Abstract
Neuronal nicotinic receptors, encoded by nine genes of the alpha and three of the beta type of subunits, and whose gene products assemble in distinct permutations as pentameric molecules, constitute a fertile area for structure-guided drug design. Design strategies are augmented by a wide variety of peptide, alkaloid and terpenoid toxins from various marine and terrestrial species that interact with nicotinic receptors. Also, acetylcholinebinding proteins from mollusks, as structural surrogates of the receptor that mimic its extracellular domain, provide atomic resolution templates for analysis of structure and response. Herein, we describe a structure-guided approach to nicotinic ligand design that employs crystallography of this protein as the basic template, but also takes into consideration the dynamic properties of the receptor molecules in their biological media. We present the crystallographic structures of several complexes of various agonists and antagonists that associate with the agonist site and can competitively block the action of acetylcholine. In so far as the extracellular domain is involved, we identify additional non-competitive sites at those subunit interfaces where agonists do not preferentially bind. Ligand association at these interface sites may modulate receptor function. Ligand binding is also shown by solution-based spectroscopic and spectrometric methods to affect the dynamics of discrete domains of the receptor molecule. The surrogate receptor molecules can then be employed to design ligands selective for receptor subtype through the novel methods of freeze-frame, click chemistry that uses the very structure of the target molecule as a template for synthesis of the inhibitor.
Keywords: Nicotinic receptor structure, Acetylcholine binding protein, Freeze-frame click chemistry, Crystal structure, Allosteric modulators, Competitive ligand site, Channel vestibule, Fluorescence anisotropy, Isotope exchange
INTRODUCTION
Neuronal nicotinic receptors, found in mammalian peripheral and central nervous systems, show considerable diversity within a single animal species by virtue of the multiple genes that encode the individual subunits and the various permutations of subunits that assemble as pentamers [1–3]. Moreover, if we extend our considerations beyond the mammalian species, we see even greater structural diversity, and the determinants governing ligand recognition between insect and mammalian nicotinic receptors are the basis for the selective toxicity of the neonicotinoids enjoying current use as insecticides [4, 5]
Of the nine subunits in neuronal nicotinic receptors in mammals, designated as α2 through α10 and containing a vicinal disulfide on the C loop of their principal binding face, α2, α3, α4 and α6 associate with the complementary β2, β3 and β4 subunits to form pentameric molecules. The α5 subunit behaves similarly to a β subunit in not having its principal face creating a binding site. The α7, α9 and α10 subunits do not require β subunit partners to function as homomeric pentamers (α7) or as pentameric combinations of α subunits (α9 with α10). Even though the variations in receptor subtype may have risen from differences in desensitization capacity, regulation of expression, stability, or ion channel conductance, we can capitalize on the structural differences in the ligand recognition domains for the development of selective ligands for particular receptor subtypes.
To develop a structure-guided approach to understanding the molecular basis of nicotinic receptor selectivity, we rely on the endeavors of nature in acquiring selectivity of natural products serving as toxins and in expressing variants of the nicotinic receptor. Since the nicotinic receptor in skeletal muscle forms the signal transducer for neuromuscular transmission controlling motor activity, and in the CNS, nicotinic receptors primarily mediate release of neurotransmitters from their presynaptic sites, a variety of toxins from marine snails, terrestrial plants, coral as stationary animals, and reptiles have evolved to block or modulate nicotinic receptor function. Several of these compounds, in contrast to acetylcholine, carbamylcholine, nicotine and epibatidine, show considerable selectivity towards receptor subtype [1–5]. Therefore these compounds can be considered as lead templates for developing selective ligands.
A second gift of nature from lower species has also emerged for analyzing the extracellular domain of the nicotinic receptor. Sixma and colleagues have characterized pharmacologically and structurally a soluble protein, the acetylcholine binding protein (AChBP) from the fresh water snail, Lymnaea stagnalis, whose subunits are homologous to the extracellular domain of the nicotinic receptor. This soluble pentameric protein from Lymnaea, the related proteins from Bulinus, and the marine species, Aplysia, bind nicotinic ligands with the requisite specificity [6, 7]. When the gene encoding an AChBP is coupled to a sequence encoding the transmembrane spans of the ligand-gated ion channel family, the expressed protein shows the capacity to gate ions when elicited by agonists, in a fashion expected for a receptor composed of the so derived expressed subunit chimera [8].
