Abstract
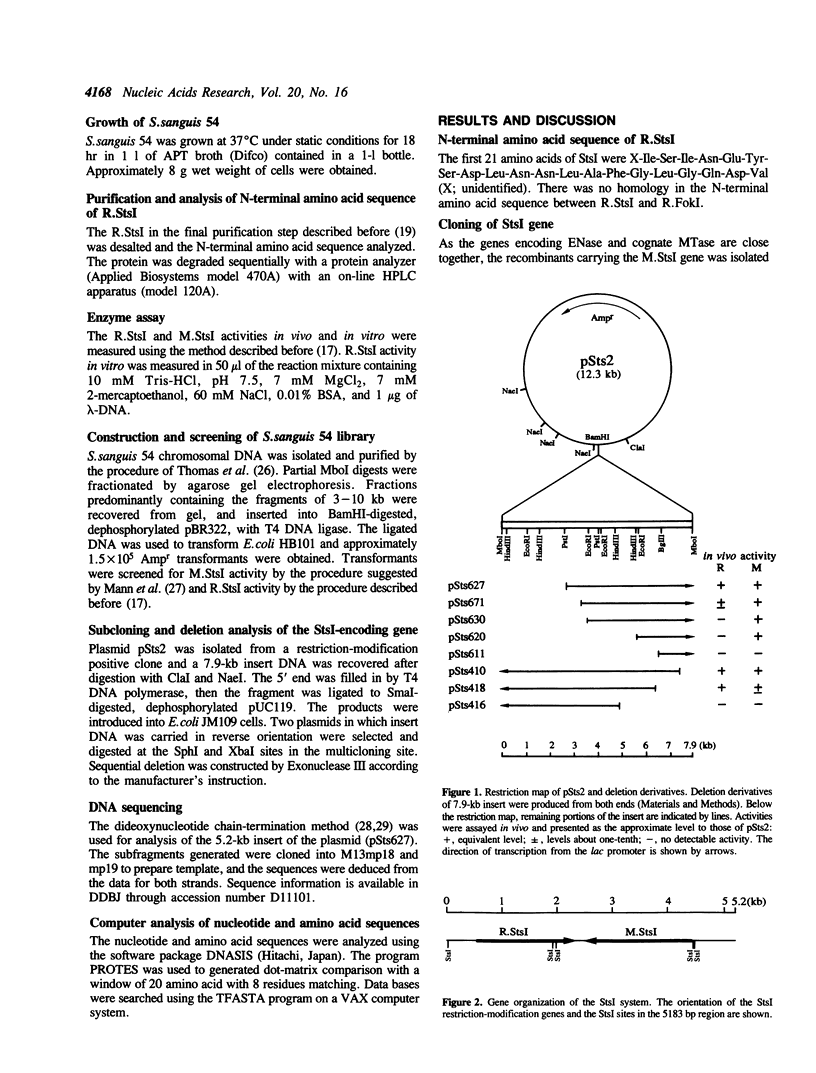
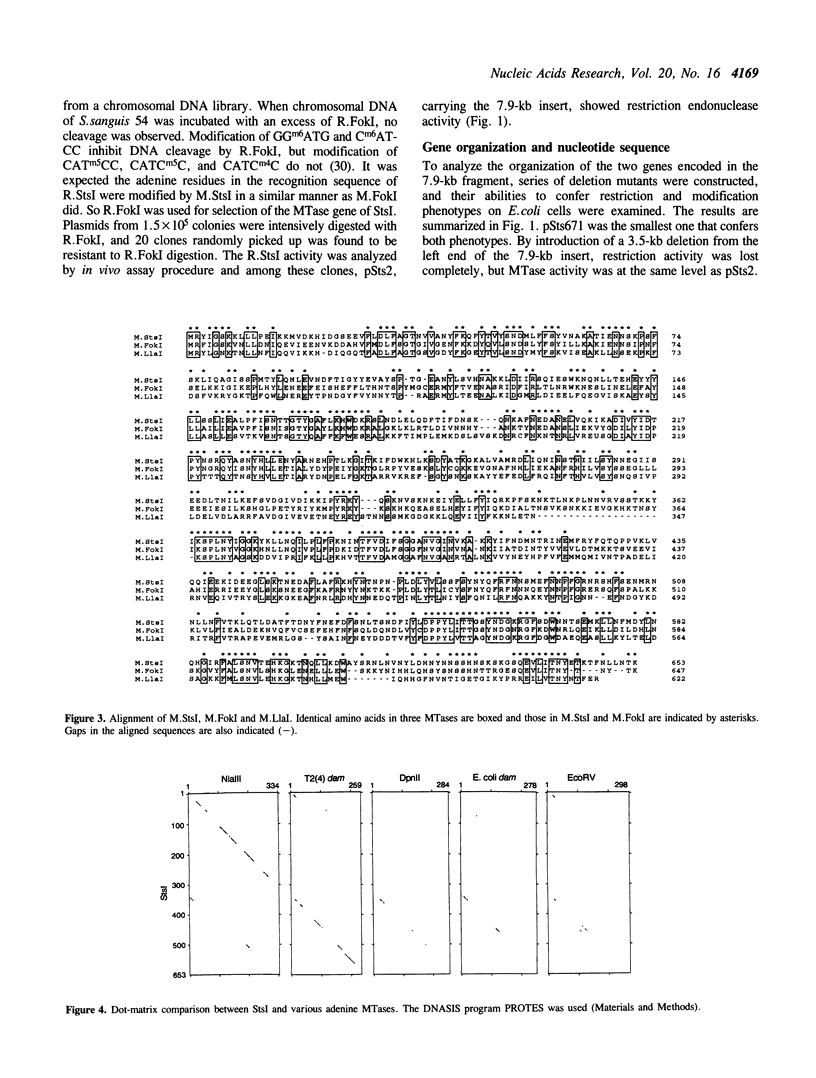
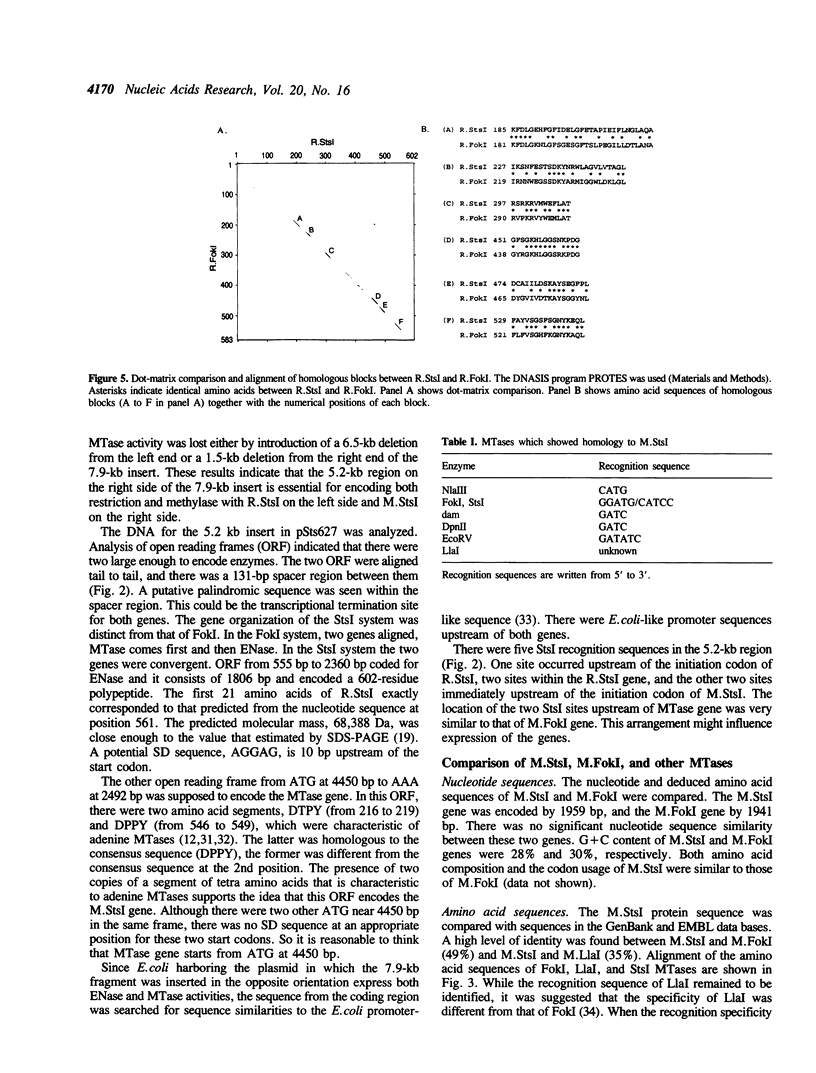
StsI endonuclease (R.StsI), a type IIs restriction endonuclease found in Streptococcus sanguis 54, recognizes the same sequence as FokI but cleaves at different positions. A DNA fragment that carried the genes for R.StsI and StsI methylase (M.StsI) was cloned from the chromosomal DNA of S.sanguis 54, and its nucleotide sequence was analyzed. The endonuclease gene was 1,806 bp long, corresponding to a protein of 602 amino acid residues (M(r) = 68,388), and the methylase gene was 1,959 bp long, corresponding to a protein of 653 amino acid residues (M(r) = 76,064). The assignment of the endonuclease gene was confirmed by analysis of the N-terminal amino acid sequence. Genes for the two proteins were in a tail-to-tail orientation, separated by a 131-nucleotide intercistronic region. The predicted amino acid sequences between the StsI system and the FokI system showed a 49% identity between the methylases and a 30% identity between the endonucleases. The sequence comparison of M.StsI with various methylases showed that the N-terminal half of M.StsI matches M.NIaIII, and the C-terminal half matches adenine methylases that recognize GATC and GATATC.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiken C. R., Fisher E. W., Gumport R. I. The specific binding, bending, and unwinding of DNA by RsrI endonuclease, an isoschizomer of EcoRI endonuclease. J Biol Chem. 1991 Oct 5;266(28):19063–19069. [PubMed] [Google Scholar]
- Aiken C. R., McLaughlin L. W., Gumport R. I. The highly homologous isoschizomers RsrI endonuclease and EcoRI endonuclease do not recognize their target sequence identically. J Biol Chem. 1991 Oct 5;266(28):19070–19078. [PubMed] [Google Scholar]
- Barany F., Danzitz M., Zebala J., Mayer A. Cloning and sequencing of genes encoding the TthHB8I restriction and modification enzymes: comparison with the isoschizomeric TaqI enzymes. Gene. 1992 Mar 1;112(1):3–12. doi: 10.1016/0378-1119(92)90296-2. [DOI] [PubMed] [Google Scholar]
