The elastic behavior of the 3D extracellular matrix determines the relative polarization of intracellular signaling and whether cells migrate using lamellipodia or lobopodia.
Abstract
We search in this paper for context-specific modes of three-dimensional (3D) cell migration using imaging for phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and active Rac1 and Cdc42 in primary fibroblasts migrating within different 3D environments. In 3D collagen, PIP3 and active Rac1 and Cdc42 were targeted to the leading edge, consistent with lamellipodia-based migration. In contrast, elongated cells migrating inside dermal explants and the cell-derived matrix (CDM) formed blunt, cylindrical protrusions, termed lobopodia, and Rac1, Cdc42, and PIP3 signaling was nonpolarized. Reducing RhoA, Rho-associated protein kinase (ROCK), or myosin II activity switched the cells to lamellipodia-based 3D migration. These modes of 3D migration were regulated by matrix physical properties. Specifically, experimentally modifying the elasticity of the CDM or collagen gels established that nonlinear elasticity supported lamellipodia-based migration, whereas linear elasticity switched cells to lobopodia-based migration. Thus, the relative polarization of intracellular signaling identifies two distinct modes of 3D cell migration governed intrinsically by RhoA, ROCK, and myosin II and extrinsically by the elastic behavior of the 3D extracellular matrix.
Introduction
How normal cells move efficiently through chemically and structurally diverse 3D environments in vivo is not well understood. In contrast, findings of metazoan cells migrating on uniform 2D surfaces in vitro have led to a comprehensive model of cell motility wherein polarized signaling orchestrates cell movement by directing lamellipodial protrusion at the leading edge, adhesion to the underlying substrate, and retraction at the trailing edge (Lauffenburger and Horwitz, 1996; Ridley et al., 2003). The second messenger phosphatidylinositol (3,4,5)-trisphosphate (PIP3) is enriched at the leading edge (Haugh et al., 2000), where it can recruit downstream effectors, such as guanine exchange factors (Côté et al., 2005) that activate the Rho family of GTPases. Rho family members Rac1, Cdc42, and RhoA are active at the leading edge and coordinate protrusion and adhesion (Kraynov et al., 2000; Nalbant et al., 2004; Pertz et al., 2006; Machacek et al., 2009).
Disrupting the subcellular localization of Rac1, Cdc42, or RhoA can lead to defects in adhesion and motility (van Hennik et al., 2003; ten Klooster et al., 2006; Bass et al., 2007), whereas the light-mediated activation of photosensitive guanine exchange factor, Rac1, or Cdc42 constructs at discrete regions of the plasma membrane triggers protrusion and directional cell migration (Levskaya et al., 2009; Wu et al., 2009). Discrepancies in the localization of Rho family GTPase activities during cell migration in vivo versus on 2D surfaces might reveal differences in the mechanisms that drive cell motility. Studies of cancer cell migration in 3D environments show that metastatic cells can switch between adhesion-dependent mesenchymal (elongated) and adhesion-independent amoeboid (rounded) cell motility (Table S1), driven by actin polymerization and actomyosin contraction, respectively (Wolf et al., 2003; Lämmermann and Sixt, 2009). Although these two different modes of cancer cell migration have specific requirements for Rho family GTPase signaling, how that signaling is organized is not known. Furthermore, it is unclear how the mesenchymal–amoeboid transition relates to normal 3D cell migration (Sanz-Moreno and Marshall, 2010).
Some aspects of intracellular signaling organization during cell migration in vivo can differ from the organization seen on 2D surfaces. Chemotaxing primordial germ cells display randomly distributed regions of RhoA activity and a uniform distribution of PIP3 in the plasma membrane (Dumstrei et al., 2004; Kardash et al., 2010). However, Rac1 activity is enriched at the leading edge of migrating border cells and primordial germ cells during development, and PIP3 is abundant at the leading edge of neutrophils during interstitial migration toward wounded tissue (Kardash et al., 2010; Wang et al., 2010; Yoo et al., 2010). The reason for these differences is not clear, but they may result from structural differences in the surrounding ECM (Friedl and Wolf, 2010). Two structural parameters that characterize the ECM are stiffness, defined by the elastic or Young’s modulus (E; Engler et al., 2006), and strain stiffening, a measurement of how the stiffness of a material depends on the magnitude of force applied to it (here measured as Ehigh/Emed; Storm et al., 2005; Winer et al., 2009). Strain stiffening (Ehigh/Emed > 1) is a form of nonlinear elasticity; thus, materials that do not undergo strain stiffening (Ehigh/Emed = 1) are considered linearly elastic.
Tissue explants and in vitro models of the 3D ECM, such as the cell-derived matrix (CDM) and type I collagen, can closely mimic different complex tissue environments (Elsdale and Bard, 1972; Cukierman et al., 2001; Even-Ram and Yamada, 2005; Ahlfors and Billiar, 2007; Wolf et al., 2009) and permit high-resolution live-cell imaging to visualize intracellular signaling. We used primary human fibroblasts in these models to test the hypothesis that structurally distinct 3D ECM environments support different modes of normal cell migration. We find that the degree of polarization of PIP3 and Rho family GTPase signaling at the leading edge identifies two distinct modes of normal cell motility governed intrinsically by RhoA, Rho-associated protein kinase (ROCK), and myosin II and extrinsically by the elastic behavior of the ECM.
Results
Lamellipodia-independent 3D migration in dermal explants
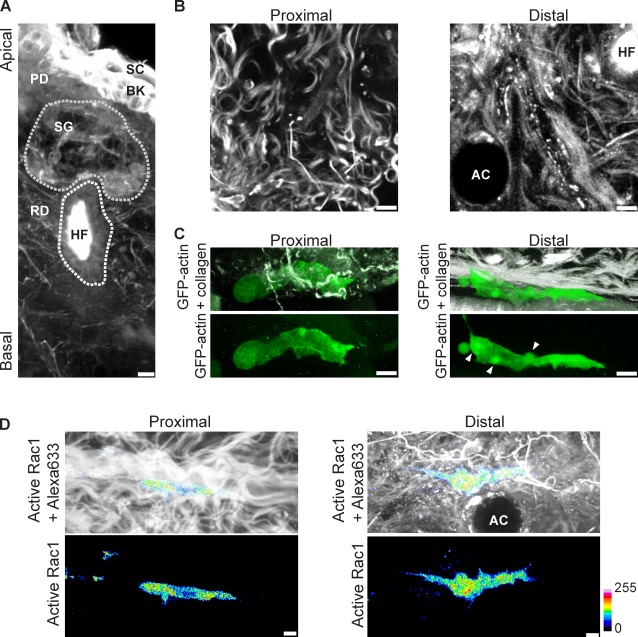
Dermal tissue explants derived from mouse ears contain many of the structural features found in the human dermis (Fig. 1 A; Montagna et al., 1992; Lämmermann et al., 2008). Thick bundles of collagen fibers are proximal to the basal explant surface, whereas adipocytes and hair follicles embedded within reticular collagen and elastic fibers tend to be more distal (Fig. 1 B). Human fibroblasts migrating in both proximal and distal regions of the explant were predominantly uniaxial, consistent with cells migrating inside 3D in vitro models of the ECM (Fig. 1 C; Bard and Hay, 1975; Cukierman et al., 2001). The prominent lamellipodia and flat lamellae of cells migrating in 2D were not apparent, and many cells instead featured large blunt, cylindrical protrusions characteristic of lobopodia (Kudo, 1977), an intracellular pressure-driven protrusion (Fig. 1 C and Video 1; Yanai et al., 1996). This morphology of human foreskin fibroblasts (HFFs) in dermal explants was similar to that of cells in wounded tissue (Singer et al., 1984). In addition, cells often displayed small blebs along their sides that we termed lateral blebs (Fig. 1 C, arrowheads). Imaging a dual-chain Rac1 biosensor based on intermolecular fluorescence resonance energy transfer (FRET) revealed that, contrary to findings using 2D surfaces (Kraynov et al., 2000), active Rac1 was not targeted to the leading edge during HFF migration in both proximal or distal regions of the explant (85%, n = 26; Rac1 polarization index [PI] = −0.04 ± 0.09, in which 1 = forward polarization, 0 = nonpolarization, and −1 = rear polarization; Figs. 1 D, S1, and S2). Thus, Rac1 activity was not polarized in elongated HFFs migrating within the structurally heterogeneous, physiological 3D environment of dermal explants. To study the mechanistic basis of this apparently novel mode of lamellipodia-independent 3D cell motility (hereafter referred to as lobopodia-based migration), we recapitulated it using 3D in vitro models of the ECM.
Figure 1.
Lobopodia-based 3D migration occurs in the mammalian dermis. (A) A 3D reconstruction of a mouse ear dermal explant labeled with Alexa Fluor 633 (grayscale). Stratum corneum (SC), basal keratinocytes (BK), papillary dermis (PD), and reticular dermis (RD) are indicated. Sebaceous gland (SG) and hair follicle (HF) are outlined in gray. (B) Examples of ECM structures proximal (left) and distal (right) to the basal surface of a dermal explant labeled with Alexa Fluor 633. Images are from the same confocal stack, 9 µm (left) and 30 µm (right) from the basal surface. AC, adipocyte. (C) 3D reconstructions of lobopodia-bearing HFFs migrating in proximal and distal collagen; GFP-actin is shown in green, and second harmonic imaging of collagen appears in grayscale. Arrowheads indicate lateral blebs. (D) Active Rac1 is not targeted to the leading edge of HFFs migrating in the mammalian dermis. Rac1 activity was imaged in HFFs migrating in proximal or distal ECMs; active Rac1, representing the Fc image, was pseudocolored according to the 16-color scale shown to the right of the figure, and the explant was labeled with Alexa Fluor 633 (grayscale). All cells are oriented with the leading edge toward the right of the figure. Bars, 5 µm.