In our studies, we have capitalized on these advances to elucidate further structures of the AChBPs. Although the sequences from fresh water, Lymnaea, and salt water, Aplysia, snails differ substantially (Fig. 1), all form pentamers and bind nicotinic ligands. As found for the different receptor subtypes, there are signature ligands that show considerable selectivity for the AChBPs from different species (Table I). Having soluble proteins expressed from multiple species also facilitates studies of structure owing to differences in physical properties affecting the ease of crystallization and amenability to spectroscopic or spectrometric studies in solution.
Figure 1.
Alignment of Amino Acid Residues in the Three Acetylcholine Binding Proteins from Lymnaea stagnalis (Ls), Aplysia californica (Ac) and Bulinus truncatus (Bt) with the α7 nicotinic receptor from humans. Residue numbering corresponds to Aplysia californica. The black and gray backgrounds correspond to identical and homologous residues in the binding proteins and receptor, respectively, the embolden letters denote identical residues in the three binding proteins. Residues on the principal and complementary faces are denoted.
Table I.
Classes of Ligands Studied that Interact with AChBP
| Class of Ligands | Ligandsa | Lymnaea | Aplysia | (Ac/Ls)b | PDB ID | Reference |
|---|---|---|---|---|---|---|
| Alkaloids | Kd (nM) | |||||
| Nicotine | 100 | 260 | 2.6 | 1UW6 | [10, 19, 29] | |
| Epibatidine | 0.3 | 8.6 | 28.7 | 2BYQ | [11, 19, 29] | |
| Lobeline | 130 | 0.5 | 0.0038 | 2BYS | [11] | |
| Anabaseine | 240 | 9,000 | 37.5 | [29] | ||
| d-Tubocurarine | 67 | 420 | 6.3 | [18, 31] | ||
| Metocurine | 120 | 570 | 4.8 | [31] | ||
| Methyllycaconitine | 0.4 | 2.8 | 7 | 2BYR | [11, 19] | |
| Terpenes | ||||||
| Bipinnatin H | ||||||
| Bipinnatin K | ||||||
| Conus Peptides | ||||||
| α-ImI | 14,000 | 2.1 | 0.00015 | 2BYP 2C9T |
[11, 33] | |
| α-OmIA | 1.7 | 6.7 | 3.9 | [21] | ||
| α-PnIA (A10L D14K) | 19 | 21 | 1.1 | 2BR8 | [32] | |
| α-PnIA (A10L) | 200 | 28 | 0.1 | [32] | ||
| α-MI | 2,800 | 1,000 | 0.4 | [11] | ||
| Elapid Toxins | ||||||
| α-Bungarotoxin | 1.8 | 170 | 94.4 | [11] | ||
| α-Cobratoxin | 15 | 320 | 21.3 | 1YI52 | [11] | |
| Neonicotinoids | ||||||
| Imidacloprid | 180 | [5] | ||||
| Thiacloprid | 8.4 | [5] | ||||
| Acetamiprid | 19 | [5] | ||||
| Nitenpyram | 186 | [5] | ||||
| Clothianidin | 574 | [5] | ||||
| Nicotinoids | ||||||
| Desnitroimidacloprid | 44 | [5] | ||||
| Descyanothiacloprid | 8.5 | [5] | ||||
| Allosteric Ligands | ||||||
| Galanthamine | 3,000 | 16,000 | 5.3 | 2PH9 | [12] | |
| Cocaine | 6,700 | 4,500 | 0.7 | 2PGZ | [12] | |
Signature ligands have ~ 100 fold or greater differences between the two binding proteins and are noted by the embolden font.
Ratio of affinities: Aplysia californica/Lymnaea stagnalis
Crystal Structures of Competitive Agonists and Antagonists
Several structures of agonist (epibatidine, nicotine, lobeline, carbamylcholine) and antagonist (α-conotoxin, methyllycaconitine, α-bungarotoxin) bound to AChBP have been reported [6, 9–12], and other structures have been resolved at high resolution or are under study. Hence, one now has a fairly comprehensive perspective on structures of the AChBP complexes. Also, a structure approaching that of the non-liganded protein (the apo-AChBP) has been resolved [11]. Accordingly, several predictions might be made.
