Abstract
During meiosis in many organisms, homologous chromosomes engage in numerous recombination events initiated by DNA double-strand breaks (DSBs) formed by the Spo11 protein. DSBs are distributed nonrandomly, which governs how recombination influences inheritance and genome evolution. The chromosomal features that shape DSB distribution are not well understood. In the budding yeast Saccharomyces cerevisiae, trimethylation of lysine 4 of histone H3 (H3K4me3) has been suggested to play a causal role in targeting Spo11 activity to small regions of preferred DSB formation called hotspots. The link between H3K4me3 and DSBs is supported in part by a genome-wide spatial correlation between the two. However, this correlation has only been evaluated using relatively low-resolution maps of DSBs, H3K4me3 or both. These maps illuminate chromosomal features that influence DSB distributions on a large scale (several kb and greater) but do not adequately resolve features, such as chromatin structure, that act on finer scales (kb and shorter). Using recent nucleotide-resolution maps of DSBs and meiotic chromatin structure, we find that the previously described spatial correlation between H3K4me3 and DSB hotspots is principally attributable to coincident localization of both to gene promoters. Once proximity to the nucleosome-depleted regions in promoters is accounted for, H3K4me3 status has only modest predictive power for determining DSB frequency or location. This analysis provides a cautionary tale about the importance of scale in genome-wide analyses of DSB and recombination patterns.
Keywords: chromatin structure, double-strand breaks, H3K4 trimethylation, meiosis, recombination, Set1, Spo11
Introduction
During meiosis, many species carry out recombination at multiple positions throughout the genome in order for homologous maternal and paternal chromosomes to pair with one another and become physically connected, ensuring proper chromosome segregation.1-3 Recombination also alters genome structure by disrupting genetic linkage groups, thereby contributing to genome diversity and evolution.4
Meiotic recombination initiates with DNA double-strand breaks (DSBs) made by the topoisomerase-like Spo11 protein, which cleaves DNA via a covalent protein-DNA intermediate.5 DSBs and the resulting recombination events are not randomly distributed throughout the genome.4,6 In the budding yeast Saccharomyces cerevisiae, the DSB “landscape” is organized into alternating hot and cold domains that are relatively large (tens to hundreds of kb). These domains, in turn, comprise smaller regions called hotspots (generally < 300 bp wide) separated by stretches of DNA (typically several kb long) that are only infrequently targeted for DSBs.7-10
This landscape is shaped by a hierarchical combination of many factors, including higher order chromosome structure, local chromatin structure and binding of sequence-specific transcription factors.6,10-13 Importantly, different factors operate over different size scales. For example, binding of sister chromatid cohesion proteins is strongly anticorrelated with DSB patterns at ~5–10 kb scales.9,10,13,14 Conversely, a dominant factor at sub-kb scales is the strong tendency of DSBs to form preferentially in gene promoters, a reflection of the relatively open, nucleosome-depleted structure of chromatin in these locations.10,15-20
One feature that has received considerable attention as a potential determinant of DSB location in yeast is the posttranslational trimethylation of lysine 4 of histone H3 (H3K4me3).21-25 The Set1 lysine methyltransferase is responsible for all mono-, di- and trimethylation of H3K4 in S. cerevisiae,26,27 and set1 mutants display altered DSB distributions.21,23 Moreover, a strong spatial correlation has been observed, in which regions with high DSB levels also display significant H3K4me3 enrichment,21-25 leading to the hypothesis that H3K4me3 is a “prominent and preexisting mark” of meiotic recombination sites, independent of transcript level of nearby genes.21 This idea is made more attractive by the recent discovery that PRDM9, a sequence-specific Zn finger-containing protein that can trimethylate H3K4,28 exerts a profound influence on the distribution of meiotic recombination in mice and humans,29-33 suggesting that H3K4me3 might have a widely conserved role in targeting Spo11 activity.
