al., 2005), a receptor that requires direct ubiquitination and ESCRTs for degradation. We found that the association of ALIX with activated PAR1 coincided with PAR1 sorting to ILVs at the MVB. Moreover, degradation of endogenous and ectopically expressed PAR1 was significantly impaired in ALIX-deficient cells and was restored by expression of an siRNA-resistant ALIX WT variant, indicating that ALIX function is specific for this pathway. ALIX also mediated degradation of ubiquitination-deficient PAR1, but not the related PAR2, which requires ubiquitination for lysosomal sorting. In addition, ALIX is required for activated PAR1 recruitment of CHMP4 subunits, the critical components of ESCRT-III, and the ALIX Bro domain important for CHMP4 interaction is necessary for the rescue of agonist-promoted PAR1 degradation in cells deficient in endogenous ALIX expression. Thus, ALIX has the capacity to mediate cargo sorting at the MVB in mammalian cells through a pathway that requires ESCRT-III recruitment. ALIX-mediated MVB sorting of PAR1 also appears to be distinctly regulated, as PAR1 does not accumulate at MVBs when ILV sorting is blocked, compared with ubiquitinated cargo that use canonical ESCRT components. The mechanistic basis for this observation is not known but may involve trafficking of PAR1 from the MVB to a different compartment and/or recycling back to the cell surface.
This study is the first to demonstrate ALIX-mediated MVB/lysosomal sorting of a mammalian cell host protein via a YPXnL motif. The ALIX central V domain has been shown to bind to YPXnL motifs of retroviral Gag proteins (Fisher et al., 2007; Zhai et al., 2008). We found that ALIX directly interacts with a highly conserved YPX3L motif within the PAR1’s intracellular loop via its V domain to facilitate MVB/lysosomal degradation. Previous studies showed that the HIV-1 and EIAV viruses recruit the ESCRT machinery through interaction with ALIX (Strack et al., 2003; Zhai et al., 2008) The HIV-1 structural Gag protein harbors a tetrapeptide motif that binds to Tsg101 (Garrus et al., 2001; Martin-Serrano et al., 2001) and a YPXnL motif that interacts with the V domain of ALIX (Strack et al., 2003; Fisher et al., 2007) to mediate ESCRT-III recruitment. In contrast, EIAV Gag proteins only possess an ALIX-binding YPXnL motif that is critical for ESCRT-III recruitment (Martin-Serrano et al., 2003) and viral budding (Tanzi et al., 2003). Similar to HIV-1 and EIAV (Strack et al., 2003), mutation of the critical tyrosine within the YPX3L motif of PAR1 disrupted interaction with ALIX and impaired PAR1 MVB/lysosomal degradation. In vitro binding assays confirmed that the V domain of ALIX binds directly to the YPX3L motif of PAR1. Thus, ALIX directly interacts with PAR1 and mediates MVB/lysosomal sorting via an ESCRT-III–dependent pathway.
In summary, this study provides the first molecular insight into a mechanism by which a signaling receptor can sort to ILVs of MVBs independent of ubiquitination and ubiquitin-binding ESCRTs. Unlike most ubiquitinated cargo, we show that PAR1 sorts to ILVs of MVBs through a distinct ubiquitin-independent pathway that requires ALIX and ESCRT-III function but not the canonical ubiquitin-binding ESCRT components. These findings unveil a new mechanism that enriches the diversity of MVB sorting pathways for signaling receptors. This work also suggests that ubiquitination of mammalian GPCRs is likely to have additional functions besides mediating receptor sorting at the MVB. ALIX has also been shown to bind to the cytoplasmic tail domains of the transferrin receptor (Géminard et al., 2004), the GPCR vasopressin V2R (Yi et al., 2007), and D1-like and D3 dopamine GPCRs (Zhan et al., 2008) and appears to regulate various aspects of receptor trafficking. However, these cargo lack the ALIX-binding YPXnL motif, and, thus, mechanistically how ALIX functions in these pathways is not known. Remarkably, we discovered seven other mammalian GPCRs that contained conserved YPXnL motifs within their second intracellular loop, raising the intriguing possibility that ALIX mediates MVB/lysosomal sorting of a subset of GPCRs in mammalian cells.