First, it appears that the critical C loop on the outer perimeter of the protein envelops the bound agonists, where the complex has a closed C loop in the vast majority of the cases (Fig. 2). Since AChBP has five identical sites for occupation, one can compare C loop closure and occupation for the various ligand-AChBP crystal structures available. By contrast, loop C closure is not evident for antagonists, and this raises the question as to whether the C loop configuration is simply accommodating the molecular volume of the occupying ligand. Some of the potential agonists, such as lobeline, approach or exceed the molecular weight of the antagonists. Hence, molecular weight or volume is not likely to be a deciding factor. More likely factors governing loop closure may be the overall flexibility of the ligand and whether it can be accommodated within the confines of a site where C loop closure is possible. Hence, agonists have the requisite flexibility and steric features to allow C loop closure. Antagonists appear to keep the C loop in its open position or extend the C loop in a more radial direction [9, 11].
Figure 2.
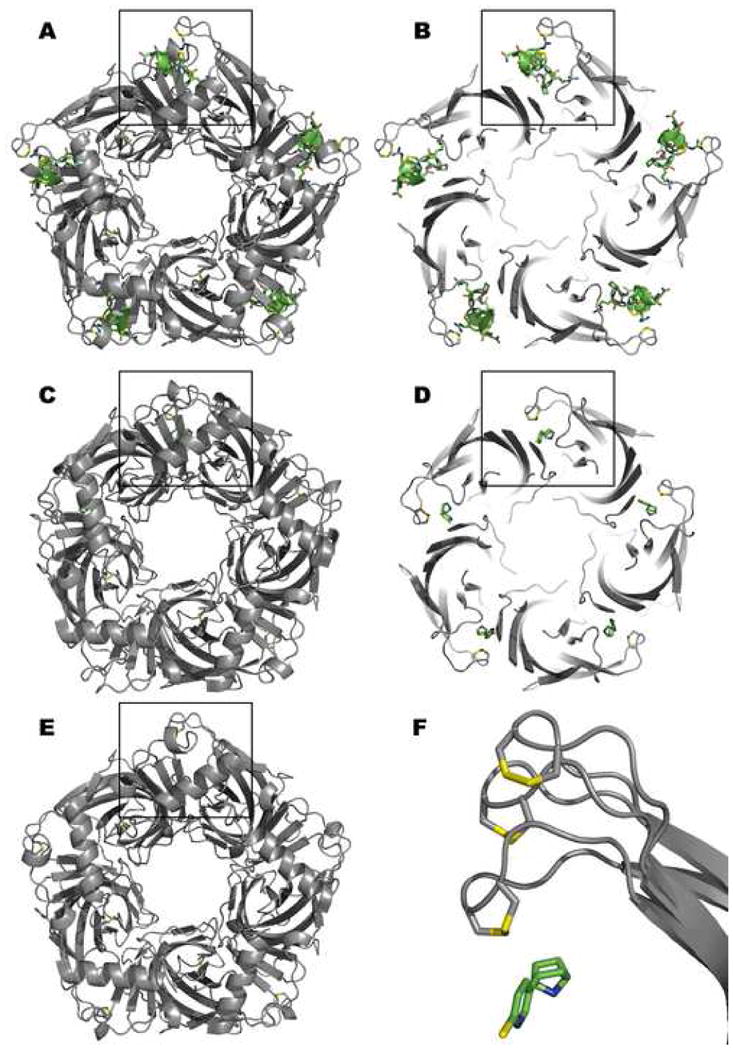
Structure of the Aplysia californica Acetylcholine Binding Protein with α-Conotoxin-IM1 (A and B), Epibatidine (C and D), and in the Absence Ligand (E). Note the different extensions in the positions of the C loop at the outer perimeter in each case. This is the only apparent structural change arising from ligand association. Panels A and C show the view from the apical (synaptic) surface of the receptor. Panels B and D show an axial cutaway segments of 20Å thickness at the level of the C loop for the conotoxin and epibatidine complexes. Panel F is a magnified inset to show the three C loop positions from the boxed areas in relation to bound epibatidine. The most extended C loop is for the α-conotoxin complex followed by the apo form and epibatidine complex. For the ligands, carbon is shown in green, nitrogen in blue, oxygen in red, chloride in yellow. The vicinal disulfide bond sulfurs are shown in yellow.
Some uncertainty surrounds the position of the C loop in the absence of ligand. The initial crystal structure showed the amine containing buffer, HEPES at ~100mM concentration, to occupy the agonist site [6]. Crystallization in the presence of polyethylene glycol shows low occupation of the cryoprotectant in the agonist site and radial extension of the C loop from the core backbone of the molecule [11] (Fig. 2).