Lobopodia and lamellipodia in 3D in vitro models of the ECM
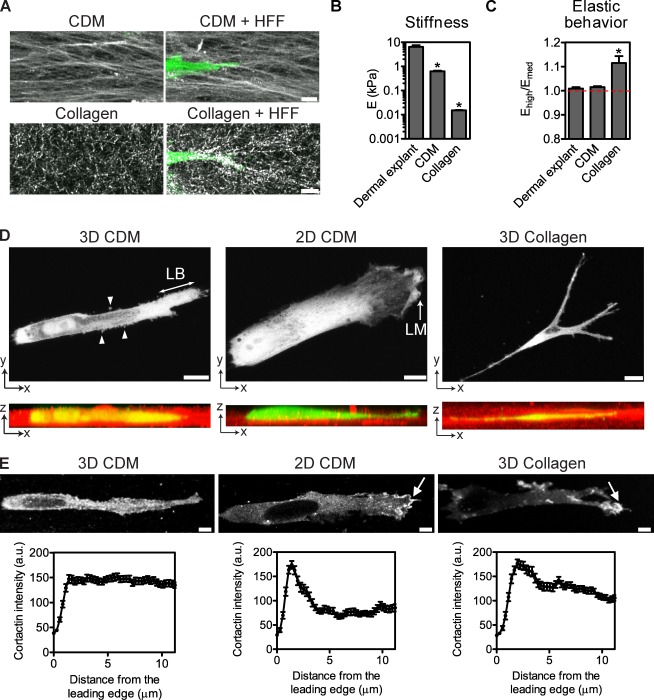
The CDM contains fibronectin, collagen I and III, hyaluronic acid, heparan sulfate proteoglycan, and thrombospondin in parallel fibers 50–500 nm thick (Fig. 2 A; Hedman et al., 1979; Allio and McKeown-Longo, 1988). Polymerized collagen I forms a nonaligned 3D meshwork (Fig. 2 A) of single or bundled collagen fibers 2–500 nm in diameter (Elsdale and Bard, 1972; Gelman et al., 1979). We compared the mechanical properties of dermal explants, the CDM, and collagen by characterizing their stiffness and elastic behavior. Dermal explants (6,427 Pa, range of 277–19,400 Pa) and CDMs (627 Pa, range of 224–2,454 Pa) were stiffer than 1.7 mg/ml collagen (15 Pa, range of 11–21 Pa), did not undergo strain stiffening (Ehigh/Emed = 1.01), and were thus linearly elastic (Fig. 2, B and C). In contrast, 1.7 mg/ml collagen displayed the strain-stiffening behavior (Ehigh/Emed = 1.12) characteristic of nonlinear elastic materials as previously established (Storm et al., 2005). Cells migrating in the soft 1.7 mg/ml of 3D collagen deformed the collagen fibers as described in a previous study of collagen remodeling by motile cells (Grinnell and Lamke, 1984), whereas the arrangement of fibers within the stiffer CDM was largely unaffected by migrating HFFs (Fig. 2 A).
Figure 2.
CDM and type I collagen support lobopodia- and lamellipodia-based 3D migration, respectively. (A) HFF-generated CDM has an aligned, fibrillar structure (top left), whereas polymerized 1.7 mg/ml type I collagen forms a random meshwork (bottom left). Both images are maximum projections of 30-µm confocal stacks. Collagen is remodeled by migrating HFFs (GFP, green) along the axis of migration (bottom right), whereas the organization of the CDM is unaffected by migrating HFFs (top right). The CDM was labeled with Alexa Fluor 633, and collagen was visualized by reflection microscopy. (B) Matrix stiffness (Young’s modulus [E]) of the indicated 3D matrices. (C) Strain-stiffening (Ehigh/Emed) behavior of the indicated 3D matrices. Ehigh/Emed > 1 indicates nonlinear elasticity, whereas Ehigh/Emed = 1 indicates linear elasticity (dashed red line). (D) Collagen and 2D CDM support lamellipodia-based migration, whereas 3D CDM triggers lobopodia-based motility. (top) Maximum projections of HFFs expressing GFP migrating inside the 3D CDM, on top of the 2D CDM, or inside type I collagen. LM, lamellipodium; LB, lobopodium. (bottom) The orthogonal views of the corresponding panel above, with the CDM (Alexa Fluor 633) and type I collagen (reflection microscopy) in red. Arrowheads indicate lateral blebs. (E) Cortactin is not enriched at the leading edge during lobopodia-based migration. HFFs migrating in the indicated ECM were fixed and immunostained for cortactin. Arrows indicate the local accumulation of cortactin at the leading edge. Bottom graphs correspond with their respective top images and represent the mean cortactin intensity measured from the leading edge (0 µm) toward the cell center. Each cortactin intensity profile was averaged from 13 cells, with three measurements per cell. Bars: (A and E) 5 µm; (D, top left and middle) 10 µm; (D, top right) 20 µm. All cells are oriented with the leading edge toward the right of the figure. Error bars show means ± SEM. *, P < 0.001 versus the dermal explant. a.u., arbitrary unit.
Consistent with the similar stiffness and elastic behavior of the CDM and dermal explants, the majority of HFFs migrating inside the CDM also lacked obvious lamellipodia and instead displayed the blunt cylindrical protrusions and lateral blebs observed during lamellipodia-independent migration in dermal explants (77%, n = 31; Fig. 2 D and Video 2). Lobopodia-based migration appeared specific to 3D because HFFs migrating on top of the CDM (2D CDM) formed broad lamellipodia with prominent ruffles (100%, n = 33; Fig. 2 D and Video 2). In contrast to cells migrating in the 3D CDM, cells migrating in 1.7 mg/ml of 3D collagen formed multiple branched protrusions tipped with small lamellipodia (100%, n = 12; Fig. 2 D, right panels; and Video 3), consistent with a previous study (Bard and Hay, 1975). To confirm that these morphological differences corresponded to distinct cellular structures, we compared the distribution of the actin-binding protein cortactin (Kaksonen et al., 2000) in HFFs migrating in collagen and the CDM. Although cortactin was enriched at the leading edge during migration on the 2D CDM and in 3D collagen, no such enrichment was detected during migration in the 3D CDM (Fig. 2 E). Similar distributions were observed for vasodilator-stimulated phosphoprotein (VASP; Bear et al., 2002) and F-actin (by phalloidin staining; Small, 1981). Together, these data indicate that normal fibroblasts use at least two distinct modes of 3D cell migration: lobopodia-based migration in the stiff, linear elastic environment of dermal explants and the 3D CDM and lamellipodia-based migration in soft, nonlinear elastic 3D collagen.
Nonpolarized signaling during lobopodia-based migration
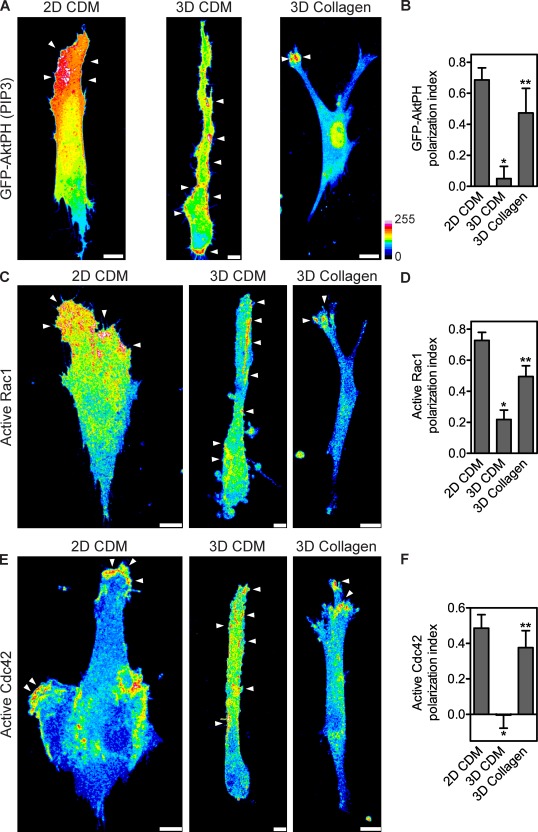
We investigated whether the distinct morphological features of lobopodia- versus lamellipodia-based 3D migration were linked to different signaling mechanisms. We compared the canonical polarization of PIP3, Rac1, and Cdc42 signaling during 2D lamellipodia-based migration (Haugh et al., 2000; Kraynov et al., 2000; Nalbant et al., 2004) by imaging the relative distribution of PIP3 and active Rac1 and Cdc42 in live HFFs migrating in the CDM and collagen. The intracellular localization of PIP3 was imaged by binding of the pleckstrin homology (PH) domain of Akt fused to GFP (Haugh et al., 2000). PIP3 was highly enriched near the leading edge of the elongated cells migrating on top of the CDM (PI = 0.69, n = 13; Fig. 3, A and B), as reported for cells migrating on 2D surfaces using lamellipodia (Haugh et al., 2000). This enrichment was specific compared with localization of phosphatidylinositol (4,5)-bisphosphate (PIP2) using GFP-PLC-δPH and was confirmed by immunolocalization of PIP3 (Fig. S3; Várnai and Balla, 1998; van Rheenen and Jalink, 2002). Importantly, PIP3 was not polarized toward the leading edge of HFFs using lobopodia-based motility in the CDM but was instead distributed around the cell in clusters at the plasma membrane in a pattern indistinguishable from PIP2 (PI = 0.05, n = 18; Figs. 3, A and B; and S3). In contrast, PIP3 was concentrated at the leading edge of small lamellipodia during migration in 3D collagen (PI = 0.47, n = 13; Fig. 3, A and B). Thus, lobopodia-based migration in the 3D CDM does not require the polarization of PIP3 at the leading edge.
Figure 3.
Nonpolarized PIP3, Rac1, and Cdc42 signaling during lobopodia-based 3D migration. (A) Maximally projected confocal stacks of HFFs expressing GFP-AktPH to detect PIP3, migrating on the 2D CDM, in the 3D CDM, or inside 1.7 mg/ml of 3D collagen. Images were pseudocolored according to the 16-color scale. Bars, 10 µm. (B) Mean PI of GFP-AktPH in HFFs migrating in the indicated ECM environments. (C–F) Localization of active Rac1 and Cdc42 away from the leading edge during lobopodia-based 3D migration inside the CDM. (C and E) Maximally projected confocal stacks of HFFs expressing Rac1 (C) or Cdc42 (E) biosensors migrating on the 2D CDM (left), in the 3D CDM (middle), or in 3D collagen (right). The Fc images, representing the total activity of each GTPase, were pseudocolored according to the 16-color scale. Arrowheads indicate regions of intracellular signaling. Bars: (3D CDM) 5 µm; (collagen and 2D CDM) 10 µm. (D and F) Mean PI of active Rac1 (D) and active Cdc42 (F) in HFFs migrating in the indicated ECM environments. All cells are oriented with the leading edge toward the top of the figure. Error bars show means ± SEM. *, P < 0.05 versus the 2D CDM; **, P < 0.05 versus the 3D CDM.
Active Rac1 and Cdc42 were imaged using dual-chain FRET-based biosensors (Kraynov et al., 2000; Picard et al., 2009). Both the single- and dual-chain versions of these biosensors have been used to image the distribution of Rho family GTPase activities in living cells (Pertz and Hahn, 2004). To validate the FRET-based biosensors, we determined that their expression in HFFs did not affect the amount or activity of the corresponding endogenous GTPase (Boulter et al., 2010), and the magnitude of the corrected FRET (Fc) signal imaged in cells expressing constitutively active, nonfunctional, or wild-type versions of the biosensors correlated with the amounts of activity detected by a GTPase pull-down assay (Fig. S1, A, B, D, and E; Benard et al., 1999). Plotting Fc versus donor intensity for the constitutively active and nonfunctional versions of each biosensor showed the signal from the wild-type Rac1 and Cdc42 biosensors could be considered positive when Fc > 500 arbitrary units (Fig. S1, C and F). We confirmed that their expression did not significantly affect cell velocity or downstream signaling (Fig. S2, A and B). Additionally, the subcellular localization of the CFP-tagged Rac1 and Cdc42 was predominantly cytosolic and consistent with the corresponding endogenous protein (Fig. S2 C); excessive GTPase expression would have resulted in inappropriate membrane targeting (Michaelson et al., 2001).