However, the precise connection between histone methylation and DSB location in yeast remains unclear. H3K4me3 is strongly enriched in nucleosomes near promoters of active genes,24,34‑37 so a spatial correlation with DSBs would be predicted in yeast, even if there were no functional connection, simply because both H3K4me3 and DSBs are often found in or near promoters. As discussed below, previously available genome-wide data for DSBs and histone modifications had insufficient spatial resolution to rigorously evaluate the DSB-H3K4me3 correlation and to control for promoter proximity. We addressed this issue using recent nucleotide-resolution maps of DSBs10 and modified nucleosomes during meiosis.24
The Importance of Scale
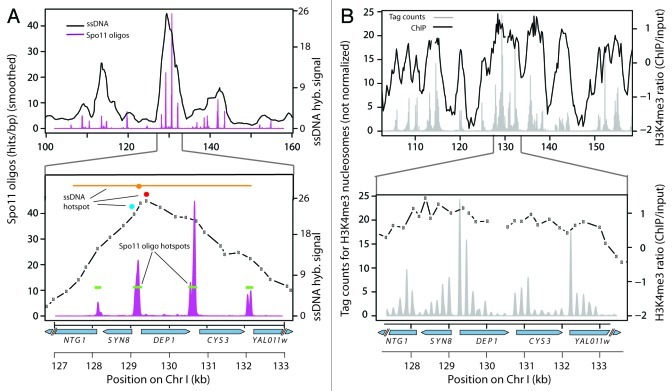
To understand how DSB distributions are shaped, it is imperative that maps of DSBs and of the relevant chromosomal features have sufficient resolution, i.e., matching or exceeding the scale over which the DSB-modulating factors operate. This issue can be illustrated by comparison of genome-wide data sets that differ in spatial resolution (Fig. 1). Early studies mapped DSBs using covalent Spo11-DSB complexes that accumulate in rad50S-like mutants or ssDNA generated by DSB resection as microarray hybridization probes.7-9,38 Resolution of these studies was inherently limited to a range of several kb because of microarray probe spacing and, more importantly, the large size of the enriched DNA fragments hybridized to the arrays. Recently, a new method was developed for determining genome-wide DSB distributions by purification and sequencing of the short DNA oligonucleotides (oligos) that remain covalently attached to Spo11 as a byproduct of DSB formation and early processing.10,39 Spo11 oligo sequencing faithfully represents quantitative DSB profiles at or near single-nucleotide resolution, easily resolving the prominent clustering of Spo11 cleavage sites within promoters (Fig. 1A).10
Figure 1. Resolution of maps around CYS3 DSB hotspot. Top panels show a 60 kb window on chromosome I; bottom panels show a zoomed-in view of the region around CYS3. (A) DSB maps from Spo11 oligo10 and ssDNA8 studies. Green bars are hotspot calls from Spo11 oligo mapping.10 Hotspots from ssDNA mapping are also indicated (red and teal circles, peaks only8,21; orange circle and line, peak and region with signal over threshold7). (B) Single-nucleosome resolution tag counts24 and ChIP-chip enrichment ratios21 for H3K4me3. Gaps in the ChIP-chip plots reflect probes with missing data.
With lower resolution DSB maps, general trends could be deduced from genome-wide averages (e.g., refs. 20 and 21), but sub-kb-scale features of the DSB landscape could not be distinguished, and the “hotspots” identified (usually defined as peaks in hybridization signal) represented relatively large domains that cannot be assigned to individual DSB hotspots (Fig. 1A). Indeed, hot regions often merged overlapping hybridization signals from multiple DSB hotspots (Fig. 1A).10 This limitation was recognized by Buhler et al., who explicitly cautioned that low-resolution DSB maps are inadequate to evaluate correlations with fine-scale chromosomal features, such as chromatin structure.8
This issue is particularly acute for a recent computational analysis that attempted to identify features that could distinguish DSB-hot from DSB-cold regions.25 Because the regions chosen for analysis were compiled from published8 DSB data that had relatively low resolution, the collection of hot regions was substantially enriched for sites close to promoters, while the collection of cold regions was depleted for promoters (data not shown). This systematic difference in promoter presence probably accounts for why the hotspot-associated features identified in this study,25 including H3K4me3, are already known to be associated with transcription units. Indeed, it is not clear that any of the identified features are predictive of DSB activity at all.