Deuterium-hydrogen exchange studies also point to an exposed C loop in the absence of bound ligand. Of the 17 backbone amide hydrogens in the C loop peptide, 12 are available to solvent (H2O) for exchange in the absence of bound ligand revealing the unique exposure of this region of the molecule [13]. Upon binding of agonist, 5 of the amides are no longer available, likely reflecting the closure of the C loop. Similar behavior is seen with the larger antagonists, where they also protect the amide hydrogens from exposure to solvent [13]. Since the kinetics of isotope exchange differs among the antagonist complexes, it is likely that the larger ligands directly occlude solvent access.
Other studies employing fluorescence anisotropy decay reveal independent segmental motions of individual domains of the acetylcholine binding protein [14, 15]. However, studies of the C loop by these methods are complicated by additional interactions conferred by the conjugated fluorophore. Studying dynamics of the molecule adds another dimension to structural studies, and owing to the multiple conformational states of the AChBP and presumably the nicotinic receptor, this approach becomes an essential prelude to understanding function [13–15].
Crystal structures for a second set of ligands, thought in many receptor systems to act non-competitively or allosterically, have also been investigated with AChBP [12]. Both galanthamine and cocaine are found to bind to AChBP in the micromolar range, but in contrast to agonists, their binding constants are not affected by modifications of C loop structure, such as elimination of the vicinal disulfide bonds [12]. Hence, the structures derived from the cocaine and galanthamine complexes of AChBP provide a framework for understanding potential sites where non-competitive activators or inhibitors of heteromeric forms of the acetylcholine receptor bind. From these studies of affinity and structure, the interfacial site formed by the principal face of the non-alpha subunits becomes a candidate location for these ligands. A non-competitive site at non-agonist binding interfaces may resemble the locus for benzodiazepines with the γ-aminobutyric acid receptor [16, 17].
Other sites common to AChBP and the receptor, where ligands may reside, include the vestibule of the ion channel that harbors several interesting sequence conservations with the pentameric, ligand-gated anionic channels (S. B. Hansen and P. Taylor, in preparation). However, still other candidate, non-competitive sites obviously cannot be examined through the AChBP; these include internal transmembrane channel and annular membrane sites. Also, molecular recognition by AChBP is likely to yield limited information on the coupling between allosteric sites. Cooperativity has not been observed with fractional site occupation on the AChBP with either a primary agonist alone or a primary agonist combined with potential heterotropic modifier [12, 18]. Accordingly, cooperative occupation and responses in the intact receptor may well require coupling into the transmembrane span of the occupied subunit or subunit interactions at the membrane span level.
Structural Determinants of Specificity
AChBP with its known three-dimensional structure allows one to conduct extensive structure-activity studies and relate ligand selectivity to overall structural changes in the molecule. An extensive variety of ligands have been studied through measurements of ligand binding at equilibrium and by transient kinetics [18]. Some of the features of the binding studies are highlighted below.
Kinetic Studies
Quenching of the native tryptophan fluorescence in AChBP provides a convenient means for following the overall kinetics of ligand association with the binding protein [18]. Agonist binding approaches the diffusion controlled limit, and acetylcholine association and dissociation rates can just be caught by stopped-flow methods [18]. By contrast, the peptide antagonists, particularly α-bungarotoxin and α-conotoxin have rates of association up to three orders of magnitude slower, implicating a significant conformational barrier for association. In turn, locking the peptide ligand into its binding site in a distinct conformation may contribute to its slow rate of dissociation.
The Binding Surface of the Ligands
Ligands with measurably high affinities bind with 1:1 stoichiometry per subunit in the pentamer as ascertained by stoichiometric titration and ligand occupancies in the crystal structures. Interestingly, several of the naturally occurring peptide or alkaloid toxins have evolved to interact with different regions of the subunit interface. For example, the α-conotoxins bind apical to the C loop, while the alkaloid methyllycaconitine binds largely on the membrane side of the loop (Fig. 3). In fact, the selective surfaces of the two sites constitute two regions from which building blocks for the novel synthetic chemistry presented in a subsequent section are being developed.
Figure 3.