Active Rac1 and Cdc42 were both polarized near the leading edge of cells on the 2D CDM (Rac1: PI = 0.73, n = 27; Cdc42: PI = 0.49, n = 14; Fig. 3, C–F) and in 3D collagen (Rac1: PI = 0.49, n = 34; Cdc42: PI = 0.38, n = 21; Fig. 3, C–F), as previously reported for lamellipodia-based migration on 2D surfaces (Kraynov et al., 2000; Nalbant et al., 2004). However, during lobopodia-based migration in the 3D CDM, active Rac1 and Cdc42 were no longer restricted to the leading edge; instead, they were localized to patches of unknown function around the perimeter of the cell (Rac1: PI = 0.22, n = 44; Cdc42: PI = 0, n = 21; Fig. 3, C–F), consistent with the nonpolarized distribution of active Rac1 in HFFs in dermal explants (Fig. 1 D). The distribution of Rac1 and Cdc42 activity in lobopodia could not be attributed to failure of the biosensors because nonfunctional versions reported uniform Fc patterns in HFFs on 2D and in 3D CDMs (100%; Fig. S2 D). Together, these data show that the canonical polarizations of PIP3, Rac1, and Cdc42 signaling during 2D migration are not necessary for lobopodia-based 3D migration in dermal explants and the CDM but are observed during lamellipodia-based migration in 3D collagen.
RhoA, ROCK, and myosin II are required for lobopodia-based 3D migration
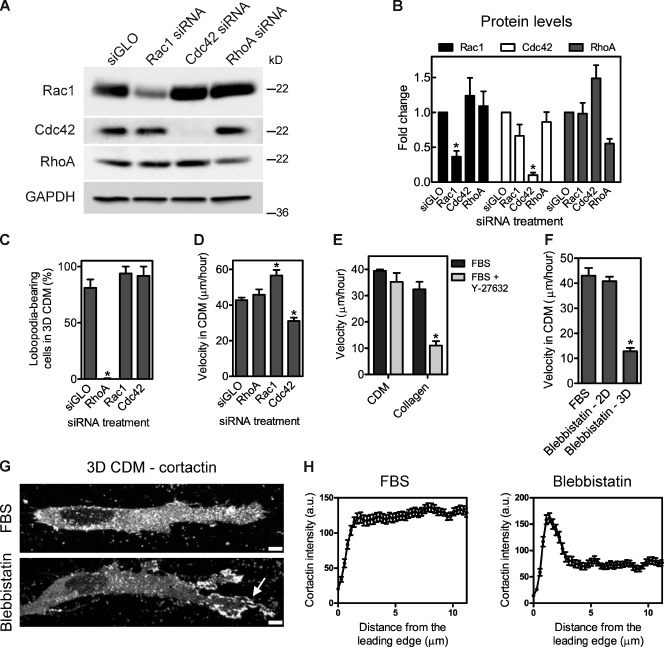
To define further the mechanistic basis of lobopodia-based 3D migration, the roles of Rac1, Cdc42, and RhoA in motility in the 3D CDM were tested by specific siRNA-mediated knockdown (Figs. 4, A and B; and S4). Reducing Rac1 or Cdc42 protein levels moderately increased or decreased the velocity of HFFs migrating in and on the CDM, respectively, without affecting the mode of 3D cell motility (Fig. 4, C and D). Cells that were inside the 3D CDM continued to migrate without lamellipodia (94% for Rac1 siRNA and 92% for Cdc42 siRNA), and many of the cells retained lateral blebs. In contrast to Rac1 and Cdc42 siRNA, knockdown of RhoA protein and activity dramatically switched the mode of migration in the 3D CDM from lobopodia to lamellipodia based. RhoA-depleted cells migrated in the 3D CDM with distinct lamellipodia at the leading edge (100%; Figs. 4 C and S4 A and Video 4) and without any lateral blebs. The role of RhoA in regulating the mode of migration was confirmed with independent single siRNAs (Fig. S4, B–D) and a single siRNA from the original RhoA siRNA pool (Fig. S4, E–G). Additionally, treating cells migrating in the 3D CDM with a RhoA inhibitor switched the cells to lamellipodia-based motility (Fig. S4, H and I). Despite switching the mode of 3D cell migration in the CDM, knocking down RhoA did not affect the velocity of HFF migration in or on the CDM (Fig. 4 D).
Figure 4.
RhoA, ROCK, and myosin II are required for lobopodia-based 3D migration inside CDM. (A) A representative Western blot demonstrating the specificity of siRNA-mediated knockdown of Rac1, Cdc42, or RhoA. HFFs were transfected with the indicated siRNAs and lysed 72 h after transfection, and the lysates were blotted with the indicated antibodies. (B) Quantification of Western blots represented in A. (C) RhoA siRNA treatment switches HFFs to lamellipodia-based 3D migration in the CDM. The percentage of lobopodia-bearing HFFs migrating inside the CDM after the indicated treatments. (A and B) *, P < 0.001 versus the siGLO control. 48 h after siRNA treatment, HFFs were transfected with GFP-actin and imaged migrating in the 3D CDM. (D) Quantification of the velocity of siRNA-treated HFFs in the CDM 72–84 h after transfection. *, P < 0.05 versus the siGLO control. (E) ROCK dependence distinguishes HFF migration in collagen from the CDM. Quantification of the velocity of HFFs migrating in the CDM or 1.7 mg/ml collagen treated with FBS or FBS + 10 µM Y-27632. *, P < 0.03 versus the FBS control. (F) Quantification of HFF velocity in the CDM when treated with FBS or FBS + 25 µM blebbistatin. Blebbistatin treatment resulted in two subsets of HFFs, rapidly moving spread cells on top of the CDM (2D), and slowly moving elongated cells inside the CDM (3D). *, P < 0.001 versus the FBS control. (G and H) Myosin II is required for 3D lobopodia-based migration. (G) Cortactin localization in HFFs in the 3D CDM, either untreated or treated with 25 µM blebbistatin. The arrow indicates the local accumulation of cortactin at the leading edge. Bars, 5 µm. (H) The mean cortactin intensity profile, measured from the leading edge (0 µm) toward the cell center, of cells treated with FBS or FBS + 25 µM blebbistatin. Each cortactin intensity profile was averaged from 13 cells, with three measurements per cell. All cells are oriented with the leading edge toward the right of the figure. Error bars show means ± SEM. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; a.u. arbitrary unit.
Chemical inhibition of the RhoA effector ROCK also switched the mode of HFF migration in the 3D CDM, transitioning cells from lobopodia- to lamellipodia-based migration within 20 min of treatment (50%, n = 10; Video 5). Similar to RhoA siRNA, ROCK inhibition switched the mode without affecting the velocity of cells migrating in or on the CDM (Fig. 4 E). In contrast, ROCK inhibition significantly reduced HFF migration in 3D collagen, as previously observed for epithelial cells (Provenzano et al., 2008). Thus, ROCK inhibition functionally distinguishes lobopodia- from lamellipodia-based 3D motility. Inhibition of the ROCK target myosin II by blebbistatin also switched the mode to lamellipodia-based migration in the 3D CDM (Fig. 4, G and H). In contrast to RhoA and ROCK inhibition, myosin II inhibition significantly reduced the velocity of elongated cells migrating in the 3D CDM, apparently impeding translocation of the nucleus through the matrix, whereas the migration of cells spread on top of the matrix was unaffected (Fig. 4 F and Video 6) and was consistent with the effect of blebbistatin on 2D fibroblast migration (Even-Ram et al., 2007). Therefore, RhoA, ROCK, and myosin II form part of the mechanistic basis of lobopodia-based migration. RhoA and ROCK inhibition switched the mode of migration without affecting the efficiency of migration, whereas myosin II activity was required for both lobopodia formation and efficient migration in the 3D CDM.
3D matrix adhesions and lobopodia-based migration
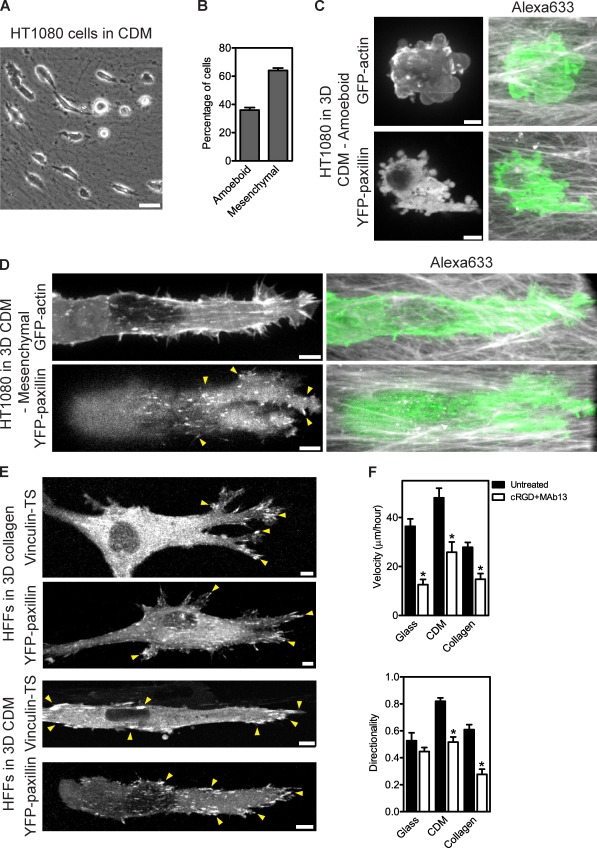
To determine that amoeboid cancer cell motility (Table S1) was distinct from the lobopodia-based migration of normal cells, despite their shared requirement for RhoA activity (Sahai and Marshall, 2003), we imaged the HT1080 human fibrosarcoma cell line within the CDM (Wolf et al., 2003). HT1080 cells adopted a mixture of amoeboid and mesenchymal morphologies in aligned, fibrillar regions of the CDM (Fig. 5, A–D) similar to those reported for breast cancer cells (Deakin and Turner, 2011). Round amoeboid cells in the CDM lacked 3D matrix adhesions (90%, n = 20) but had large prominent blebs (100%, n = 20; Fig. 5 C). Elongated mesenchymal cells exhibited actin stress fibers and fan-shaped protrusions consistent with lamellipodia (100%, n = 20; Fig. 5 D). These cells also formed 3D matrix adhesions (100%, n = 20; Fig. 5 D). In contrast to primary fibroblasts in the same 3D environment, no lobopodia-bearing HT1080 cells were observed migrating in the CDM. Unlike the two modes of cancer cell motility, HFFs formed 3D matrix adhesions containing paxillin and vinculin during both lamellipodia (100%, n = 18)- and lobopodia (100%, n = 20)-based 3D migration (Fig. 5 E). Blocking αvβ3 and β1 integrins significantly decreased the velocity and directionality of HFFs migrating in and on the CDM and collagen, indicating that integrin-mediated adhesion contributed to the efficient directional migration of these cell populations (Fig. 5 F). Therefore, lobopodia-based 3D migration was distinct from amoeboid and mesenchymal cancer cell motility, based on the formation of 3D matrix adhesions, its integrin dependence, morphology, and regulation by the ECM.
Figure 5.