Scale also matters for maps of chromatin structure, including those that measure chromatin accessibility as well as those that assess specific histone modifications. Relatively low-resolution ChIP-on-chip data reveal large-scale trends but cannot resolve fine structure at individual genes (Fig. 1B).20,21 In contrast, high-resolution maps of chromatin structure have been generated by deep sequencing of mononucleosome-sized DNA fragments liberated by digesting chromatin with micrococcal nuclease.10,24,40-42 These maps capture many features of the chromatin landscape, including nucleosome-depleted regions (NDRs) at promoters and preferential enrichment of H3K4me3 on promoter-proximal nucleosomes (Fig. 1B). For the analyses described below, we used results of a comprehensive study of histone modifications during meiosis in the same S. cerevisiae strain background (SK1) in which DSBs were mapped.24
Hotspot-Centric Assessment of the DSB-H3K4me3 Correlation
Uniquely mapped Spo11 oligo sequences were assigned to the position of the 5'-most nucleotide of each mapped read, as described in reference 10. Tag counts from deep sequencing of anti-H3 or anti-H3K4me3 immunoprecipitates were assigned to theoretical mononucleosome midpoints, as previously described in reference 24, (data generously provided by L. Zhang and B.F. Pugh). Normalized H3K4me3 levels were calculated by dividing H3K4me3 tag counts by H3 tag counts in 3-bp bins. H3K4me3 levels change relatively little over the first few hours in meiosis when DSB formation occurs,24 so values from cultures immediately prior to meiotic induction were used.
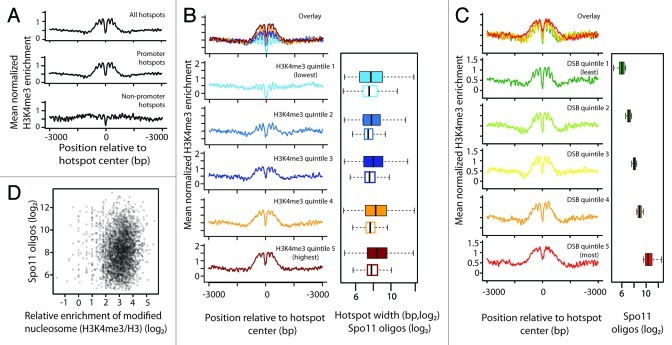
Pan et al. identified 3,600 DSB hotspots, not including those within selectable markers and other DNA sequences from exogenous sources integrated into the chromosomes of the mapped strain.10 When normalized H3K4me3 levels were averaged across these hotspots centered on their midpoints, substantial enrichment was observed over a zone of ~500 bp total (Fig. 2A). This agrees with patterns previously observed with low-resolution data21 or combinations of high- and low-resolution data.24,25 Of note, however, the high resolution of maps used here reveals additional fine-scale features: lack of H3K4me3 enrichment within a zone of ~100 bp at hotspot centers, attributable to low nucleosome density,10 and the regularly spaced peaks in the average profile, which reflect a propensity for nucleosomes with high H3K4me3 levels to be well positioned, immediately flanking hotspots (Fig. 2A).
Figure 2. H3K4me3 around DSB hotspots. (A) Mean normalized H3K4me3 enrichment (H3K4me3/H3) around all DSB hotspots, or around hotspots that did or did not overlap with promoters. Data are averaged in 3 bp bins and smoothed using a kernel regression smoother (“ksmooth” function in R). (B) H3K4me3 enrichment at promoter hotspots divided into quintiles by H3K4me3 enrichment. Data are plotted in 3 bp bins and smoothed. Box plots indicate distribution of Spo11 oligo counts (filled boxes) and hotspot widths (open boxes) within each quintile. The thick vertical line in each box is the median, the left and right edges of the boxes are the lower and upper quartiles, and the whiskers indicate the lowest and highest values within 1.5x of the inter-quartile range. Outliers are not shown. (C) H3K4me3 enrichment for promoter hotspots divided into quintiles by Spo11 oligo count. Data are plotted in 3 bp bins and smoothed. Box plots show distribution of Spo11 oligo counts within each quintile. (D) Scatter plot of total Spo11 oligo count vs. H3K4me3 enrichment for promoter hotspots.
To determine the contribution of colocalization of DSB hotspots with promoters, we divided hotspots into two classes: those overlapping (3,182) or not (418) with promoters (defined as the region 400 bp upstream from transcription start sites (TSS) annotated in ref. 43). On average, non-promoter hotspots showed no obvious enrichment for H3K4me3 (Fig. 2A). Thus, the spatial correlation seen when aggregating all hotspots is attributable to those that overlap promoters.