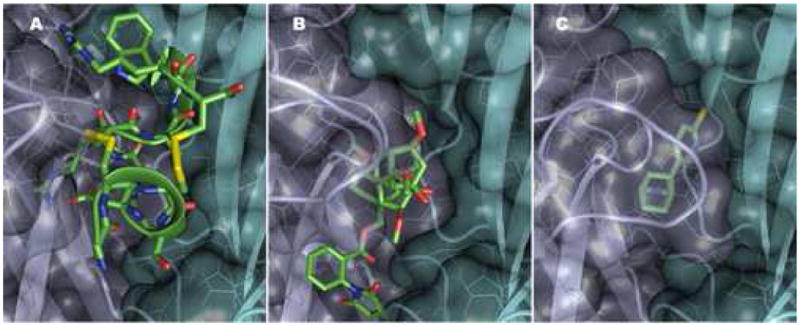
α-Conotoxin Im1, Methyllycaconitine and Epibatidine Association with the Subunit Interface of the Aplysia Acetylcholine Binding Protein. Panels A–C shows the occupation by conotoxin, methyllycaconitine and epibatidine with a common binding surface for the three ligands. The additional interactive surfaces of conotoxin and methyllycaconitine extend in the apical and membrane directions, respectively, along the subunit interface. The ligands are in the positions found in the crystal structure putting the channel core in identical positions. With each ligand yellow signifies chlorine, red oxygen, blue nitrogen and green carbon. Yellow residues denote the disulfide of the C loop. Residues occluded from view by the Connolly surface of AChBP are seen behind the partially opaque surfaces. The contributing subunits are shown in gray (principal or C loop containing face, turquoise (complementary subunit). Residue colors are the same as in Figure 2. The opening of subunit interface from radial extension of the C loop shows less burying of the α-Conotoxin Structure.
Peptide Toxins
Studies of snake (Elapidae and Viperidae) and cone snail (Conus) venoms with the AChBP are particularly interesting owing to their historical significance in receptor characterization and their exquisite selectivity toward receptor subtype. Accordingly, we have examined several members of the large family of conus snail toxins for selectivity to the Lymnaea, Aplysia and Bulinus binding proteins [11, 19–21]. Many toxins also show considerable selectivity for the three binding proteins (Table I) [22, 23].
Natural Alkaloids and Terpenoids
Studies with cholinergic systems largely originated nearly 150 years ago with the endeavors of Claude Bernard in the study of alkaloids of the curare family. These ligands, by virtue of their historical interest warrant a description of their structure with the nicotinic receptor. A comparison within the agonist and antagonist alkaloids shows a base region to which all of the alkaloids bind, and this can be identified by the carbonyl oxygen of Trp 147 in Aplysia on the principal face and the immediate surrounding nest of aromatic side chains, defined by principal face residues Trp 147, Tyr 93, Tyr 188, Tyr 195 and complementary face residue Tyr 55 (Fig. 1). It also appears that quaternary ligands rely more heavily on the aromatic nest, whereas a hydrogen bond between the protonated amine hydrogen and the carbonyl oxygen of Trp 147 plays a greater stabilization role for the secondary and tertiary amine nicotinic agonists, such as nicotine and anabaseine. The family of antagonists, being of greater size, typically employ a larger cross section of the subunit interfacial areas of the partnering subunits for their binding than agonists.
The terpenoid inhibitors of nicotinic receptors warrant a more complete investigation. Those related to lophotoxin react covalently with the receptor interacting with Tyr 190 in the α1 subunit [24] (corresponding to Tyr 188 in Aplysia), whereas the cembranoids [25] bind reversibly and appear to modulate the actions of agonists. It will be of interest to ascertain whether these cembranoid compounds, lacking a basic residue, bind at non-α interfaces as do cocaine and galanthamine.
Nicotinoids and Neonicotinoids
These compounds present an interesting series owing to their selectivity for the insect receptor and their commercial applications as insecticides [4, 5]. Radiolabeling experiments show that these ligands bind to the agonist site and with the pyridine ring system positioned similar to that in epibatidine or nicotine [4, 5]. The unique characteristics of the neonicotinoids stem from substitution of the cationic nitrogen of the tertiary or quaternary agonists that stimulate the mammalian receptor. The substitution renders the neonicotinoid nitrogen position neutral to anionic [4, 5].