- Bougueleret L., Schwarzstein M., Tsugita A., Zabeau M. Characterization of the genes coding for the Eco RV restriction and modification system of Escherichia coli. Nucleic Acids Res. 1984 Apr 25;12(8):3659–3676. doi: 10.1093/nar/12.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasegaran S., Lunnen K. D., Smith H. O., Wilson G. G. Cloning and sequencing the HinfI restriction and modification genes. Gene. 1988 Oct 30;70(2):387–392. doi: 10.1016/0378-1119(88)90210-7. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Gupta M., Boyer H. W., Brown W. E., Rosenberg J. M. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J Biol Chem. 1981 Mar 10;256(5):2143–2153. [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991 Jul;173(14):4363–4370. doi: 10.1128/jb.173.14.4363-4370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K., Kotani H., Hiraoka N., Nakamura T., Yonaha K. Overproduction and crystallization of FokI restriction endonuclease. Nucleic Acids Res. 1989 Nov 11;17(21):8741–8753. doi: 10.1093/nar/17.21.8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K., Kotani H., Ohta H., Yanase H., Kato N. StsI, a new FokI isoschizomer from Streptococcus sanguis 54, cleaves 5' GGATG(N)10/14 3'. Nucleic Acids Res. 1992 Feb 11;20(3):618–618. doi: 10.1093/nar/20.3.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K., Kotani H., Sugisaki H., Takanami M. The fokI restriction-modification system. I. Organization and nucleotide sequences of the restriction and modification genes. J Biol Chem. 1989 Apr 5;264(10):5751–5756. [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé D., Höltke H. J., Lau P. C. Cloning and characterization of two tandemly arranged DNA methyltransferase genes of Neisseria lactamica: an adenine-specific M.NlaIII and a cytosine-type methylase. Mol Gen Genet. 1990 Oct;224(1):101–110. doi: 10.1007/BF00259456. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Mannarelli B. M., Springhorn S. S., Greenberg B. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell. 1986 Sep 26;46(7):993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- Lauster R. Evolution of type II DNA methyltransferases. A gene duplication model. J Mol Biol. 1989 Mar 20;206(2):313–321. doi: 10.1016/0022-2836(89)90481-6. [DOI] [PubMed] [Google Scholar]