Since the profiles obtained from genome-wide averaging might obscure potential variation among hotspots, we asked whether all promoter hotspots were enriched for H3K4me3. We subdivided promoter hotspots into quintiles according to local H3K4me3 levels, which we defined by averaging the upstream and downstream maxima of normalized H3K4me3 values within 500 bp of each hotspot center. Hotspots in the lowest quintile showed little if any H3K4me3 enrichment (Fig. 2B), similar to non-promoter hotspots (Fig. 2A), demonstrating that a substantial fraction of promoters that lack significant levels of this histone mark are nonetheless targeted by Spo11. We previously showed that low nucleosome occupancy is a universal feature of hotspots, regardless of promoter proximity.10 Thus, we infer that targeting of Spo11 to specific regions reflects the open chromatin structure in these regions but does not require that H3K4 trimethylation be enriched locally in the population of cells.
Spo11 oligo counts for non-promoter hotspots were ~2.6-fold lower than for promoter hotspots on average (p < 2.2 x 10-16, Wilcoxon rank sum test), raising the possibility that H3K4me3 might tend to mark stronger hotspots. However, this correlation appears to reflect promoter vs. non-promoter hotspot differences rather than H3K4me3 enrichment, because there was only a modest difference in DSB activity between the different quintiles of promoter hotspots despite large differences in histone methylation (Fig. 2B). The median Spo11 oligo count was only ~50% greater in the highest H3K4me3 quintile than the lowest (337 vs. 223 oligos; p = 4.8 x 10-9), and all quintiles showed nearly complete overlap for the distribution of hotspot strengths (Fig. 2B). Of note, the highest H3K4me3 quintile was enriched for wider hotspots relative to the lowest quintile (p = 6.1 x 10-6) (Fig. 2B). Hotspots with more Spo11 oligos also tend to be wider,10 so it is not clear whether hotspot heat or width (or some other factor) is driving the weak correlation with H3K4me3 levels.
We further evaluated whether there is a quantitative relationship between DSB activity and H3K4me3 levels by subdividing promoter hotspots into quintiles based on the number of Spo11 oligos (Fig. 2C). Average H3K4me3 profiles showed only minor differences among the lowest four quintiles, but the hottest quintile differed qualitatively from the others, displaying a wider and less distinct NDR and less well-defined signatures of positioned flanking nucleosomes. These differences parallel those previously noted from total nucleosome profiles: stronger hotspots are more often wider and show more variable arrangement of nucleosomes as well, yielding a less distinct profile when many are averaged together.10 Importantly, the modestly higher H3K4me3 enrichment in the hottest quintile relative to the coldest contrasts with the extreme difference in Spo11 oligo counts (~20-fold difference in medians) (Fig. 2C). Indeed, when H3K4me3 and Spo11 oligo counts for all promoter hotspots were considered individually, we observed a statistically significant but extremely small correlation, accounting for just 1% of the variation (R2 = 0.011; p = 1.4 x 10-9) (Fig. 2D). Non-promoter hotspots subdivided by Spo11 oligo count did not show appreciable differences (data not shown).
A Promoter-Centric View
Although it is often useful to catalog DSB hotspots, at their most fundamental level they are best viewed as a cluster of phosphodiester bonds that share a high likelihood of cleavage by Spo11. In other words, rather than being discrete functional entities, hotspots are merely one among many levels of spatial organization of the DSB landscape.10 Because hotspot definition is inherently arbitrary, with little in the way of obvious, biologically meaningful boundaries between what is and is not a hotspot,10 we also sought to evaluate correlations between DSBs and H3K4me3 from a non-hotspot-centric perspective.
Most S. cerevisiae promoters have a short NDR flanked by well-positioned nucleosomes, with the TSS often lying within the first (+1) nucleosome.43,44 H3K4me3 is enriched preferentially within the first few nucleosomes and typically decreases progressively along a transcription unit away from the promoter24,36 (see examples in Fig. 1B, bottom part). For all genes considered together, we found only a weak correlation between the number of Spo11 oligos mapping within promoters and H3K4me3 level (scored as the normalized enrichment averaged over the position of the +1 nucleosome; R2 = 0.072, p < 2.2 x 10-16).