Future Structure-Guided Ligand Syntheses
Despite a host of nicotinic agonists and antagonists that have been developed, the multiple potential sites and the subunit diversity in the assembled neuronal nicotinic receptors continue to present new platforms for drug development. Most agonist and partial agonist development to date has relied on various heterocylic compounds with a localized cationic charge from an aliphatic nitrogen. A new departure would be to consider templates that more closely resemble the soft or more diffuse cation of arginine, and we have begun work with amidines as a possible basic anchor [26]. These compounds carry the potential advantage of having pKa values in the vicinity of 8 to 9, therein potentially crossing the blood-brain barrier.
With the great structural resolution of the extracellular domain of the nicotinic receptor validated from a soluble surrogate, the mollusk AChBP, structure-guided drug design can be realistically conducted. We are applying the approach of freeze-frame, click chemistry, used successfully for acetylcholinesterase inhibition [27, 28], to this end. By employing building blocks that anchor at the apical and membrane side of the C loop (cf: Figure 3), and extending the mutually reactive azide and acetylene groups underneath the C loop, candidate ligands can be constructed to serve as agonists or partial agonists for the receptor. Through mutagenesis, residues can be substituted to develop a modified AChBP whose sequences at the subunit interfaces resemble more closely those found with various nicotine receptor subtypes. These constructs can then serve as templates for synthesis of presumably directed receptor subtype selective ligands. Our initial synthetic building blocks are directed to positioning the triazole formed by the biorthogonal reaction between the azide and acetylene behind the C loop, with their tethering points mimicking interaction positions of known agonists and antagonists.
Physical Properties of AChBP from Solution based Studies
For AChBP to serve as a template for nicotinic ligand design, a more complete understanding of structural fluctuations and conformational states of the unliganded and various liganded AChBPs would be helpful. Below are examples of studies undertaken by our laboratory in conjunction with several collaborators.
Information from Spectroscopy
These studies require a spectral change in either an attached probe on the AChBP or intrinsic spectroscopic properties of the ligand that can be distinguished from the baseline signal of the larger macromolecule. For example, the benzylidene anabaseines have sufficient conjugation that their absorption and fluorescence emission spectra lie outside of those of the protein envelope [29]. Through the use of absorption difference spectra, we established that the protonated form of the secondary or tertiary amine ligands is bound, a long held assumption from modeling and pH dependence of binding studies. For the benzylidene anabaseines, the change in protonation state of the tetrahydropyridine moiety gives rise to the observed spectral changes. Titrations over a physiologic pH range show an isosbestic point with the counterbalancing of high and low wavelength peaks. By contrast, changes in the polarizability and relative permuttivity (dielectric constant) of the surrounding media cause bathochromic or hypsochromic shifts in the respective peaks. Difference spectra between bound and free benzylidene anabaseines show the bound species to be protonated, and the bathochromic shift is consistent with the extensive polarizability of the “aromatic nest” in which the ligand resides [29].
Changes in the fluorescence spectra of the bound benzylidene anabaseine ligand reflect electron transition energies of the excited state of the molecule. The enhancement of quantum yields of the bound anabaseines likely arises from the restricted environment for torsional motion of the ring systems. Rigidity or constrained motion diminish the opportunity for depopulation of the excited state by heat energy and enhances loss through fluorescence emission [29].
Fluorescence Spectroscopy
Steady-state fluorescence emission of conjugated fluorophores on the protein also yields distinct information about particular residue locations. To achieve labeling, free cysteines are introduced by substitution at strategic locations in AChBP through mutagenesis; the endogenous cysteines all exist as unreactive disulfide residues. Sulfhydryl reactive fluorescent labels are selected to provide specific information. For example, fluorescence emission of the acrylodan label is exquisitely sensitive to the dielectric environment of the fluorophore, and its emission spectrum reveals the extent of solvent exposure. Accordingly, changes in conformation associated with ligand occupation can then be reported for the acrylodan conjugated AChBP molecule [30]. Other probes such as fluorescein or naphthalene sulfonates have longer lifetimes, and by following their decay of anisotropy, torsional and segmental motions of side chains can be monitored [14, 15]. Some ligands may also be tagged, and the fluorescent labeling of α-cobratoxin at various residue positions has shown that the core hand portion of the toxin, when bound, can move independently of its fingers, thereby establishing that the toxin molecule exhibits distinct segmental motion independent of its target [14].
Intrinsic fluorescence of the AChBP yielded another surprise for it was observed that various ligands produce substantial enhancement or quenching of AChBP tryptophan fluorescence when bound to the protein [18]. Given that four tryptophans are found in each subunit and most ligands themselves do not have the capacity to quench fluorescence by resonance energy transfer, it is likely that the entering ligand substantially changes the aromatic connectivity of side chains residing within the binding site. Hence, the unique positioning of these aromatic side chains confers a practical advantage of direct fluorescence measurements of ligand binding without requiring structural modification.