Lobopodia-based migration is distinct from cancer cell motility. (A) Amoeboid and mesenchymal HT1080 cells in the CDM. Phase-contrast image showing amoeboid (rounded) and mesenchymal (elongated) HT1080 cells in the CDM. (B) The percentage of amoeboid and mesenchymal HT1080 cells in the CDM (n = 1,001). (C and D) HT1080 cells do not form lobopodia in the 3D CDM. Round amoeboid cells lack matrix adhesions, whereas elongated mesenchymal cells have prominent lamellipodia and matrix adhesions. Images show maximally projected confocal stacks of amoeboid (C) and mesenchymal (D) HT1080 cells, expressing GFP-actin or YFP-paxillin, migrating inside the CDM (Alexa Fluor 633, grayscale). Arrowheads indicate matrix adhesions. (E) Matrix adhesions are present during both lamellipodia- and lobopodia-based 3D migration of elongated normal fibroblasts. Maximally projected confocal stacks of HFFs expressing YFP-paxillin or vinculin–tension sensor (TS) migrating inside 3D collagen or the 3D CDM. (F) Matrix adhesions contribute to both lamellipodia- and lobopodia-based migration of normal fibroblasts. Blocking αvβ3 and β1 integrins significantly decreased the velocity and directionality of HFFs migrating in 3D collagen and CDM, indicating that integrin-mediated adhesion contributed to the efficient directional migration of both cell populations. Quantification of the velocity (top) and directionality (bottom) of HFFs migrating on glass, CDM, or 3D collagen, either in media or in media with 100 µM cyclic RGD (cRGD; an αVβ3-blocking peptide) plus 500 µg/ml β1 integrin–blocking antibody (mAb13). *, P < 0.05 versus the untreated control. All cells are oriented with the leading edge toward the right of the figure. Bars: (A) 50 µm; (C–E) 5 µm.
Matrix elastic behavior dictates the mode of 3D migration
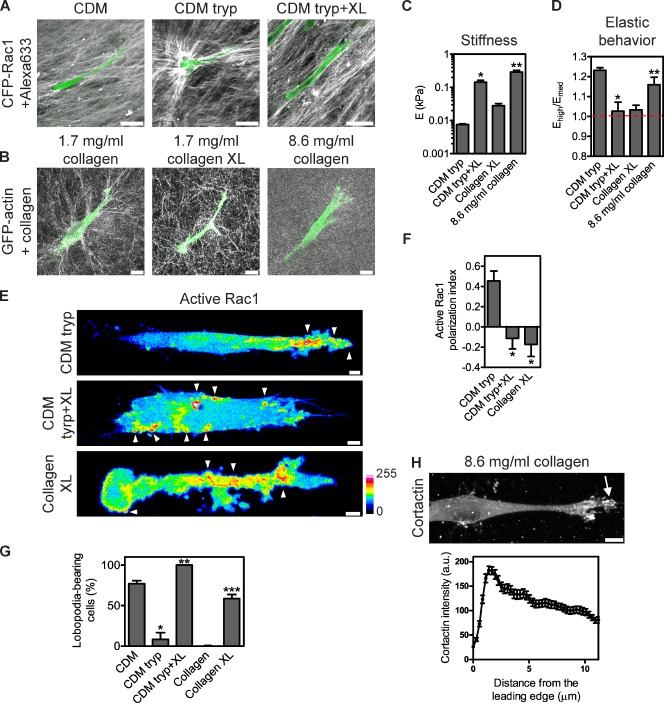
To define the relationship of matrix stiffness and/or elastic behavior to the mode of normal 3D cell migration, we manipulated these physical characteristics without affecting the organization of the matrices (Fig. 6, A–D). Trypsinization decreased the stiffness of the CDM (8 Pa, range of 4–10 Pa) and rendered it nonlinearly elastic (Ehigh/Emed = 1.23) compared with the untreated CDM (Fig. 2, B and C) and permitted fiber remodeling by migrating cells. Cross-linking the trypsinized CDM increased the stiffness (143 Pa, range of 19–243 Pa; Fig. 6, B and C), restored the linear elasticity of the matrix (Ehigh/Emed = 1.03; Fig. 6 D), and prevented cells from remodeling the fibers. Although cross-linking the 1.7 mg/ml collagen gels only marginally increased their stiffness (28 Pa, range of 12–201 Pa [Fig. 6 C] versus 15 Pa, range of 11–21 Pa for uncross-linked 1.7 mg/ml collagen [Fig. 2 B]), it rendered the material linearly elastic (Ehigh/Emed = 1.03; Fig. 6 D) compared with uncross-linked 1.7 mg/ml collagen (Fig. 2 C). Elongated fibroblasts used lamellipodia-based migration inside the trypsinized CDM (92 versus 23% in the untreated CDM; Fig. 6 G and Video 7), with Rac1 activity polarized toward the leading edge (PI = 0.45, n = 17; Fig. 6, E and F). Cells migrated inside the trypsinized and cross-linked CDM without polarization of active Rac1 at the leading edge (PI = −0.11, n = 16; Fig. 6, E and F), consistent with lobopodia-based motility (100%; Fig. 6 G and Video 8). Importantly, cross-linked 1.7 mg/ml collagen supported lobopodia-based migration of the majority of cells (59 versus 0% in untreated collagen; Fig. 6 G and Video 9), with a loss of polarized Rac1 activity in leading protrusions (PI = −0.17, n = 6; Fig. 6, E and F).
Figure 6.
Matrix elastic behavior governs the mode of normal 3D migration. (A) HFFs can remodel trypsinized CDM (middle), whereas the structure of the untreated (left) or the trypsinized (tryp) and cross-linked CDM (XL; right) is unaffected by HFF migration. CFP-Rac1 is shown in green, and Alexa Fluor 633–labeled matrix is in gray. Bars, 40 µm. (B) Cross-linked 1.7 mg/ml collagen (middle) and 8.6 mg/ml collagen (right) are not remodeled during HFF migration. GFP-actin is shown in green, and collagen is shown in gray (reflection). Bars, 10 µm. (C) Matrix stiffness (Young’s modulus [E]) of the indicated modified matrices. (D) Strain-stiffening (Ehigh/Emed) behavior of the indicated native and modified matrices. The dashed red line indicates a value of Ehigh/Emed corresponding to 1 (linear elasticity). (C and D) *, P < 0.05 versus trypsinized CDM; **, P < 0.05 versus cross-linked collagen. (E–G) Trypsinization and chemical cross-linking redistribute Rac1 activity and switch the mode of cell migration. (E) Maximally projected confocal stacks of HFFs expressing the Rac1 biosensor migrating in the trypsinized CDM (top), trypsinized and cross-linked CDMs (middle), or cross-linked collagen (bottom). The Fc images, representing the total activity of each GTPase, were pseudocolored according to the 16-color scale. Arrowheads indicate regions of intracellular signaling. Bars, 5 µm. (F) Mean PI of active Rac1 in HFFs migrating in the indicated ECM environments. *, P < 0.05 versus 3D trypsinized CDM. (G) The percentage of lobopodia-bearing HFFs migrating inside the CDM or collagen treated as indicated. *, P < 0.001 versus untreated CDM; **, P < 0.001 versus trypsinized CDM; ***, P < 0.007 versus untreated collagen. (H) 8.6 mg/ml collagen supports lamellipodia-based 3D cell migration. (top) A representative image of cortactin enrichment at the leading edge of cells migrating in 8.6 mg/ml collagen (arrow). (bottom) The mean cortactin intensity profile measured from 13 cells, with three measurements per cell. Cells are oriented with their leading edge toward the top right (A and B) or the right (E and H) of the figure. a.u., arbitrary unit.
To determine whether matrix stiffness, capacity for remodeling, or elastic behavior govern lobopodia formation, we examined HFF migration in 8.6 mg/ml collagen. Cells in 8.6 mg/ml collagen underwent lamellipodia-based 3D migration, including enrichment of cortactin at the leading edge (Fig. 6 H). The 8.6 mg/ml collagen was substantially stiffer than 1.7 mg/ml collagen (∼19×, P < 0.001) and was not remodeled during cell migration, as expected (Fig. 6, B and C; and Video 10; Miron-Mendoza et al., 2010), similar to the CDM. Significantly, like 1.7 mg/ml collagen, 8.6 mg/ml collagen exhibited nonlinear elastic behavior, undergoing strain stiffening (Ehigh/Emed = 1.2; Fig. 6 D). Together, these data show that 3D matrix linear elasticity is a key structural property necessary for nonpolarized Rac1 activity during lobopodia-based migration.
Lobopodia and efficient migration in the CDM
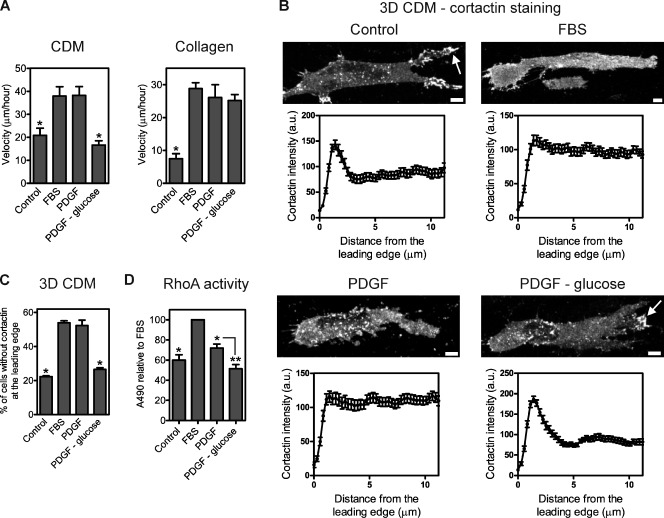
Given that RhoA or ROCK inhibition in 10% FBS switched the mode of HFF migration in the CDM without affecting velocity, it was not clear why more than one mode of migration was necessary. To test the hypothesis that lobopodial cells migrated more efficiently in the linear elastic ECM under suboptimal environmental conditions, we assessed HFF migration driven by PDGF in the presence of different levels of glucose. 10 ng/ml PDGF with 25 mM glucose was sufficient to replace 10% FBS for rapid migration in and on the CDM and collagen compared with DME alone, with the cells in the CDM forming lobopodia (Fig. 7, A–C). However, treatment of cells with PDGF in glucose-deficient medium reduced cell velocity in and on the CDM without affecting the velocity of lamellipodia-based migration in collagen (Fig. 7 A). This loss of migration efficiency in and on the CDM was associated with a switch to lamellipodia-based 3D motility (Fig. 7, B and C) and a reduction of RhoA activity to 51% of control, significantly less than in cells treated with PDGF with glucose (P < 0.05; Fig. 7 D). Therefore, lobopodia may be required for efficient motility in the CDM under low-glucose conditions. Thus, the external signaling environment can regulate the mode and efficiency of normal cell migration in the CDM as well as the relative activation of RhoA.
Figure 7.
Regulation of the mode and efficiency of normal 3D cell migration by extracellular soluble factors. (A) 10 ng/ml PDGF in glucose-deficient media specifically reduces cell velocity in the CDM versus collagen. Quantification of cell velocity in the CDM or collagen in response to DME with 25 mM glucose (control), 10% FBS in DME with 25 mM glucose (FBS), 10 ng/ml PDGF in DME with 25 mM glucose (PDGF), or 10 ng/ml PDGF in glucose-deficient DME (PDGF − glucose). *, P < 0.05 versus FBS. (B) Representative images of cortactin localization in HFFs in the 3D CDM treated as indicated and quantified in C. Arrows indicate the local accumulation of cortactin at the leading edge. Bottom graphs correspond with their respective top images and represent the mean cortactin intensity measured from the leading edge (0 µm) toward the cell center. Each cortactin intensity profile was averaged from 13 cells, with three measurements per cell. (C) Quantification of the percentage of cells without enrichment of cortactin at the leading edge in B. *, P < 0.001 versus FBS. (D) Treatment of HFFs with 10 ng/ml PDGF in glucose-deficient media reduces cellular RhoA activity by ∼50%. RhoA activities in HFFs treated on tissue-culture plastic as indicated were measured using G-LISA activation assays. Absorbance values were normalized to the relative amount of actin in each sample before comparison with the FBS treatment. *, P < 0.01 versus FBS; **, P < 0.05 versus PDGF. Cells are oriented with their leading edge toward the right of the figure. Bars, 5 µm. a.u., arbitrary unit.