The S. cerevisiae genome is gene-dense, with numerous divergent transcription units sharing a single NDR with relatively high H3K4me3 on both sides (e.g., the SYN8-DEP1 pair in Fig. 1B). Other transcription units are oriented in tandem. Because H3K4me3 is low at 3' ends of genes, tandem promoters tend to show asymmetric H3K4me3 enrichment (e.g., the CYS3-YAL011w pair in Fig. 1B). To control for the difference in disposition of H3K4me3, we divided genes according to orientation of the upstream gene (divergent vs. tandem). Genes within 20 kb of telomeres or 5 kb from centromeres were omitted, because these regions are generally suppressed for DSB formation by larger-scale position effects that might obscure correlations with fine-scale features.7,8,10 Intergenic regions between convergent gene pairs are not considered here, because they rarely experience DSBs.10,19
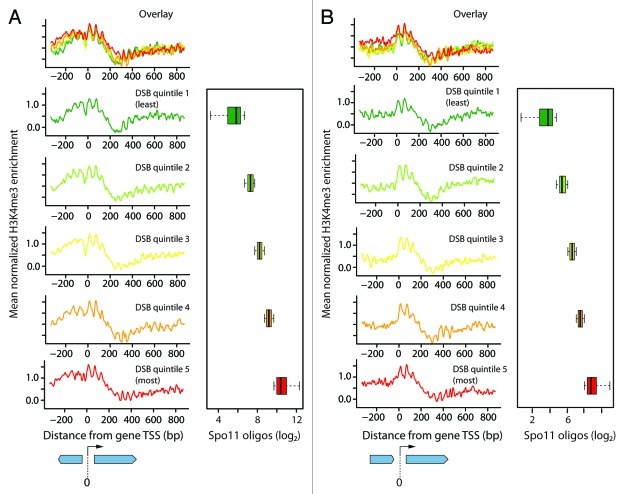
To look for quantitative DSB-H3K4me3 relationships, we further subdivided each group into quintiles by the number of Spo11 oligos in each promoter (Fig. 3). As expected from previous analyses of these and other genome-wide data,24,36,37 the average H3K4me3 profile for each quintile showed enrichment near the TSS, with prominent peaks for the +1 and +2 nucleosomes and with signal decreasing progressively away from the promoter. Also as expected, both the divergent (Fig. 3A) and tandem (Fig. 3B) gene groups showed H3K4me3 enrichment on average, but with a greater degree of additional enrichment upstream of TSSs for divergent genes. However, there were only modest differences between the quintiles for H3K4me3 enrichment despite very large differences in Spo11 oligo counts (Fig. 3). This suggests that while H3K4me3 is clearly a mark of transcribed genes, it is not a strong independent indicator for meiotic recombination initiation sites.
Figure 3. Histone marks around transcription start sites of genes. Mean normalized H3K4me3 enrichment at non-regulated genes with upstream genes in divergent orientation (A) or tandem orientation (B) (n = 1668 and 1491, respectively). Data are plotted in 3 bp bins and smoothed. Genes were divided into quintiles by Spo11 oligo count in their promoter. TSS is at 0 bp. Box plots show distribution of Spo11 oligo counts for each quintile.