Deuterium/Hydrogen Exchange
Measuring deuterium incorporation from D2O into the amide backbone hydrogen positions in AChBP provides an alternative means of assessing solvent exposure of various segments of the protein. As alluded to in a previous section, the C loop of AChBP shows substantial solvent accessibility that is lost upon closure of the loop by agonists and occlusion from solvent by the larger antagonists [13]. Accordingly, this approach has enabled us to detect localized conformational changes in solution associated with ligand binding or other interventions.
Summary
We describe here a variety of techniques amenable to studying structure of the soluble AChBP that are not possible to exploit fully or successfully with the membrane-associated, intact receptor. The approaches extend from high through-put analysis of ligand specificity, fluorescence and difference absorption spectroscopy, D/H exchange, decay of fluorescence anisotropy, chemical modification with identification of the conjugated residues and crystallization for X-ray analysis. Solution-based physical studies offer an additional dimension to crystallography in yielding essential information on fluctuations in structure and conformational changes attendant to ligand binding.
Acknowledgments
Supported by USPHS grants: U01 DA019372, and R37 GM18360
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Changeux JP, Edelstein SJ. Nicotinic Acetylcholine Receptors: From Molecular Biology to Cognition. New York: Editions Odile Jacob/Johns Hopkins University Press; 2005. [Google Scholar]
- 2.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci. 2002;3:102–14. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 3.Taylor P, Osaka H, Molles B, Keller SH, Malany S. Contributions of studies of the nicotinic receptor from muscle to defining structural and functional properties of ligand-gated ion channels. In: Clementi F, Fornasari D, Gotti C, editors. Neuronal Nicotinic Receptors. Berlin; New York: Springer-Verlag; 2000. pp. 79–100. [Google Scholar]
- 4.Tomizawa M, Casida JE. Neonicotinoid insecticide toxicology: mechanisms of selective action. Annu Rev Pharmacol Toxicol. 2005;45:247–68. doi: 10.1146/annurev.pharmtox.45.120403.095930. [DOI] [PubMed] [Google Scholar]
- 5.Tomizawa M, Talley TT, Maltby D, Durkin KA, Medzihradszky KF, Burlingame AL, et al. Mapping the elusive neonicotinoid binding site. Proc Natl Acad Sci U S A. 2007;104:9075–80. doi: 10.1073/pnas.0703309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–76. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 7.Smit AB, Syed NI, Schaap D, van Minnen J, Klumperman J, Kits KS, et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411:261–8. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 8.Bouzat C, Barrantes F, Sine S. Nicotinic receptor fourth transmembrane domain: hydrogen bonding by conserved threonine contributes to channel gating kinetics. J Gen Physiol. 2000;115:663–72. doi: 10.1085/jgp.115.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. Embo J. 2005;24:1512–22. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–14. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 11.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. Embo J. 2005;24:3635–46. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen SB, Taylor P. Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors. J Mol Biol. 2007;369:895–901. doi: 10.1016/j.jmb.2007.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J, Koeppe JR, Komives EA, Taylor P. Ligand-induced conformational changes in the acetylcholine-binding protein analyzed by hydrogen-deuterium exchange mass spectrometry. J Biol Chem. 2006;281:12170–7. doi: 10.1074/jbc.M600154200. [DOI] [PubMed] [Google Scholar]
- 14.Hibbs RE, Johnson DA, Shi J, Hansen SB, Taylor P. Structural dynamics of the alpha-neurotoxin-acetylcholine-binding protein complex: hydrodynamic and fluorescence anisotropy decay analyses. Biochemistry. 2005;44:16602–11. doi: 10.1021/bi051735p. [DOI] [PubMed] [Google Scholar]
- 15.Hibbs RE, Radic Z, Taylor P, Johnson DA. Influence of agonists and antagonists on the segmental motion of residues near the agonist binding pocket of the acetylcholine-binding protein. J Biol Chem. 2006;281:39708–18. doi: 10.1074/jbc.M604752200. [DOI] [PubMed] [Google Scholar]