- Looney M. C., Moran L. S., Jack W. E., Feehery G. R., Benner J. S., Slatko B. E., Wilson G. G. Nucleotide sequence of the FokI restriction-modification system: separate strand-specificity domains in the methyltransferase. Gene. 1989 Aug 15;80(2):193–208. doi: 10.1016/0378-1119(89)90284-9. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Rao R. N., Smith H. O. Cloning of restriction and modification genes in E. coli: the HbaII system from Haemophilus haemolyticus. Gene. 1978 Apr;3(2):97–112. doi: 10.1016/0378-1119(78)90054-9. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- McClelland M., Nelson M. Effect of site-specific methylation on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1992 May 11;20 (Suppl):2145–2157. doi: 10.1093/nar/20.suppl.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miner Z., Hattman S. Molecular cloning, sequencing, and mapping of the bacteriophage T2 dam gene. J Bacteriol. 1988 Nov;170(11):5177–5184. doi: 10.1128/jb.170.11.5177-5184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagman S. L., Hattman S. Molecular cloning of a functional dam+ gene coding for phage T4 DNA adenine methylase. Gene. 1983 May-Jun;22(2-3):139–156. doi: 10.1016/0378-1119(83)90098-7. [DOI] [PubMed] [Google Scholar]
- Schoner B., Kelly S., Smith H. O. The nucleotide sequence of the HhaII restriction and modification genes from Haemophilus haemolyticus. Gene. 1983 Oct;24(2-3):227–236. doi: 10.1016/0378-1119(83)90083-5. [DOI] [PubMed] [Google Scholar]
- Slatko B. E., Benner J. S., Jager-Quinton T., Moran L. S., Simcox T. G., Van Cott E. M., Wilson G. G. Cloning, sequencing and expression of the Taq I restriction-modification system. Nucleic Acids Res. 1987 Dec 10;15(23):9781–9796. doi: 10.1093/nar/15.23.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Annau T. M., Chandrasegaran S. Finding sequence motifs in groups of functionally related proteins. Proc Natl Acad Sci U S A. 1990 Jan;87(2):826–830. doi: 10.1073/pnas.87.2.