Materials and methods
Antibodies and reagents
PAR1 peptide agonists (SFLLRN and TFLLRNPNDK) and PAR2 peptide agonist (SLIGKV) were synthesized and purified by reverse-phase high-pressure liquid chromatography at the Tufts University Core Facility. Polyclonal anti-FLAG and -HA antibodies were obtained from Rockland Immunochemicals. Anti-LAMP1 and -CD63 antibodies were obtained from the Developmental Studies Hybridoma Bank. Anti-EEA1 antibody was obtained from BD. Monoclonal anti-PAR1 WEDE antibody was purchased from Beckman Coulter. Polyclonal CHMP4A and CHMP4B antibodies as well as monoclonal anti-ALIX (3A9) and anti-Vps4 antibodies were obtained from Santa Cruz Biotechnology, Inc. The monoclonal anti-HA and -GFP antibodies were purchased from Covance. The M2 anti-FLAG and -actin antibodies were obtained from Sigma-Aldrich. The HA antibody conjugated to HRP was purchased from Roche. HRP-conjugated goat–anti rabbit and goat–anti mouse antibodies were purchased from Bio-Rad Laboratories. Alexa Fluor 488, 594, and 647 secondary antibodies were purchased from Invitrogen.
Plasmids and cell lines
The full-length human N-terminal FLAG-tagged PAR1 WT, FLAG-PAR1 0K mutant cDNAs cloned into the mammalian expression vector pBJ, and full-length FLAG-tagged PAR2 cloned into pBJ vector were previously described (Trejo et al., 2000; Wolfe et al., 2007; Ricks and Trejo, 2009). The FLAG-PAR1 Y206A mutant cloned into pBJ vector was generated by QuikChange mutagenesis (Agilent Technologies) and confirmed by dideoxy sequencing. The full-length FLAG-tagged β2AR cloned into pcDNA3.1 was provided by M. von Zastrow (University of California San Francisco, San Francisco, CA). The GFP-tagged Vps4 WT and E228Q mutants were cloned into pEGFP vector and provided by A. Marchese (Loyola University, Maywood, IL). FLAG-tagged CHMP4A construct was provided by P. Hanson (Washington University, St. Louis, MO). The cDNAs encoding HA-CHMP4B, HA-CHMP4C, HA-ALIX WT, and HA-ALIX F676D and Bro domain mutants were provided by J. Martin-Serrano (King’s College, London, England, UK). GST–ALIX V, ALIX V F676D, and ALIX Bro1 constructs were provided by W. Sundquist (University of Utah, Salt Lake City, UT). HeLa cells stably expressing FLAG-PAR1 or FLAG-PAR2 cloned into pBJ vectors under the control of cytomegalovirus promoter were generated by cotransfection with a plasmid encoding a hygromycin resistance gene, selected in 250 µg/ml hygromycin, and screened by cell surface ELISA. Cells were grown and maintained as previously described (Trejo et al., 2000). Human endothelial EA.hy926 cells were grown and maintained as previously described (Russo et al., 2009).
siRNA and cDNA transfections
HeLa cells or endothelial cells were transfected with 100 nM siRNA using either Lipofectamine 2000 (Invitrogen) or Oligofectamine (Invitrogen), respectively, and examined after 48 h. The CHMP4A, CHMP4B, CHMP4C, Vps4A, Vps4B, and ALIX siRNAs were purchased as SMARTpools from Thermo Fisher Scientific. The single CHMP4B siRNAs used were 1 (5′-AGAAGAGUUUGACGAGGAU-3′) and 2 (5′-CGGAAGAGAUGUUAACGAA-3′). Nonspecific siRNA (5′-GGCUACGUCCAGGAGCGCACC-3′) was used as a negative control. The ALIX knockdown rescue experiment was performed by cotransfecting HEK293 cells with ALIX-specific siRNAs (5′-GUACCUCAGUCUAUAUUGA-3′ and 5′-UCGAGACGCUCCUGAGAUA-3′; Thermo Fisher Scientific) together with cDNAs containing HA-ALIX WT or HA-ALIX Bro domain mutant harboring silent mutations within the siRNA-targeted region using Lipofectamine 2000 reagent.