Discussion
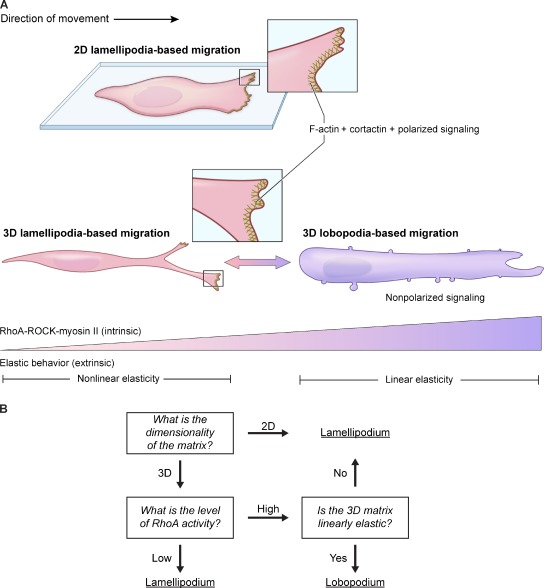
The major differences in polarization of active Rac1, Cdc42, and PIP3 identified in this study reveal lobopodial and lamellipodial migration as two distinct modes of normal 3D cell motility governed intrinsically by RhoA, ROCK, and myosin II and extrinsically by the elastic behavior of the ECM (Fig. 8 A). The lobopodia we find associated with 3D mesenchymal cell motility appear to be distinct from other cellular protrusions in addition to lamellipodia, including filopodia (Nobes and Hall, 1995), podosomes (Tarone et al., 1985), invadopodia (Mueller et al., 1992), eupodia (Fukui and Inoué, 1997), and membrane blebs (Charras et al., 2005). The decision between lamellipodia- and lobopodia-based motility during normal mesenchymal cell migration can be conceptualized as a series of three binary questions evaluated by migrating cells (Fig. 8 B): what is the dimensionality of the matrix, what is the level of RhoA activity, and is the 3D matrix linearly elastic? This study also provides the first description of lobopodia-based movement coupled to 3D matrix adhesions and the first demonstration of a transition between lamellipodia-dependent and -independent migration in normal mesenchymal cells as well as its regulation.
Figure 8.
Dimensionality, matrix elastic behavior, and RhoA–ROCK–myosin II govern the mode of normal cell migration. (A) During cell migration on flat or fibrillar 2D surfaces, lamellipodia form the leading edge of adherent fibroblasts. The presence of lamellipodia was confirmed by a prominent rim of F-actin and cortactin, along with active Rac1 and Cdc42 with PIP3 at the leading edge. In a 3D ECM, HFFs can use either lobopodia- or lamellipodia-based 3D migration. (B) The choice to migrate using lobopodia- or lamellipodia-based migration can be represented by a decision tree consisting of three questions: what is the dimensionality of the matrix, what is the level of RhoA activity, and is the 3D matrix linearly elastic? Lobopodia-based 3D migration predominates in linear elastic ECM and may use high actomyosin contraction downstream of RhoA–ROCK–myosin II to increase intracellular pressure and push the leading edge forward in combination with integrin-mediated adhesion. When RhoA–ROCK–myosin II activity is diminished, either in nonlinear ECM or through treatment of cells with RhoA siRNA or specific inhibitors, cells form lamellipodia with actin polymerization to advance the leading edge. Both modes of migration involve elongated cells that form 3D matrix adhesions, but the distribution of active Rac1, Cdc42, and PIP3 distinguishes the two modes.
We demonstrate that the canonical polarization of PIP3, Rac1, and Cdc42 activity during 2D migration (Ridley et al., 2003) is not required for normal mesenchymal cells to migrate equally efficiently in the 3D ECM. During lobopodia-based migration in the 3D CDM, nonpolarized Rac1 and Cdc42 activity can modulate the velocity of migration independently of lamellipodia formation. Enrichment of active Rac1 and Cdc42, along with PIP3, at the tips of fan-shaped protrusions during 3D collagen migration identified these structures as lamellipodia, consistent with a previous morphological study (Small, 1981). These features in 3D collagen mimic the 2D signaling and lamellipodial dynamics that control directional cell movement (Haugh et al., 2000; Kraynov et al., 2000; Nalbant et al., 2004; Pertz et al., 2006; Petrie et al., 2009). Despite the nonpolarized distribution of Rac1, Cdc42, and PIP3 intracellular signaling, lobopodial cells can still be considered polarized based on their elongated morphology and directionally persistent migration in the 3D CDM. Although the mechanism that sustains this polarity is not clear, the polarization of the microtubule-organizing center, Golgi apparatus, and/or membrane trafficking might be maintained in these cells independent of the distribution of PIP3, Rac1, and Cdc42 activity (Kupfer et al., 1982; Bergmann et al., 1983; Gundersen and Bulinski, 1988).
Lobopodia formation was governed intrinsically by RhoA, ROCK, and myosin II, consistent with other contractility-based modes of cell migration (Sahai and Marshall, 2003; Klopocka and Redowicz, 2004; Yoshida and Soldati, 2006; Lämmermann et al., 2008). In fact, the lateral blebs we often observed during lobopodia-based motility, which were absent after RhoA or ROCK inhibition, may be a manifestation of increased cytoplasmic pressure (Charras et al., 2005) because of RhoA-mediated actomyosin contraction (Chrzanowska-Wodnicka and Burridge, 1996; Lämmermann and Sixt, 2009). Lobopodial migration was controlled extrinsically by soluble signals and the elastic behavior of the ECM. Cells in the CDM without motogens present formed lamellipodia, but these structures were not associated with significant cell movement. Thus, normal fibroblasts require a motogenic signal, e.g., 10% FBS or 10 ng/ml PDGF with 25 mM glucose, in combination with the linear elastic 3D ECM to undergo lobopodia-based migration. Stimulation with PDGF in glucose-deficient medium or inhibition of RhoA, ROCK, or myosin II switched the mode of migration independently of the elastic behavior of the matrix. Interestingly, we found that lobopodia were associated with efficient migration in the CDM when motility was triggered in low glucose, a condition that did not affect lamellipodia-based migration in 3D collagen. This suggests that lobopodia are required for rapid movement in the linear elastic environment of the CDM under certain environmental conditions. However, rapid motility in the CDM did not depend on a single mode of migration in 25 mM glucose, suggesting that lobopodia might also be involved in other aspects of normal fibroblast function, such as matrix production and remodeling.
An example of migration mode regulation is the amoeboid–mesenchymal transition during cancer cell migration (Wolf et al., 2003). Importantly, lobopodia-bearing HFFs differ from amoeboid cancer cells on the basis of morphology, the use of 3D matrix adhesions, and their regulation by the ECM (Table S1; Wolf et al., 2003; Sabeh et al., 2009). Determining whether the transition between lobopodia- and lamellipodia-based integrin-dependent migration is restricted to normal cells may help us to understand what signaling pathways are subverted to promote metastasis through the mesenchymal–amoeboid transition (Bissell, 1981; Sanz-Moreno and Marshall, 2010). For example, the integrin independence of amoeboid cancer cell migration could represent a regulatory defect promoting metastatic disease.
In summary, the differential organization of intracellular signaling in lobopodia and lamellipodia identifies these cellular structures as mediating two distinct modes of normal 3D cell migration. The transition between these modes can be regulated externally by the elastic behavior of the ECM via intracellular RhoA, ROCK, and myosin II, and it may represent a universal property of 3D cell motility.
Materials and methods
Reagents, cell culture, and transfection
The following reagents were used in this study: rhodamine-phalloidin (Invitrogen), Y-27632 (EMD), blebbistatin (EMD), cell-permeable C3 transferase (Cytoskeleton), human recombinant PDGF-BB (Sigma-Aldrich), and glucose-free DME (Invitrogen). HFFs (used at passages 8–20) and HT1080 cells were maintained in phenol red–free DME (HyClone) containing 10% FBS (HyClone), 4.5 g/liter glucose, 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen), and 2 mM l-glutamine (Invitrogen) at 37°C and in 10% CO2. All cDNA constructs were transfected into cells with the Nucleofector system (Lonza) using the human dermal fibroblast kit (Lonza) according to the manufacturer’s instructions. siRNA was transfected into HFFs using Lipofectamine 2000 (Invitrogen) as previously described (Pankov et al., 2005).
cDNA constructs and siRNAs
The pECFP-Rac1 and -Cdc42 constructs (Picard et al., 2009) were generated by subcloning the full-length sequence into the EcoRI–BamH1 sites of pECFP-C1 (Takara Bio Inc.). pYPet–p21-binding domain (PBD) was generated by subcloning the sequence encoding amino acids 65–150 of human Pak1 into the EcoRI–BamH1 sites of pYPet-C1. pYPet-C1 was constructed by subcloning the sequence corresponding to the fluorescent protein YPet into the AgeI–XhoI sites of pEGFP-C1 (Takara Bio Inc.). The YPet sequence was amplified from pCEP4YPet-MAMM (plasmid 14032; Addgene; Nguyen and Daugherty, 2005). pGFP-actin was purchased from Takara Bio Inc. pGFP-PLC-δPH was a gift from T. Balla (National Institute of Child Health and Human Development, Bethesda, MD), who generated it by subcloning the cDNA sequence corresponding to the PH domain of PLC-δ1 (amino acids 1–170) into pEGFP-N1 (Takara Bio Inc.; Várnai and Balla, 1998). To create pGFP-AktPH, the Akt PH domain (amino acids 1–148) was subcloned into pEGFP-C1 (Pankov et al., 2005). Vinculin–tension sensor (plasmid 26019; Addgene) was generated by Grashoff et al. (2010) by inserting a FRET-based sensor module between the vinculin head and vinculin tail domains of vinculin. pYFP-paxillin was generated by subcloning the sequence encoding human paxillin into the HindIII–XbaI sites of pEYFP-C1 (Takara Bio Inc.) that contained a modified multiple cloning site.