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson F. H., Ballard B. T., Boyer H. W., Rosenberg J. M., Greene P. J. Comparison of the nucleotide and amino acid sequences of the RsrI and EcoRI restriction endonucleases. Gene. 1989 Dec 21;85(1):1–13. doi: 10.1016/0378-1119(89)90458-7. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Kanazawa S. New restriction endonucleases from Flavobacterium okeanokoites (FokI) and Micrococcus luteus (MluI). Gene. 1981 Dec;16(1-3):73–78. doi: 10.1016/0378-1119(81)90062-7. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Kita K., Takanami M. The FokI restriction-modification system. II. Presence of two domains in FokI methylase responsible for modification of different DNA strands. J Biol Chem. 1989 Apr 5;264(10):5757–5761. [PubMed] [Google Scholar]
- Sugisaki H., Yamamoto K., Takanami M. The HgaI restriction-modification system contains two cytosine methylase genes responsible for modification of different DNA strands. J Biol Chem. 1991 Jul 25;266(21):13952–13957. [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Wolfes H., Pieper U., Geiger R., Urbanke C., Winkler F. K., Pingoud A. Site-directed mutagenesis studies with EcoRV restriction endonuclease to identify regions involved in recognition and catalysis. Biochemistry. 1991 Jul 2;30(26):6416–6422. doi: 10.1021/bi00240a011. [DOI] [PubMed] [Google Scholar]
- Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988 Oct 30;70(2):399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Wolfes H., Alves J., Fliess A., Geiger R., Pingoud A. Site directed mutagenesis experiments suggest that Glu 111, Glu 144 and Arg 145 are essential for endonucleolytic activity of EcoRI. Nucleic Acids Res. 1986 Nov 25;14(22):9063–9080. doi: 10.1093/nar/14.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. Y., Schildkraut I. Isolation of BamHI variants with reduced cleavage activities. J Biol Chem. 1991 Mar 5;266(7):4425–4429. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yanofsky S. D., Love R., McClarin J. A., Rosenberg J. M., Boyer H. W., Greene P. J. Clustering of null mutations in the EcoRI endonuclease. Proteins. 1987;2(4):273–282. doi: 10.1002/prot.340020403. [DOI] [PubMed] [Google Scholar]