Immuno-EM
HeLa cells stably expressing full-length FLAG-PAR1 or FLAG PAR1 0K mutant were grown in 10-cm dishes (1.0 × 106 cells/dish) for 48 h. Cells were washed and preincubated on ice for 1 h with anti-FLAG antibody to label only receptors at the cell surface. Cells were washed to remove unbound antibody and treated with agonist for 20 min at 37°C, fixed with 4% PFA in 0.1 M phosphate buffer, pH 7.4, overnight at room temperature, washed with 0.15 M glycine/phosphate buffer, and then embedded in 10% gelatin/phosphate buffer infused with 2.3 M sucrose/phosphate buffer overnight at 4°C. 1-mm3 cell blocks were mounted onto specimen holders and snap frozen in liquid nitrogen. Ultracryomicrotomy was performed at −100°C on an ultramicrotome with an EM FCS cryoattachment (Ultracut UCT; Leica) using a diamond knife (DiATOME US). 80–90-nm frozen sections were picked up with a 1:1 mixture of 2.3 M sucrose and 2% methyl cellulose, as previously described (Lavoie et al., 2002), and transferred onto Formvar and carbon-coated copper grids. Immunolabeling was performed by placing grids on 2% gelatin at 37°C for 20 min and rinsed with 0.15 M glycine/PBS, and the sections were blocked using 1% cold water fish skin gelatin. Previously titrated primary antibodies were diluted in 1% BSA/PBS. Incubation with primary antibodies for 1 h at room temperature was followed by gold-conjugated goat anti–mouse IgG and IgM (Jackson ImmunoResearch Laboratories, Inc.) or gold-conjugated goat anti–rabbit IgG, both diluted 1/25 in 1% BSA/PBS at room temperature for 30 min. Grids were viewed using a transmission electron microscope (1200-EX II; JEOL) and photographed using a digital camera (Gatan, Inc) at the University of California San Diego Electron Microscopy Facility.
PAR degradation assay
HeLa cells stably expressing FLAG-PAR1, FLAG-PAR1 0K, or FLAG-PAR2 were plated in 12-well dishes (1.5 × 105 cells/well) and grown overnight at 37°C and transfected with either siRNAs or cDNA plasmids. After 48 h, cells were washed and treated with or without 100 µM SFLLRN (PAR1-specific agonist) or 100 µM SLIGKV (PAR2-specific agonist) for various times at 37°C. Cells were placed on ice, washed with PBS, and lysed in Triton X-100 lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, 50 mM NaF, 10 mM NaPP, and 1% Triton X-100) supplemented with protease inhibitors. Cell lysates were collected and sonicated for 10 s at 10% amplitude (model 450 sonifier; Branson), and protein concentrations were determined by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). Equivalent amounts of lysates were used for analysis by immunoblotting. Endothelial cells were plated in 6-cm dishes (3.0 × 106 cells/plate), grown for 2 d, stimulated with 100 µM TFLLRNPNDK (PAR1-specific agonist) for various times at 37°C, and lysed with radioimmunoprecipitation assay lysis buffer (5 mM EDTA, 50 mM Tris-HCl, 150 mM NaCl, 0.5% [wt/vol] Na-deoxycholate, 1% [volume/volume] NP-40, and 0.1% [wt/vol] SDS) supplemented with protease inhibitors, and equivalent amounts of lysates were immunoprecipitated using the anti-PAR1 WEDE antibody. PAR1 degradation was assessed as previously described (Russo et al., 2009).
Coimmunoprecipitation assay
HeLa cells stably expressing FLAG-PAR1 were grown in six-well plates (4.0 × 105 cells/well) overnight and transiently transfected with either HA-CHMP4 or HA-ALIX. Alternatively, HeLa cells plated in six-well dishes were transiently cotransfected with PAR1 WT or Y206A mutant together with HA-ALIX. After 48 h, cells were treated with 100 µM SFLLRN (PAR1-specific agonist) for varying times at 37°C, washed, and lysed with 0.5% NP-40 lysis buffer containing 20 mM Tris-HCl, pH 7.4, and 150 mM NaCl. Cell lysates were sonicated and cleared by centrifugation, and equivalent amounts of lysates determined by BCA assay were precleared with protein A Sepharose CL-4B for 1 h at 4°C. Precleared lysates were then immunoprecipitated with M2 anti-FLAG antibody or IgG overnight at 4°C. Immunoprecipitates were washed, resuspended in 2× sample buffer, resolved by SDS-PAGE, transferred to membranes, and examined by immunoblotting. For PAR1 coimmunoprecipitation of endogenous CHMP4 and ALIX, HeLa cells expressing FLAG-PAR1 WT or FLAG-PAR1 0K mutant were grown in six-well plates (4.0 × 105 cells/well). After 48 h, cells were treated with agonist and lysed in buffer, as previously stated. Equivalent amounts of lysates were then immunoprecipitated with anti-PAR1 or polyclonal anti-FLAG antibodies, and the coassociation of CHMP4 or ALIX in immunoprecipitates was examined by immunoblotting.