Rac1, 5′-GAACUGCUAUUUCCUCUAA-3′, 5′-AUGAAAGUGUCACGGGUAA-3′, 5′-GUAGUUCUCAGAUGCGUAA-3′, and 5′-GUGAUUUCAUAGCGAGUUU-3′; Cdc42, 5′-CGGAAUAUGUACCGACUGU-3′, 5′-GACGUCACAGUUAUGAUUG-3′, 5′-GAUGACCCCUCUACUAUUG-3′, and 5′-CUGCAGGGCAAGAGGAUUA-3′; and RhoA, 5′-CGACAGCCCUGAUAGUUUA-3′, 5′-GACCAAAGAUGGAGUGAGA-3′, 5′-GCAGAGAUAUGGCAAACAG-3′, and 5′-GGAAUGAUGAGCACACAAG-3′ ON-TARGETplus SMARTpool siRNAs, along with siGLO RNA-induced silencing complex–free control siRNA, were purchased from Thermo Fisher Scientific. The specificities of the siRNA pools were confirmed with the following independent siRNAs: Rac1, 5′-GGAACUAAACUUGAUCUUATT-3′; Cdc42, 5′-UGAGAUAACUCACCACUGUTT-3′; RhoA, 5′-CACAGUGUUUGAGAACUAUTT-3′ (Silencer Select; Invitrogen); and the individual RhoA SMARTpool siRNA, 5′-CGACAGCCCUGAUAGUUUA-3′.
Live-cell imaging in dermal explants
Dermal explants (Lämmermann et al., 2008) were prepared by separating the dorsal and ventral halves of ears from euthanized ICR (Institute of Cancer Research) mice. The cartilage-free halves were washed extensively in PBS plus 100 U/ml penicillin and 100 µg/ml streptomycin. For some experiments, the explant was labeled as previously described for the CDM (Hakkinen et al., 2011). In brief, explants were incubated with 20 µg of the succinimidyl ester of Alexa Fluor 633 (Invitrogen) in 2 ml of 50-mM NaHCO3 for 20 min. The labeled explant was washed with PBS plus 100 U/ml penicillin and 100 µg/ml streptomycin, and any unreacted Alexa Fluor 633 dye was quenched with 200 mM Tris, pH 7.4, for 20 min and washed with PBS. 5 × 105 HFFs were transfected with the indicated constructs and plated in a 60-mm dish. 18 h later, the transfected cells were added to the unlabeled or Alexa Fluor 633–labeled dermal explants immediately after explant preparation. The next day, the explants with adherent HFFs were transferred to a new dish and imaged.
Image stacks of dermal explants in media at 37°C and 10% CO2 were captured using a confocal microscope (LSM 510 NLO META with Axiovert 200M; Carl Zeiss) with a Plan Apochromat 63×, 1.4 NA oil objective lens (Carl Zeiss). A 488-nm argon and 633-nm HeNe2 laser (Carl Zeiss) excited GFP-actin and Alexa Fluor 633, respectively. The two-photon laser was set to 800 nm for visualizing collagen through second harmonic generation (Friedl et al., 2007). Image stacks were imported into Volocity software (PerkinElmer) to generate the 3D reconstructions.
Time-lapse fluorescence imaging of GFP-actin–transfected HFFs in dermal explants in media at 37°C and 10% CO2 was performed by spinning-disc confocal microscopy using a microscope (Axiovert 200M) equipped with a confocal scanning unit (CSU-X1; Yokogawa), a Plan Apochromat 100×, 1.4 NA oil objective (Carl Zeiss), and an EM charge-coupled device camera (C9100; Hamamatsu Photonics). Hardware control, image acquisition, and linear brightness and contrast adjustments were performed using MetaMorph software (Molecular Devices). GFP-actin was imaged with a 488-nm argon laser, and a 647-nm krypton-argon laser was used for Alexa Fluor 633.
For FRET imaging, HFFs were transfected with pYPet-PBD and pCFP-Rac1 and plated on Alexa Fluor 633–labeled dermal explants. The next day, the explants with HFFs were transferred to a new dish and imaged in media at 37°C and 10% CO2 using a confocal microscope (LSM 710 Axio Examiner.Z1; Carl Zeiss) equipped with a C Achroplan 40×, 0.8 NA water objective lens (Carl Zeiss). A 457- and 514-nm argon laser was used to excite the FRET pair, and a 633-nm HeNe laser excited Alexa Fluor 633. FRET images were processed as described in the Live-cell FRET imaging section.
GTPase activation assays
pECFP-Rac1 and -Cdc42 were mutated using the site-directed mutagenesis kit (QuikChange II; Agilent Technologies) to create the following constructs: pECFP-Rac1-Q61L (constitutively active) and -Rac1-Y40C (nonfunctional) and pECFP-Cdc42-Q61L (constitutively active) and -Cdc42-Y40C (nonfunctional). HFFs were either untransfected or transfected with the indicated constructs using transfection reagent (PolyFect; QIAGEN) according to the manufacturer’s instructions. Pull-down or G-LISA assays were performed to determine Rac1 or Cdc42 activity using the appropriate activation assay kit from Cytoskeleton following the manufacturer’s instructions.
Immunofluorescence labeling of fixed cells
The following antibodies were used: mouse anticortactin (Millipore), mouse anti-VASP (BD), mouse anti-PIP2 (Echelon), and mouse anti-PIP3 (Echelon). Mouse anti-Rac1 (Millipore) and rabbit anti-Cdc42 (Abcam) were used for immunofluorescence analysis of endogenous GTPases. To localize endogenous Rho family GTPases, PIP2, or PIP3, HFFs were fixed and stained following a published protocol for preservation of the plasma membrane during immunocytochemistry (Hammond et al., 2009) but with slight modifications. HFFs were fixed with 4% formaldehyde and 0.2% glutaraldehyde in PBS for 15 min at RT, rinsed 3× with 50 mM NH4Cl in PBS, placed in an aluminum foil–lined tissue-culture dish on ice, and chilled for 2 min, with all subsequent steps performed on ice with prechilled solutions. Fixed HFFs were treated with buffer A (1% BSA, 50 mM NH4Cl, 0.5% saponin, and 0.005% Triton X-100 in PBS) for 45 min. Antibodies were applied in buffer B (1% BSA, 50 mM NH4Cl, and 0.1% saponin in PBS) for 1 h. Cells were washed 2× with buffer C (1% BSA, 50 mM NH4Cl, and 0.5% saponin in PBS). IgG secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were applied in buffer B for 45 min. Cells were washed 4× with buffer C and postfixed with 2% formaldehyde in PBS for 10 min on ice and then 5 min at RT. Cells were washed 3× in 50 mM NH4Cl in PBS and 2× in distilled H2O before drying and mounting with mounting media (Gel/Mount; BioMeda). For VASP, cortactin, or rhodamine-phalloidin labeling, cells were fixed with 4% formaldehyde in PBS, permeabilized with 0.25% Triton X-100 in PBS, and blocked with 0.2% BSA in PBS. Rhodamine-phalloidin and primary and secondary antibodies were applied in 0.2% BSA in PBS and washed with PBS between each treatment. The fixed cells were imaged using the microscope (510 NLO META Axiovert 200M) with a Plan Apochromat 63×, 1.4 NA oil objective. Brightness and contrast were linearly adjusted using ImageJ 1.40g (National Institutes of Health).
Live-cell imaging in 3D in vitro models
CDMs were prepared from HFFs as follows (Cukierman et al., 2001). MatTek dishes were coated with 0.2% gelatin for 1 h at 37°C, treated with 1% glutaraldehyde for 30 min at RT, and incubated with DME for 30 min at RT. Three washes with PBS followed each treatment. 4 × 105 HFFs were plated per MatTek dish, which were maintained for 10 d, adding fresh media with 50 µg/ml ascorbic acid every other day. The cells were removed from the CDM with extraction buffer (20 mM NH4OH and 0.5% Triton X-100 in PBS) for 5 min at RT and washed with PBS. The cell-free CDM was treated with 10 U/ml DNase (Roche) for 30 min at 37°C, washed, and stored at 4°C in PBS with 100 U/ml penicillin and 100 µg/ml streptomycin. 105 HFFs, transfected with the indicated constructs, were plated on the CDM and blocked with 1% heat-denatured BSA (MP Biomedicals) for imaging the next day. For some experiments, the CDM was labeled with 5 µg Alexa Fluor 633 in 2 ml of 50-mM NaHCO3 as for dermal explants immediately before the addition of HFFs. 1.7 mg/ml collagen gel was prepared by combining 3.32 mg/ml rat tail type I collagen (BD) with 10× reconstitution buffer (0.26 M NaHCO3 and 0.2 M Hepes) and 10× DME (Sigma-Aldrich) at an 8:1:1 ratio, the pH was adjusted to 7.5, and the collagen was diluted to 1.7 mg/ml with media. MatTek dishes were coated with 50 µl of 1.7-mg/ml collagen, which was then polymerized at 37°C for 30 min. 200 µl of 1.7-mg/ml collagen containing 105 HFFs, transfected the previous day with the indicated constructs, was added to the first layer and polymerized at 37°C for 30 min. 2 ml of medium was added to the dish, and cells were imaged the next day.
Time-lapse fluorescence imaging of GFP-actin was performed as described for dermal explants. Cells were determined to be fully within the 3D CDM when covered with labeled matrix and within 3D collagen when above the glass surface of the MatTek dish. When determining the percentage of amoeboid and mesenchymal HT1080 cells in the CDM, a cell was considered mesenchymal when its length was at least twice its width, and amoeboid was determined when less (Sanz-Moreno et al., 2008).
Confocal stacks were captured using the microscope (LSM 510 NLO META Axiovert 200M) with a Plan Apochromat 40×, 1.0 NA oil iris objective as described in the Live-cell imaging in dermal explants section, except reflection of the 514-nm argon laser was used to visualize polymerized type I collagen, and a 457- and 514-nm argon laser was used to excite the FRET pair. Maximum projections of the confocal stacks were generated using Zen software (Carl Zeiss).
Live-cell FRET imaging
The binding of active CFP-Rac1 or -Cdc42 to YPet-PBD was detected by imaging the FRET-dependent sensitized emission of the acceptor fluorophore (YPet) in the presence of the donor fluorophore (CFP; Kraynov et al., 2000). Optimal FRET acquisition settings were independently determined for each microscope and strictly maintained during all subsequent FRET imaging. The spectral bleed-through ratios were determined for each microscope by imaging cells expressing donor or acceptor alone using the microscope’s optimized acquisition settings. All FRET image processing was performed using the LSM FRET Tool macro (Carl Zeiss). The Fc image was generated from the raw FRET image using
in which Ff is the raw FRET image, Df is the CFP image, Af is the YPet image, Fd_Dd is the proportion of CFP emission in the FRET image, Fa_Aa is the proportion of YPet emission in the FRET image, Da_Aa is the proportion of YPet emission in the donor image, Da_Fa is the cross talk coefficient determined by dividing the donor image by the FRET image of cells expressing YPet alone, and G is the system constant (set to 1; Gordon et al., 1998). Each channel image was thresholded before processing, and saturated and zero intensity pixels were excluded from the calculations. Maximum projections of the Fc confocal z stacks were generated using Zen software. Although the output of the Fc image was linearly adjusted using ImageJ 1.40g before application of the 16-color spectrum to fill the display range and show relative differences in FRET intensity, positive Rac1 and Cdc42 Fc images (12-bit; 0–4,096 fluorescence intensity range [arbitrary units]) were typically between 700 and 1,300.
FRET quantification
HFFs were transfected as indicated using PolyFect according to the manufacturer’s instructions. The next day, FRET images were captured of the live cells, and the Fc image was calculated as described in the Live-cell FRET imaging section of Materials and methods. The magnitude of the Fc signal in the transfected cells was measured in unadjusted Fc images using the LSM FRET Tool macro.