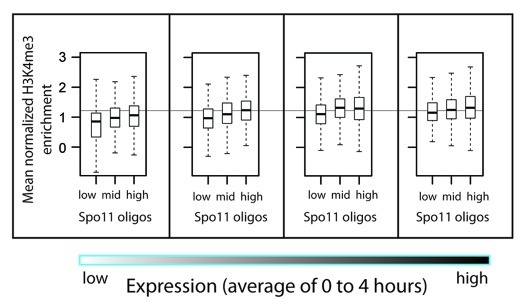
In previous studies, there appeared to be a quantitative correlation between H3K4me3 enrichment and presence vs. absence of a DSB hotspot nearby when gene expression levels were taken into account.21 However, the low-resolution data used could not unambiguously assign observed DSB or H3K4me3 signals to specific nearby genes (Fig. 1), so it is not clear that prior analyses were able to accurately evaluate the contribution of gene expression. We revisited this question with high-resolution data, following the general outline of the approach used by Borde et al.21 Meiotic transcriptome analysis identified 4,361 genes whose transcript levels changed little during meiosis.21 These were filtered to remove dubious genes, genes within 20 kb of telomeres or 5 kb of centromeres and genes lacking an annotated +1 nucleosome.43 The remaining 3,232 genes were subdivided into quartiles, using steady-state RNA levels from another study45 as a proxy for transcription activity. Then, we subdivided each quartile into thirds based on the number of Spo11 oligos mapping within each gene’s promoter region. Figure 4 shows the H3K4me3 levels of +1 nucleosomes. For all expression quartiles, there was a trend that genes with higher DSB frequency also showed higher median H3K4me3 levels, qualitatively similar to previous findings.21 However, the difference in H3K4me3 level between the highest and lowest DSB thirds in each category was small compared with the ~12–15-fold difference in Spo11 oligo counts (not shown), and there was nearly complete overlap in the range of H3K4me3 levels (Fig. 4). In linear regression analysis, factoring both gene expression and H3K4me3 level accounts for only 7.2% of the variation in Spo11 oligo count. Thus, as with other ways of subdividing the genome (see above), H3K4me3 levels have only modest predictive power for explaining DSB frequency.
Figure 4. H3K4me3 enrichment for promoter hotspots grouped by steady-state RNA levels. Non-regulated genes were divided into quartiles by average transcript level, then further divided into thirds based on Spo11 oligo counts within the promoters (see text). Box plots show the distribution of H3K4me3 enrichment for the +1 nucleosomes of the genes in each group. For all quartiles, median H3K4me3 levels for the hottest third were significantly higher than for the coldest third (Wilcoxon rank sum test, p < 6.4 x 10−5.)
Conclusions
We show here that, although there is a spatial correlation between DSBs and H3K4me3 globally, data of suitable resolution show that this correlation is driven largely by coincident localization to promoters. Many DSB hotspots lack H3K4me3 enrichment entirely, including both non-promoter hotspots and a significant subset of promoter-overlapping hotspots. Thus, local population average H3K4me3 enrichment is not necessary for robust hotspot activity at many natural DSB hotspots. Moreover, while there are modest correlations if promoter orientation and RNA levels are taken into consideration, neither H3K4me3 presence nor quantitative measures of its abundance provide much explanatory power to account for variation in DSB frequency between different promoters.
It remains the case that ablating H3K4 methylation by set1 mutation causes a drastic change in DSB distribution.21,23 We envision three general ways to account for available data. First, it may be that H3K4me3 (and/or the Set1 complex) directly targets Spo11 activity at many hotspots but fails to show strong correlations in the types of analysis done here. For example, quantitative relationships between methylation and DSBs might be obscured by uncorrelated variation in other factors, including local elements, like suites of different histone modifications or large-scale features such as cohesin binding or higher order chromosome structure. Alternatively, it is possible that signatures of direct methylation-DSB connections simply do not show up in population-averaged maps of DSBs and H3K4me3. Since only a small fraction of cells in a population make a DSB at any given hotspot, and in most cases only one of the four chromatids is broken in any one cell,46 it is plausible to envision a scenario in which every DSB occurs on a chromatid containing at least one trimethylated H3K4 nearby, without strong enrichment for H3K4me3 across a population.
Second, H3K4me3 may act locally but indirectly to influence Spo11 preference for particular promoters. H3K4me3 facilitates recruitment or stabilization on chromatin of numerous effector proteins that modulate, among other things, RNA polymerase II transcription kinetics and other types of histone modification (reviewed in ref. 47). Thus, lack of H3K4me3 may cause alterations in chromatin state around hotspots that then change Spo11 accessibility or activity.