Immunofluorescence confocal microscopy
HeLa cells stably expressing FLAG-PAR1 or FLAG-PAR2 were plated on coverslips in 12-well dishes and grown overnight. Cells were washed, stimulated with agonists, 100 µM SFLLRN (PAR1 specific), or 100 µM SLIGKV (PAR2 specific) for various times at 37°C, fixed in 4% PFA, permeabilized in 100% methanol, and processed for microscopy, as previously described (Trejo et al., 2000). Cells were then immunostained with an anti-FLAG polyclonal rabbit antibody to detect either PAR1 or PAR2 followed by monoclonal anti-EEA1, -LAMP1, or -CD63 antibodies and then incubated with species-specific secondary antibodies conjugated to Alexa Fluors and imaged by confocal microscopy. To specifically track activated PAR1 or PAR2 from the cell surface, cells were prelabeled with anti-FLAG antibody for 1 h at 4°C to ensure only surface FLAG-PAR1 or FLAG-PAR2–bound antibody, stimulated with agonist for various times at 37°C, and then processed for microscopy. Fixed cells were mounted in FluorSave reagent (EMD). The fluorochromes used were Alexa Fluor 488, 594, and 647 and GFP. Images were acquired with a disk-spinning unit confocal system (Olympus) configured with a microscope (IX81; Olympus) fitted with a PlanApo 60× oil objective (1.4 NA; Olympus) and a digital camera (ORCA-ER; Hamamatsu Photonics). Fluorescent images of 0.28-µm-thick X–Y sections were acquired at room temperature using SlideBook software (version 4.2; Intelligent Imaging Innovations, Inc.). Pearson’s correlation coefficients for quantifying colocalization of PAR1 WT and mutants with either EEA1 or LAMP1 were calculated for six independent cells from multiple experiments using SlideBook software (version 4.2).
Protein sequence alignment
The following protein sequences were acquired from the National Center for Biotechnology Information protein database: Rattus norvegicus (available from GenBank under accession no. AAA42274.1), Xenopus laevis (GenBank accession no. AAA18498.1), Mus musculus (GenBank accession no. AAA40438.1), Papio hamadryas (GenBank accession no. AAB84191.1), Cricetulus longicaudatus (GenBank accession no. AAA86747.1), Bos taurus (NCBI Protein database accession no. NP_001096567.1), Equus caballus (NCBI Protein database accession no. XP_001504007.2), Taeniopygia guttata (NCBI Protein database accession no. XP_002189106.1), Danio rerio (GenBank accession no. CAM13317.1), and Homo sapiens PAR1 (GenBank accession no. AAA36743.1) and PAR2 (GenBank accession no. AAP97012.1). Sequences were aligned using ClustalX 2.1 software (Larkin et al., 2007).
In vitro binding assay
A biotinylated PAR1 ICL2 peptide representing the second intracellular loop sequence LAVVYPMQSLSWRTLG (the segment in italics denotes the YPXL motif within the peptide fragment, which is required for binding to ALIX in the context of full-length PAR1) was synthesized and purified by the Tufts University Core Facility. Biotinylated PAR1 ICL2 peptide was immobilized on streptavidin beads (Sigma-Aldrich). GST vector, GST–ALIX V, GST–ALIX V F676D, and GST-ALIX Bro1 cDNA plasmids were transformed into BL21 (DE3) Escherichia coli cells (Agilent Technologies). E. coli were grown in ZYP-5052 media (1% tryptone, 0.5% yeast extract, 0.025 M (NH4)2SO4, 0.05 M KH2PO4, 0.05 M Na2HPO4, 0.5% glycerol, 0.05% glucose, and 0.2% α-lactose) and lysed, and GST fusion proteins were purified on GST resin (EMD). GST fusion proteins were eluted, and protein concentrations were determined using BCA assay and GST fusion proteins (10 µg) and then incubated with immobilized biotinylated PAR1 ICL2 peptide for 1 h at 4°C. After washing, samples were eluted in 2× sample buffer resolved by SDS-PAGE and analyzed by immunoblotting to detect GST–ALIX association. The input, 25%, was run on an SDS-PAGE gel and stained with Coomassie to visualize proteins.