PI
To determine the PI of a cell, its center of mass was placed at the origin of an x–y axis, with the leading edge perpendicular to the positive x axis. The positions of discrete regions of Fc (considered positive when >500 but typically between 700 and 1,300) or GFP-AktPH intensity (considered positive when 1.5× greater than the cytoplasmic background fluorescence) were plotted relative to the cell’s center of mass. The PI for each cell was calculated using
in which x is the distance of the region from the origin along the x axis, SLD is the straight line distance of the region from the origin, and n is the number of regions per cell. In this scheme, a PI of 1 = forward polarization (all regions lie on the positive x axis), 0 = nonpolarization (regions are uniformly distributed), and −1 = rearward polarization (all regions lie on the negative x axis).
siRNA treatment, Western blotting, and motility assays
2 × 105 HFFs were plated in each of three 60-mm dishes and transfected the next day with a 20-nM solution of the indicated siRNA preparation. 48 h later, 105 siRNA-treated cells from the first 60-mm dish were transfected with pGFP-actin, plated on the CDM labeled with Alexa Fluor 633, and imaged the next day using a spinning-disc confocal microscope. 104 HFFs from the second 60-mm dish were plated in 10% FBS in DME on the CDM. The next day, time-lapse sequences were captured at 37°C and 10% CO2 using a 5×, 0.12 NA A-Plan objective on a microscope (Axiovert 40C; Carl Zeiss) with a charge-coupled device camera (INFINITY2; Lumenera). Cells were tracked every 26 min for 12 h using the Manual Tracking plugin (F. Cordelieres, Institut Curie, Paris, France) with ImageJ 1.40g. Velocity and directionality (the ratio of Euclidean to accumulated distance traveled, in which 1 = a straight line) were calculated from the tracking data using the Chemotaxis and Migration Tool plugin (ibidi) with ImageJ. For the integrin, ROCK, or myosin II inhibition experiments or PDGF ± glucose motility assays, 104 HFFs were plated on glass, the CDM, or in 1.7 mg/ml collagen in 35-mm MatTek dishes and treated the following day. Imaging, cell tracking (every 26 min for 12 h), and analysis were performed as described for the siRNA-treated cells.
72 h after transfection with siRNA, HFFs from the third 60-mm dish were lysed in lysis buffer (2% IGEPAL, 40 mM NaCl, 10 mM MgCl2, and 50 mM Tris, pH 7.5) plus protease inhibitors (Cytoskeleton). Cleared lysates were combined with an equal volume of 2× sample buffer (Invitrogen), heated to 95°C for 5 min, resolved by SDS-PAGE on a 4–12% Tris-glycine polyacrylamide gel (Invitrogen), and transferred to nitrocellulose (0.2-µm pores; Invitrogen). Antibodies used for Western blotting were mouse anti-Rac1 (Millipore), mouse anti-Cdc42 (BD), mouse anti-RhoA (Abcam), mouse antiactin (Sigma-Aldrich), and mouse anti–glyceraldehyde 3-phosphate dehydrogenase (Fitzgerald Industries). Blots were developed using ECL reagents (GE Healthcare) and visualized on a luminescent image analyzer (LAS-4000; Fujifilm). Western blots were quantified by normalizing the GTPase intensity to the corresponding glyceraldehyde 3-phosphate dehydrogenase signal and determining the change in expression relative to the siGLO-treated control.
Measurement of Young’s modulus by atomic force microscopy
Experiments were performed using an atomic force microscope (Catalyst; Bruker AXS) mounted on the stage of an inverted microscope (Axiovert 200) placed on a vibration isolation table (IsoStation). Matrices were kept at 37°C and buffered with PBS throughout the experiments. Force displacement curves were obtained in contact mode using pyramidal tip cantilevers of nominal stiffness, k = 0.03 N/m. For each cantilever used, stiffness was obtained by the thermal fluctuations method (Butt and Jaschke, 1995). Force displacement curves were acquired by ramping the cantilever 12 µm at 1 Hz. To prevent damage to the samples, the maximum force applied was ∼0.5 nN for soft matrices and ∼3 nN for stiff matrices. Young’s modulus (E) was computed by fitting force displacement curves with Sneddon’s formula for an indenting cone using
in which F is the applied force, ν is Poisson’s ratio and was assumed to be 0.5, δ is the indentation, and α is the half-opening angle of the pyramidal tip (Sneddon, 1965). To measure strain stiffening, E was computed fitting only regions of the force displacement curves corresponding to large indentations or medium indentations. The ratio of Ehigh/Emed was reported as a measure of the matrix’s strain-hardening behavior. All computations were performed using custom-built code written in Matlab (MathWorks).
Modification of the 3D in vitro models
Alexa Fluor 633–labeled CDM was treated with 0.05% trypsin + 0.5 mM EDTA (Invitrogen) for 10 min at 37°C and then washed extensively with DME with 10% FBS. 1.7 mg/ml collagen and trypsinized CDM were cross-linked with 25 mM BS(PEG)9 (bis-N-succinimidyl-(nonaethylene glycol) ester) and BS(PEG)5 (Thermo Fisher Scientific), respectively, in PBS for 30 min at RT followed by extensive washing with DME with 10% FBS. 105 HFFs transfected with pCFP-Rac1 and pYPet-PBD or pGFP-actin were plated on the trypsinized and/or cross-linked matrix and imaged the next day. GFP-actin dynamics were analyzed by spinning-disc confocal microscopy, and Rac1 activity was visualized using the confocal microscope (LSM 710 Axio Examiner.Z1) as described in Live-cell imaging in dermal explants.
Statistical analysis
Results are reported as the mean ± SEM. One-way analysis of variance with Tukey posttests was used to compare three or more variables; otherwise, unpaired, two-tailed Student’s t tests were performed. Nonparametric Kruskal–Wallis tests were used with a Dunn’s multiple comparisons posttest when comparing more than two groups of measurements with bounded values when analyzing the PI. All comparisons were performed using Prism5 (GraphPad Software). Differences were considered statistically significant at P < 0.05.
Online supplemental material
Fig. S1 and S2 detail the validation of the Rac1 and Cdc42 FRET-based biosensors. Fig. S3 shows the relative distribution of GFP-PLC-δPH in HFFs in different environments, along with endogenous PIP2 and PIP3. Fig. S4 provides additional evidence that RhoA activity is required for lobopodia formation in the 3D CDM. Table S1 compares the lobopodia- and lamellipodia-based 3D migration of normal cells with previously published work examining cancer cell motility. Video 1 shows lobopodia-based 3D migration in the mammalian dermis. Video 2 shows lobopodia-based 3D migration in the CDM and lamellipodia-based 2D migration on top of the CDM. Video 3 that shows HFFs use lamellipodia-based 3D migration in type I collagen. Video 4 shows that reduction of RhoA protein switches the mode to lamellipodia-based 3D migration in the CDM. Video 5 shows that ROCK activity is required for lobopodia-based migration. Video 6 is a phase-contrast video of HFFs migrating in and on the CDM during blebbistatin treatment. Video 7 shows that HFFs use lamellipodia-based 3D migration in a pliable CDM. Video 8 shows that cross-linking the trypsinized CDM restores lobopodia-based 3D migration. Video 9 shows that HFFs use lobopodia-based 3D migration in cross-linked collagen. Video 10 shows lamellipodia-based 3D HFF migration in 8.6 mg/ml collagen. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201124/DC1.
Acknowledgments
We thank Myungso Chung for helping to develop the glucose-deficient migration assays, Dr. Carole Parent for helpful discussions, and Dr. Andrew Doyle, Dr. Sarah Knox, and Matthew Kutys for critical comments on the manuscript.
This work was supported by the Intramural Research Programs of the National Institute of Dental and Craniofacial Research and the National Institute on Deafness and Other Communication Disorders at the National Institutes of Health.
Footnotes
Abbreviations used in this paper:
- CDM
- cell-derived matrix
- ERK
- extracellular signal–regulated kinase
- Fc
- corrected FRET
- FRET
- fluorescence resonance energy transfer
- HFF
- human foreskin fibroblast
- PBD
- p21-binding domain
- PH
- pleckstrin homology
- PI
- polarization index
- PIP3
- phosphatidylinositol (3,4,5)-trisphosphate
- PIP2
- phosphatidylinositol (4,5)-bisphosphate
- ROCK
- Rho-associated protein kinase
- VASP
- vasodilator-stimulated phosphoprotein
References
- Ahlfors J.-E.W., Billiar K.L. 2007. Biomechanical and biochemical characteristics of a human fibroblast-produced and remodeled matrix. Biomaterials. 28:2183–2191 10.1016/j.biomaterials.2006.12.030 [DOI] [PubMed] [Google Scholar]
- Allio A.E., McKeown-Longo P.J. 1988. Extracellular matrix assembly of cell-derived and plasma-derived fibronectins by substrate-attached fibroblasts. J. Cell. Physiol. 135:459–466 10.1002/jcp.1041350313 [DOI] [PubMed] [Google Scholar]
- Bard J.B., Hay E.D. 1975. The behavior of fibroblasts from the developing avian cornea. Morphology and movement in situ and in vitro. J. Cell Biol. 67:400–418 10.1083/jcb.67.2.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass M.D., Roach K.A., Morgan M.R., Mostafavi-Pour Z., Schoen T., Muramatsu T., Mayer U., Ballestrem C., Spatz J.P., Humphries M.J. 2007. Syndecan-4–dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J. Cell Biol. 177:527–538 10.1083/jcb.200610076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J.E., Svitkina T.M., Krause M., Schafer D.A., Loureiro J.J., Strasser G.A., Maly I.V., Chaga O.Y., Cooper J.A., Borisy G.G., Gertler F.B. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- Benard V., Bohl B.P., Bokoch G.M. 1999. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 274:13198–13204 10.1074/jbc.274.19.13198 [DOI] [PubMed] [Google Scholar]
- Bergmann J.E., Kupfer A., Singer S.J. 1983. Membrane insertion at the leading edge of motile fibroblasts. Proc. Natl. Acad. Sci. USA. 80:1367–1371 10.1073/pnas.80.5.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell M.J. 1981. The differentiated state of normal and malignant cells or how to define a “normal” cell in culture. Int. Rev. Cytol. 70:27–100 10.1016/S0074-7696(08)61130-4 [DOI] [PubMed] [Google Scholar]
- Boulter E., Garcia-Mata R., Guilluy C., Dubash A., Rossi G., Brennwald P.J., Burridge K. 2010. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 12:477–483 10.1038/ncb2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H.-J., Jaschke M. 1995. Calculation of thermal noise in atomic force microscopy. Nanotechnology. 6:1–7 10.1088/0957-4484/6/1/001 [DOI] [Google Scholar]
- Charras G.T., Yarrow J.C., Horton M.A., Mahadevan L., Mitchison T.J. 2005. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 435:365–369 10.1038/nature03550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., Burridge K. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J.-F., Motoyama A.B., Bush J.A., Vuori K. 2005. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 7:797–807 10.1038/ncb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D.R., Yamada K.M. 2001. Taking cell-matrix adhesions to the third dimension. Science. 294:1708–1712 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Deakin N.O., Turner C.E. 2011. Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol. Biol. Cell. 22:327–341 10.1091/mbc.E10-09-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K., Mennecke R., Raz E. 2004. Signaling pathways controlling primordial germ cell migration in zebrafish. J. Cell Sci. 117:4787–4795 10.1242/jcs.01362 [DOI] [PubMed] [Google Scholar]