- 16.Campagna-Slater V, Weaver DF. Molecular modelling of the GABA-A ion channel protein. J Mol Graph Model. 2007;25:721–30. doi: 10.1016/j.jmgm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Sarto-Jackson I, Ramerstorfer J, Ernst M, Sieghart W. Identification of amino acid residues important for assembly of GABA receptor alpha1 and gamma2 subunits. J Neurochem. 2006;96:983–95. doi: 10.1111/j.1471-4159.2005.03626.x. [DOI] [PubMed] [Google Scholar]
- 18.Hansen SB, Radic Z, Talley TT, Molles BE, Deerinck T, Tsigelny I, et al. Tryptophan fluorescence reveals conformational changes in the acetylcholine binding protein. J Biol Chem. 2002;277:41299–302. doi: 10.1074/jbc.C200462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen SB, Talley TT, Radic Z, Taylor P. Structural and ligand recognition characteristics of an acetylcholine-binding protein from Aplysia californica. J Biol Chem. 2004;279:24197–202. doi: 10.1074/jbc.M402452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang TS, Radic Z, Talley TT, Jois SD, Taylor P, Kini RM. Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins. Biochemistry. 2007;46:3338–55. doi: 10.1021/bi061969o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talley TT, Olivera BM, Han KH, Christensen SB, Dowell C, Tsigelny I, et al. α-Conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as α3β2 and α7 nicotinic acetylcholine receptors. J Biol Chem. 2006;281:24678–86. doi: 10.1074/jbc.M602969200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntosh JM, Santos AD, Olivera BM. Conus peptides targeted to specific nicotinic acetylcholine receptor subtypes. Annu Rev Biochem. 1999;68:59–88. doi: 10.1146/annurev.biochem.68.1.59. [DOI] [PubMed] [Google Scholar]
- 23.Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–7. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- 24.Eterovic VA, Hann RM, Ferchmin PA, Rodriguez AD, Li L, Lee YH, et al. Diterpenoids from Caribbean gorgonians act as noncompetitive inhibitors of the nicotinic acetylcholine receptor. Cell Mol Neurobiol. 1993;13:99–110. doi: 10.1007/BF00735367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyde EG, Thornhill SM, Boyer AJ, Abramson SN. Evidence for a carbocation intermediate during conversion of bipinnatin-A and -C into irreversible inhibitors of nicotinic acetylcholine receptors. J Med Chem. 1995;38:4704–9. doi: 10.1021/jm00023a010. [DOI] [PubMed] [Google Scholar]
- 26.Lewis WG. Thesis, Chemistry. La Jolla: The Scripps Research Institute; 2005. [Google Scholar]
- 27.Bourne Y, Kolb HC, Radic Z, Sharpless KB, Taylor P, Marchot P. Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation. Proc Natl Acad Sci U S A. 2004;101:1449–54. doi: 10.1073/pnas.0308206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, et al. Click chemistry in situ: acetylcholinesterase as a reaction vessel for the selective assembly of a femtomolar inhibitor from an array of building blocks. Angew Chem Int Ed Engl. 2002;41:1053–7. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Talley TT, Yalda S, Ho KY, Tor Y, Soti FS, Kem WR, et al. Spectroscopic analysis of benzylidene anabaseine complexes with acetylcholine binding proteins as models for ligand-nicotinic receptor interactions. Biochemistry. 2006;45:8894–902. doi: 10.1021/bi060534y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hibbs RE, Talley TT, Taylor P. Acrylodan-conjugated cysteine side chains reveal conformational state and ligand site locations of the acetylcholine-binding protein. J Biol Chem. 2004;279:28483–91. doi: 10.1074/jbc.M403713200. [DOI] [PubMed] [Google Scholar]
- 31.Gao F, Bern N, Little A, Wang HL, Hansen SB, Talley TT, et al. Curariform antagonists bind in different orientations to acetylcholine-binding protein. J Biol Chem. 2003;278:23020–6. doi: 10.1074/jbc.M301151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celie PH, Kasheverov IE, Mordvintsev DY, Hogg RC, van Nierop P, van Elk R, et al. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat Struct Mol Biol. 2005;12:582–8. doi: 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
- 33.Ulens C, Hogg RC, Celie PH, Bertrand D, Tsetlin V, Smit AB, et al. Structural determinants of selective alpha-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc Natl Acad Sci U S A. 2006;103:3615–20. doi: 10.1073/pnas.0507889103. [DOI] [PMC free article] [PubMed] [Google Scholar]