Proteinase K protection assay
The proteinase K protection assay was performed as previously described (Malerød et al., 2007), with minor modifications. HeLa cells were plated in six-well culture dishes (4.0 × 105 cells/well), grown overnight at 37°C, and treated with or without agonist. Cells were placed on ice and incubated for 5 min with PBS, harvested, and gently permeabilized using 6.5 µg/ml digitonin. Membranes were collected by centrifugation and resuspended in buffer (100 mM K2HPO4/KH2PO4, 5 mM MgCl2, and 250 mM sucrose). Membranes were then divided into three aliquots either left untreated, treated with 2.5 ng/ml proteinase K, or treated with proteinase K supplemented with 0.1% Triton X-100 for 10 min at room temperature. After treatments, samples were diluted with 100 µl of 2× SDS sample buffer containing 20 mM PMSF and analyzed by immunoblotting.
RT-PCR
The first-strand cDNA was generated from mRNA extracted from either HeLa using SuperScript II reverse transcription (Invitrogen) following the manufacturer’s instructions. The RT enzyme was omitted from the cDNA synthesis reaction in the control samples. The first-strand cDNA was amplified via PCR using primer pairs specific for CHMP4A (forward 5′-GCATGAAGAAGGCCTACCAG-3′ and reverse 5′-AGGCAGGGTCACACTACCAC-3′), CHMP4B (forward 5′-CCCATGACAACATGGACATC-3′ and reverse 5′-AGCCCAGTTCTCCAATTCCT-3′), or CHMP4C (forward 5′-GAACTCCACAGCAATGAGCA-3′ and reverse 5′-AGTCATCACCAAAGCCAACC-3′). The PCR amplification products were resolved by 1.8% (wt/vol) agarose gel electrophoresis and visualized by ethidium bromide staining.
Cell surface ELISA
HeLa cells were plated at 0.5 × 105 cells/well in a 24-well culture dish, grown overnight, and transfected, as previously described. After 48 h, cells were either left untreated or treated with 100 µM SFLLRN (PAR1-specific agonist) for varying times at 37°C, and the amount of receptor remaining at the cell surface was determined, as previously described (Soto and Trejo, 2010). In brief, cells were fixed in 4% PFA, washed, and then incubated with polyclonal anti-FLAG antibody followed by secondary HRP-conjugated goat anti–rabbit antibody. The amount of antibody bound to the cell surface was determined by incubation with one-step 2,2’-azinobis-3-ethylbenzthiazoline-6-sulfonic acid (Thermo Fisher Scientific) substrate for 10–20 min at room temperature. An aliquot was removed, and the absorbance at 405 nm was determined using a microplate reader (SpectraMax Plus; Molecular Devices).
Statistics
Data were analyzed using Prism software (version 4.0; GraphPad Software). Statistical analysis was determined, as indicated, by performing either Student’s t test or two-way analysis of variance.
Online supplemental material
Fig. S1 shows the localization of PAR1 WT and 0K mutant at steady state and after stimulation with agonist for 60 min, relative to EEA1-positive early endosomes, and shows localization of surface-labeled PAR1 WT and 0K mutant relative to LAMP1 at 0 min and after a 20-min incubation with or without agonist. Fig. S2 shows PAR1 degradation in cells treated with individual siRNAs targeting CHMP4B. Fig. S3 demonstrates that Vps4 siRNA SMARTpools inhibit agonist-induced degradation of PAR1 and PAR2. Fig. S4 shows HA-ALIX coimmunoprecipitation with FLAG-PAR1 but not with the unrelated FLAG–β2 adrenergic receptor or FLAG-PAR2 and demonstrates that ALIX knockdown does not impair internalization of PAR1. Fig. S5 show that PAR1 Y206A is expressed at the cell surface and is internalized comparably with WT PAR1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110031/DC1.