- Elsdale T., Bard J. 1972. Collagen substrata for studies on cell behavior. J. Cell Biol. 54:626–637 10.1083/jcb.54.3.626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. 2006. Matrix elasticity directs stem cell lineage specification. Cell. 126:677–689 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Even-Ram S., Yamada K.M. 2005. Cell migration in 3D matrix. Curr. Opin. Cell Biol. 17:524–532 10.1016/j.ceb.2005.08.015 [DOI] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A.D., Conti M.A., Matsumoto K., Adelstein R.S., Yamada K.M. 2007. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 9:299–309 10.1038/ncb1540 [DOI] [PubMed] [Google Scholar]
- Friedl P., Wolf K. 2010. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188:11–19 10.1083/jcb.200909003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Wolf K., von Andrian U.H., Harms G. 2007. Biological second and third harmonic generation microscopy. Curr. Protoc. Cell Biol. Chapter 4:Unit 4.15. [DOI] [PubMed] [Google Scholar]
- Fukui Y., Inoué S. 1997. Amoeboid movement anchored by eupodia, new actin-rich knobby feet in Dictyostelium. Cell Motil. Cytoskeleton. 36:339–354 [DOI] [PubMed] [Google Scholar]
- Gelman R.A., Williams B.R., Piez K.A. 1979. Collagen fibril formation. Evidence for a multistep process. J. Biol. Chem. 254:180–186 [PubMed] [Google Scholar]
- Gordon G.W., Berry G., Liang X.H., Levine B., Herman B. 1998. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74:2702–2713 10.1016/S0006-3495(98)77976-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B.D., Brenner M.D., Zhou R., Parsons M., Yang M.T., McLean M.A., Sligar S.G., Chen C.S., Ha T., Schwartz M.A. 2010. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 466:263–266 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell F., Lamke C.R. 1984. Reorganization of hydrated collagen lattices by human skin fibroblasts. J. Cell Sci. 66:51–63 [DOI] [PubMed] [Google Scholar]
- Gundersen G.G., Bulinski J.C. 1988. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc. Natl. Acad. Sci. USA. 85:5946–5950 10.1073/pnas.85.16.5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkinen K.M., Harunaga J.S., Doyle A.D., Yamada K.M. 2011. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng. Part A. 17:713–724 10.1089/ten.tea.2010.0273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond G.R., Schiavo G., Irvine R.F. 2009. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem. J. 422:23–35 10.1042/BJ20090428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugh J.M., Codazzi F., Teruel M., Meyer T. 2000. Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J. Cell Biol. 151:1269–1280 10.1083/jcb.151.6.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman K., Kurkinen M., Alitalo K., Vaheri A., Johansson S., Höök M. 1979. Isolation of the pericellular matrix of human fibroblast cultures. J. Cell Biol. 81:83–91 10.1083/jcb.81.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Peng H.B., Rauvala H. 2000. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J. Cell Sci. 113:4421–4426 [DOI] [PubMed] [Google Scholar]
- Kardash E., Reichman-Fried M., Maître J.L., Boldajipour B., Papusheva E., Messerschmidt E.M., Heisenberg C.P., Raz E. 2010. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 12:47–53 10.1038/ncb2003 [DOI] [PubMed] [Google Scholar]
- Klopocka W., Redowicz M.J. 2004. Rho/Rho-dependent kinase affects locomotion and actin-myosin II activity of Amoeba proteus. Protoplasma. 224:113–121 [DOI] [PubMed] [Google Scholar]
- Kraynov V.S., Chamberlain C., Bokoch G.M., Schwartz M.A., Slabaugh S., Hahn K.M. 2000. Localized Rac activation dynamics visualized in living cells. Science. 290:333–337 10.1126/science.290.5490.333 [DOI] [PubMed] [Google Scholar]
- Kudo R.R. 1977. Protozoology. Fifth Edition Charles C. Thomas, Springfield, IL: 1,174 pp [Google Scholar]
- Kupfer A., Louvard D., Singer S.J. 1982. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc. Natl. Acad. Sci. USA. 79:2603–2607 10.1073/pnas.79.8.2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann T., Sixt M. 2009. Mechanical modes of ‘amoeboid’ cell migration. Curr. Opin. Cell Biol. 21:636–644 10.1016/j.ceb.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Lämmermann T., Bader B.L., Monkley S.J., Worbs T., Wedlich-Söldner R., Hirsch K., Keller M., Förster R., Critchley D.R., Fässler R., Sixt M. 2008. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 453:51–55 10.1038/nature06887 [DOI] [PubMed] [Google Scholar]
- Lauffenburger D.A., Horwitz A.F. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Levskaya A., Weiner O.D., Lim W.A., Voigt C.A. 2009. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 461:997–1001 10.1038/nature08446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., Danuser G. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature. 461:99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Silletti J., Murphy G., D’Eustachio P., Rush M., Philips M.R. 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152:111–126 10.1083/jcb.152.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron-Mendoza M., Seemann J., Grinnell F. 2010. The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials. 31:6425–6435 10.1016/j.biomaterials.2010.04.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna W., Kligman A.M., Carlisle K.S. 1992. Atlas of Normal Human Skin. Springer-Verlag, New York: 384 pp [Google Scholar]
- Mueller S.C., Yeh Y., Chen W.T. 1992. Tyrosine phosphorylation of membrane proteins mediates cellular invasion by transformed cells. J. Cell Biol. 119:1309–1325 10.1083/jcb.119.5.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbant P., Hodgson L., Kraynov V., Toutchkine A., Hahn K.M. 2004. Activation of endogenous Cdc42 visualized in living cells. Science. 305:1615–1619 10.1126/science.1100367 [DOI] [PubMed] [Google Scholar]
- Nguyen A.W., Daugherty P.S. 2005. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23:355–360 10.1038/nbt1066 [DOI] [PubMed] [Google Scholar]
- Nobes C.D., Hall A. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., Yamada K.M. 2005. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 170:793–802 10.1083/jcb.200503152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O., Hahn K.M. 2004. Designing biosensors for Rho family proteins—deciphering the dynamics of Rho family GTPase activation in living cells. J. Cell Sci. 117:1313–1318 10.1242/jcs.01117 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., Hahn K.M. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Petrie R.J., Doyle A.D., Yamada K.M. 2009. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10:538–549 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M., Petrie R.J., Antoine-Bertrand J., Saint-Cyr-Proulx E., Villemure J.F., Lamarche-Vane N. 2009. Spatial and temporal activation of the small GTPases RhoA and Rac1 by the netrin-1 receptor UNC5a during neurite outgrowth. Cell. Signal. 21:1961–1973 10.1016/j.cellsig.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Provenzano P.P., Inman D.R., Eliceiri K.W., Trier S.M., Keely P.J. 2008. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 95:5374–5384 10.1529/biophysj.108.133116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Sabeh F., Shimizu-Hirota R., Weiss S.J. 2009. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 185:11–19 10.1083/jcb.200807195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711–719 10.1038/ncb1019 [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V., Marshall C.J. 2010. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr. Opin. Cell Biol. 22:690–696 10.1016/j.ceb.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C.J. 2008. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 135:510–523 10.1016/j.cell.2008.09.043 [DOI] [PubMed] [Google Scholar]
- Singer I.I., Kawka D.W., Kazazis D.M., Clark R.A. 1984. In vivo co-distribution of fibronectin and actin fibers in granulation tissue: immunofluorescence and electron microscope studies of the fibronexus at the myofibroblast surface. J. Cell Biol. 98:2091–2106 10.1083/jcb.98.6.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J.V. 1981. Organization of actin in the leading edge of cultured cells: influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. J. Cell Biol. 91:695–705 10.1083/jcb.91.3.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon I.N. 1965. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 3:47–57 10.1016/0020-7225(65)90019-4 [DOI] [Google Scholar]
- Storm C., Pastore J.J., MacKintosh F.C., Lubensky T.C., Janmey P.A. 2005. Nonlinear elasticity in biological gels. Nature. 435:191–194 10.1038/nature03521 [DOI] [PubMed] [Google Scholar]
- Tarone G., Cirillo D., Giancotti F.G., Comoglio P.M., Marchisio P.C. 1985. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane called podosomes. Exp. Cell Res. 159:141–157 10.1016/S0014-4827(85)80044-6 [DOI] [PubMed] [Google Scholar]
- ten Klooster J.P., Jaffer Z.M., Chernoff J., Hordijk P.L. 2006. Targeting and activation of Rac1 are mediated by the exchange factor β-Pix. J. Cell Biol. 172:759–769 10.1083/jcb.200509096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hennik P.B., ten Klooster J.P., Halstead J.R., Voermans C., Anthony E.C., Divecha N., Hordijk P.L. 2003. The C-terminal domain of Rac1 contains two motifs that control targeting and signaling specificity. J. Biol. Chem. 278:39166–39175 10.1074/jbc.M307001200 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Jalink K. 2002. Agonist-induced PIP(2) hydrolysis inhibits cortical actin dynamics: regulation at a global but not at a micrometer scale. Mol. Biol. Cell. 13:3257–3267 10.1091/mbc.E02-04-0231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P., Balla T. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143:501–510 10.1083/jcb.143.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., He L., Wu Y.I., Hahn K.M., Montell D.J. 2010. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12:591–597 10.1038/ncb2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J.P., Oake S., Janmey P.A. 2009. Non-linear elasticity of extracellular matrices enables contractile cells to communicate local position and orientation. PLoS ONE. 4:e6382 10.1371/journal.pone.0006382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Mazo I., Leung H., Engelke K., von Andrian U.H., Deryugina E.I., Strongin A.Y., Bröcker E.B., Friedl P. 2003. Compensation mechanism in tumor cell migration: mesenchymal–amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160:267–277 10.1083/jcb.200209006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Alexander S., Schacht V., Coussens L.M., von Andrian U.H., van Rheenen J., Deryugina E., Friedl P. 2009. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 20:931–941 10.1016/j.semcdb.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.I., Frey D., Lungu O.I., Jaehrig A., Schlichting I., Kuhlman B., Hahn K.M. 2009. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 461:104–108 10.1038/nature08241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai M., Kenyon C.M., Butler J.P., Macklem P.T., Kelly S.M. 1996. Intracellular pressure is a motive force for cell motion in Amoeba proteus. Cell Motil. Cytoskeleton. 33:22–29 [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Deng Q., Cavnar P.J., Wu Y.I., Hahn K.M., Huttenlocher A. 2010. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell. 18:226–236 10.1016/j.devcel.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Soldati T. 2006. Dissection of amoeboid movement into two mechanically distinct modes. J. Cell Sci. 119:3833–3844 10.1242/jcs.03152 [DOI] [PubMed] [Google Scholar]
- Zaman M.H., Trapani L.M., Sieminski A.L., Mackellar D., Gong H., Kamm R.D., Wells A., Lauffenburger D.A., Matsudaira P. 2006. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA. 103:10889–10894 10.1073/pnas.0604460103 [DOI] [PMC free article] [PubMed] [Google Scholar]