Third, Spo11 may be entirely unaffected by local H3K4me3. Set1-deficient cells display substantial transcriptional misregulation during meiosis, including delayed and reduced expression of many genes known or suspected to control DSB locations.23 Thus, altered DSB distributions in set1 mutants might be an indirect consequence of wholesale changes in the level and/or timing of expression of proteins that influence Spo11 cleavage preferences. Indeed, there is ample precedent for such indirect effects, as it is known that DSB distributions can be substantially altered by seemingly innocuous changes in cellular physiology, such as the difference between auxotrophy and prototrophy for any of several amino acids or nucleotide bases.48,49
The correlative genomic analysis presented here cannot distinguish between these possibilities. However, the previously documented spatial correlation between these two aspects of promoter behavior—H3K4me3 enrichment and preferential DSB formation—should no longer be viewed as providing strong evidence for a functional connection. Similarly, caution is warranted in interpreting spatial correlations of DSBs with other chromatin modifications or DNA sequence features that are themselves independently tied to transcription, such as acetylation of histone H3 lysine 14 or trimethylation of histone H3 lysine 79.25 On a more general level, our findings also illustrate the importance of resolution and scale in evaluating correlations between different chromosomal processes.
Acknowledgements
We thank Valérie Borde and Nicolas Robine for discussions and sharing data; Liye Zhang and Frank Pugh for sharing data; members of the Keeney lab, especially Liisa Kauppi, Ryan Kniewel, Julian Lange and Mariko Sasaki for discussions and comments on the manuscript. This work was supported by NIH grant R01 GM058673. S.E.T. was supported in part by the Tri-Institutional Training Program in Computational Biology and Medicine. S.K. is an Investigator of the Howard Hughes Medical Institute.
Glossary
Abbreviations:
- DSB
double-strand break
- H3K4me3
trimethylated histone H3 lysine 4
- ChIP
chromatin immunoprecipitation
- oligo
oligonucleotide
- NDR
nucleosome-depleted region
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/19733
References
- 1.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–9. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 2.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, eds. Molecular Genetics of Recombination. Berlin Heidelberg: Springer-Verlag, 2007:381-442. [Google Scholar]
- 3.Bhalla N, Dernburg AF. Prelude to a division. Annu Rev Cell Dev Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–24. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 5.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Lankenau DH, ed. Recombination and Meiosis. Heidelberg: Springer-Verlag, 2008:81-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–9. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 7.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol. 2007;17:2003–12. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 8.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:11383–90. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–31. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–94. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 12.Lichten M. Meiotic chromatin: The substrate for recombination initiation. In: Egel R, Lankenau DH, eds. Recombination and Meiosis: Models, Means, and Evolution. Berlin: Springer-Verlag, 2008:165-93. [Google Scholar]
- 13.Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–83. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/S0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 15.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–63. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu T-C, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–8. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 17.Fan QQ, Petes TD. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2037–43. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas A. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc Natl Acad Sci USA. 1998;95:87–9. doi: 10.1073/pnas.95.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA. 1997;94:5213–8. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res. 2009;19:2245–57. doi: 10.1101/gr.096297.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kniewel R, Keeney S. Histone methylation sets the stage for meiotic DNA breaks. EMBO J. 2009;28:81–3. doi: 10.1038/emboj.2008.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, et al. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–67. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011;21:875–84. doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen L, Kim NK, Mariño-Ramírez L, Landsman D. Analysis of Biological Features Associated with Meiotic Recombination Hot and Cold Spots in Saccharomyces cerevisiae. PLoS ONE. 2011;6:e29711. doi: 10.1371/journal.pone.0029711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–95. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–48. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–8. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 29.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–63. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–9. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–5. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grey C, Barthès P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–9. doi: 10.1016/S1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 35.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 36.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 38.Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, Nicolas A, et al. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:1868–80. doi: 10.1128/MCB.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–7. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–52. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Dev Biol. 2010;339:258–66. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, et al. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–23. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Kleckner NE, Storlazzi A, Kim KP. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc Natl Acad Sci USA. 2011;108:20036–41. doi: 10.1073/pnas.1117937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Abdullah MF, Borts RH. Meiotic recombination frequencies are affected by nutritional states in Saccharomycescerevisiae. Proc Natl Acad Sci USA. 2001;98:14524–9. doi: 10.1073/pnas.201529598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cotton VE, Hoffmann ER, Abdullah MF, Borts RH. Interaction of genetic and environmental factors in Saccharomyces cerevisiae meiosis: the devil is in the details. Methods Mol Biol. 2009;557:3–20. doi: 10.1007/978-1-59745-527-5_1. [DOI] [PubMed] [Google Scholar]