Supplementary Material
Acknowledgments
We thank members of the Trejo laboratory. We are grateful for reagents provided by Drs. W. Sundquist, P. Hanson, M. von Zastrow, and J. Martin-Serrano and acknowledge Drs. T. Handel, J. Jones, D. Siderovski, and A. Marchese for reagents and advice.
This work was by supported by the National Institutes of Health grant GM090689 to J. Trejo. M.R. Dores is supported by a University of California Tobacco-Related Disease Research Program Postdoctoral Fellowship (UC TRDRP 19FT-0157). B. Chen is supported by an American Heart Association Postdoctoral Fellowship. U.J.K. Soh is supported by a University of California Tobacco-Related Disease Research Program Postdoctoral Fellowship.
Author contributions: M.R. Dores and B. Chen designed, performed, and analyzed the experiments and prepared the manuscript. H. Lin, U.J.K. Soh, M.M. Paing, W.A. Montagne, and T. Meerloo contributed experimental work. J. Trejo contributed to project design and manuscript preparation.
Footnotes
Abbreviations used in this paper:
- BCA
- bicinchoninic acid
- CHMP
- charged MVB protein
- DOR
- δ-opioid receptor
- EGFR
- EGF receptor
- ESCRT
- endosomal sorting complex required for transport
- GPCR
- G protein–coupled receptor
- HRS
- hepatocyte growth factor–regulated tyrosine kinase substrate
- ILV
- intraluminal vesicle
- MVB
- multivesicular body
- WT
- wild type
References
- Arora P., Ricks T.K., Trejo J.. 2007. Protease-activated receptor signalling, endocytic sorting and dysregulation in cancer. J. Cell Sci. 120:921–928. 10.1242/jcs.03409 [DOI] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E.J., Emr S.D.. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N., Woodman P.. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 11:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booden M.A., Eckert L.B., Der C.J., Trejo J.. 2004. Persistent signaling by dysregulated thrombin receptor trafficking promotes breast carcinoma cell invasion. Mol. Cell. Biol. 24:1990–1999. 10.1128/MCB.24.5.1990-1999.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas A., Bache K.G., Brech A., Stenmark H.. 2005. Alix regulates cortical actin and the spatial distribution of endosomes. J. Cell Sci. 118:2625–2635. 10.1242/jcs.02382 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Agromayor M., Martin-Serrano J.. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 105:10541–10546. 10.1073/pnas.0802008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin S.R. 2000. Thrombin signalling and protease-activated receptors. Nature. 407:258–264. 10.1038/35025229 [DOI] [PubMed] [Google Scholar]
- Fisher R.D., Chung H.-Y., Zhai Q., Robinson H., Sundquist W.I., Hill C.P.. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 128:841–852. 10.1016/j.cell.2007.01.035 [DOI] [PubMed] [Google Scholar]
- Garrus J.E., von Schwedler U.K., Pornillos O.W., Morham S.G., Zavitz K.H., Wang H.E., Wettstein D.A., Stray K.M., Côté M., Rich R.L., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 107:55–65. 10.1016/S0092-8674(01)00506-2 [DOI] [PubMed] [Google Scholar]
- Géminard C., De Gassart A., Blanc L., Vidal M.. 2004. Degradation of AP2 during reticulocyte maturation enhances binding of hsc70 and Alix to a common site on TFR for sorting into exosomes. Traffic. 5:181–193. 10.1111/j.1600-0854.2004.0167.x [DOI] [PubMed] [Google Scholar]
- Gullapalli A., Wolfe B.L., Griffin C.T., Magnuson T., Trejo J.. 2006. An essential role for SNX1 in lysosomal sorting of protease-activated receptor-1: Evidence for retromer-, Hrs-, and Tsg101-independent functions of sorting nexins. Mol. Biol. Cell. 17:1228–1238. 10.1091/mbc.E05-09-0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyaloglu A.C., von Zastrow M.. 2008. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 48:537–568. 10.1146/annurev.pharmtox.48.113006.094830 [DOI] [PubMed] [Google Scholar]
- Hasdemir B., Bunnett N.W., Cottrell G.S.. 2007. Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) mediates post-endocytic trafficking of protease-activated receptor 2 and calcitonin receptor-like receptor. J. Biol. Chem. 282:29646–29657. 10.1074/jbc.M702974200 [DOI] [PubMed] [Google Scholar]
- Henry A.G., White I.J., Marsh M., von Zastrow M., Hislop J.N.. 2011. The role of ubiquitination in lysosomal trafficking of δ-opioid receptors. Traffic. 12:170–184. 10.1111/j.1600-0854.2010.01145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hislop J.N., Marley A., Von Zastrow M.. 2004. Role of mammalian vacuolar protein-sorting proteins in endocytic trafficking of a non-ubiquitinated G protein-coupled receptor to lysosomes. J. Biol. Chem. 279:22522–22531. 10.1074/jbc.M311062200 [DOI] [PubMed] [Google Scholar]
- Hurley J.H., Hanson P.I.. 2010. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 11:556–566. 10.1038/nrm2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Sitaraman S., Hierro A., Beach B.M., Odorizzi G., Hurley J.H.. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 8:937–947. 10.1016/j.devcel.2005.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyuuma M., Kikuchi K., Kojima K., Sugawara Y., Sato M., Mano N., Goto J., Takeshita T., Yamamoto A., Sugamura K., Tanaka N.. 2007. AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct. Funct. 31:159–172. 10.1247/csf.06023 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lavoie C., Meerloo T., Lin P., Farquhar M.G.. 2002. Calnuc, an EF-hand Ca(2+)-binding protein, is stored and processed in the Golgi and secreted by the constitutive-like pathway in AtT20 cells. Mol. Endocrinol. 16:2462–2474. 10.1210/me.2002-0079 [DOI] [PubMed] [Google Scholar]
- Lu Q., Hope L.W., Brasch M., Reinhard C., Cohen S.N.. 2003. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA. 100:7626–7631. 10.1073/pnas.0932599100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerød L., Stuffers S., Brech A., Stenmark H.. 2007. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 8:1617–1629. 10.1111/j.1600-0854.2007.00630.x [DOI] [PubMed] [Google Scholar]
- Marchese A., Raiborg C., Santini F., Keen J.H., Stenmark H., Benovic J.L.. 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 5:709–722. 10.1016/S1534-5807(03)00321-6 [DOI] [PubMed] [Google Scholar]
- Marchese A., Paing M.M., Temple B.R.S., Trejo J.. 2008. G protein-coupled receptor sorting to endosomes and lysosomes. Annu. Rev. Pharmacol. Toxicol. 48:601–629. 10.1146/annurev.pharmtox.48.113006.094646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P.D.. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319. 10.1038/nm1201-1313 [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Yarovoy A., Perez-Caballero D., Bieniasz P.D.. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA. 100:12414–12419. 10.1073/pnas.2133846100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt M.W., McKittrick I., West M., Odorizzi G.. 2007. Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol. Biol. Cell. 18:697–706. 10.1091/mbc.E06-08-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.-Y., Morham S.G., Gygi S.P., Rodesch C.K., Sundquist W.I.. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G., Katzmann D.J., Babst M., Audhya A., Emr S.D.. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893–1903. 10.1242/jcs.00395 [DOI] [PubMed] [Google Scholar]
- Oestreich A.J., Aboian M., Lee J., Azmi I., Payne J., Issaka R., Davies B.A., Katzmann D.J.. 2007. Characterization of multiple multivesicular body sorting determinants within Sna3: A role for the ubiquitin ligase Rsp5. Mol. Biol. Cell. 18:707–720. 10.1091/mbc.E06-08-0680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paing M.M., Johnston C.A., Siderovski D.P., Trejo J.. 2006. Clathrin adaptor AP2 regulates thrombin receptor constitutive internalization and endothelial cell resensitization. Mol. Cell. Biol. 26:3231–3242. 10.1128/MCB.26.8.3231-3242.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricks T.K., Trejo J.. 2009. Phosphorylation of protease-activated receptor-2 differentially regulates desensitization and internalization. J. Biol. Chem. 284:34444–34457. 10.1074/jbc.M109.048942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A., Soh U.J.K., Paing M.M., Arora P., Trejo J.. 2009. Caveolae are required for protease-selective signaling by protease-activated receptor-1. Proc. Natl. Acad. Sci. USA. 106:6393–6397. 10.1073/pnas.0810687106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto A.G., Trejo J.. 2010. N-linked glycosylation of protease-activated receptor-1 second extracellular loop: A critical determinant for ligand-induced receptor activation and internalization. J. Biol. Chem. 285:18781–18793. 10.1074/jbc.M110.111088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B., Calistri A., Craig S., Popova E., Göttlinger H.G.. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 114:689–699. 10.1016/S0092-8674(03)00653-6 [DOI] [PubMed] [Google Scholar]
- Tanzi G.O., Piefer A.J., Bates P.. 2003. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J. Virol. 77:8440–8447. 10.1128/JVI.77.15.8440-8447.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos A.C., Truschel S.T., Tenza D., Hurbain I., Harper D.C., Berson J.F., Thomas P.C., Raposo G., Marks M.S.. 2006. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev. Cell. 10:343–354. 10.1016/j.devcel.2006.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M.. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 319:1244–1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Trejo J., Coughlin S.R.. 1999. The cytoplasmic tails of protease-activated receptor-1 and substance P receptor specify sorting to lysosomes versus recycling. J. Biol. Chem. 274:2216–2224. 10.1074/jbc.274.4.2216 [DOI] [PubMed] [Google Scholar]
- Trejo J., Hammes S.R., Coughlin S.R.. 1998. Termination of signaling by protease-activated receptor-1 is linked to lysosomal sorting. Proc. Natl. Acad. Sci. USA. 95:13698–13702. 10.1073/pnas.95.23.13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo J., Altschuler Y., Fu H.-W., Mostov K.E., Coughlin S.R.. 2000. Protease-activated receptor-1 down-regulation: A mutant HeLa cell line suggests novel requirements for PAR1 phosphorylation and recruitment to clathrin-coated pits. J. Biol. Chem. 275:31255–31265. 10.1074/jbc.M003770200 [DOI] [PubMed] [Google Scholar]
- Usami Y., Popov S., Göttlinger H.G.. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614–6622. 10.1128/JVI.00314-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M.S., Rubinstein E., Raposo G.. 2011. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell. 21:708–721. 10.1016/j.devcel.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroling B., Sanders M., Baakman C., Borrmann A., Verhoeven S., Klomp J., Oliveira L., de Vlieg J., Vriend G.. 2011. GPCRDB: Information system for G protein-coupled receptors. Nucleic Acids Res. 39(Database issue):D309–D319. 10.1093/nar/gkq1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T.-K.H., Hung D.T., Wheaton V.I., Coughlin S.R.. 1991. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 64:1057–1068. 10.1016/0092-8674(91)90261-V [DOI] [PubMed] [Google Scholar]
- Watson H., Bonifacino J.S.. 2007. Direct binding to Rsp5p regulates ubiquitination-independent vacuolar transport of Sna3p. Mol. Biol. Cell. 18:1781–1789. 10.1091/mbc.E06-10-0887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B.L., Marchese A., Trejo J.. 2007. Ubiquitination differentially regulates clathrin-dependent internalization of protease-activated receptor-1. J. Cell Biol. 177:905–916. 10.1083/jcb.200610154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Hurley J.H.. 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 464:864–869. 10.1038/nature08849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Bouley R., Lin H.Y., Bechoua S., Sun T.X., Del Re E., Shioda T., Raychowdhury M.K., Lu H.A.J., Abou-Samra A.B., et al. 2007. Alix (AIP1) is a vasopressin receptor (V2R)-interacting protein that increases lysosomal degradation of the V2R. Am. J. Physiol. Renal Physiol. 292:F1303–F1313. 10.1152/ajprenal.00441.2005 [DOI] [PubMed] [Google Scholar]
- Zhai Q., Fisher R.D., Chung H.-Y., Myszka D.G., Sundquist W.I., Hill C.P.. 2008. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 15:43–49. 10.1038/nsmb1319 [DOI] [PubMed] [Google Scholar]
- Zhan L., Liu B., Jose-Lafuente M., Chibalina M.V., Grierson A., Maclean A., Nasir J.. 2008. ALG-2 interacting protein AIP1: A novel link between D1 and D3 signalling. Eur. J. Neurosci. 27:1626–1633. 10.1111/j.1460-9568.2008.06135.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.