Abstract
To study meiosis, synchronous cultures are often indispensable, especially for physical analyses of DNA and proteins. A temperature-sensitive allele of the Pat1 protein kinase (pat1-114) has been widely used to induce synchronous meiosis in the fission yeast Schizosaccharomyces pombe, but pat1-114-induced meiosis differs from wild-type meiosis, and some of these abnormalities might be due to higher temperature needed to inactivate the Pat1 kinase. Here, we report an ATP analog-sensitive allele of Pat1 [Pat1(L95A), designated pat1-as2] that can be used to generate synchronous meiotic cultures at physiological temperature. In pat1-as2 meiosis, chromosomes segregate with higher fidelity, and spore viability is higher than in pat1-114 meiosis, although recombination is lower by a factor of 2–3 in these mutants than in starvation-induced pat1+ meiosis. Addition of the mat-Pc gene improved chromosome segregation and spore viability to nearly the level of starvation-induced meiosis. We conclude that pat1-as2 mat-Pc cells offer synchronous meiosis with most tested properties similar to those of wild-type meiosis.
Keywords: analog-sensitive, fission yeast, meiosis, Pat1 kinase, synchronization
Introduction
Sexual reproduction depends on formation of haploid gametes from a diploid precursor cell in a process called meiosis. The fission yeast Schizosaccharomyces pombe has been an excellent model system for studying meiosis at the molecular level. One of the advantages of using fission yeast is that highly synchronous meiosis can be induced by inactivation of a temperature-sensitive allele of the Pat1 (Ran1) protein kinase (pat1–114). Pat1 kinase is a negative regulator of meiosis, and its major target is Mei2, an RNA-binding protein crucial for meiotic entry.1-5 Highly synchronous meiotic cultures can be obtained by arresting pat1–114 cells in G1 by nitrogen starvation and subsequent inactivation of Pat1 by shifting cells to non-permissive temperature (34°C).6,7 When meiosis is induced in cells by Pat1 inactivation, those cells synchronously undergo premeiotic S phase followed by two rounds of chromosome segregation (meiosis I and meiosis II, respectively) and form spores. This allows study of meiotic events in an entire population, which is particularly important for biochemical and cytological assays.Although pat1–114-induced meiosis is in many aspects similar to wild-type (pat1+) meiosis, there are several important differences. The frequency of recombination is reduced; nuclear positioning of centromeres is aberrant; chromosomes frequently missegregate during meiosis I, and spore viability is significantly reduced in pat1–114-induced meiosis.6,8,9 At least some of these defects are due to the absence of mating pheromone signaling, because the chromosome segregation defect can be partially suppressed by ectopic expression of genes from both mat loci, which activates the mating pheromone-signaling pathway.8,10 However, a major disadvantage of using the pat1–114-induced meiosis is that it requires elevated temperature (34°C) to inactivate the Pat1. Detrimental effects of elevated temperature on meiosis have been described in numerous studies in references 6 and 11-14. Therefore, it is desirable that a physiological temperature (e.g., 25°C) be used for studying meiosis.
Recent advances in chemical genetics now allow construction of conditional ATP analog-sensitive protein kinase alleles that can be inactivated by adding an inhibitor without the need of elevated temperature.15,16,17 Here, we construct analog-sensitive alleles of the Pat1 kinase (pat1-as), and show that pat1-as2 [Pat1(L95A)] can be used to generate meiotic cultures that progress through meiosis with a high degree of synchrony at physiological temperature. Importantly, we show that using pat1-as2 improves the fidelity of chromosome segregation and spore viability to levels near those of fully wild-type meiosis while maintaining high synchrony.
Results
pat1-as2 can be used to generate synchronous meiotic cultures at physiological temperature
To inactivate Pat1 conditionally, we applied a chemical-genetic strategy for sensitizing protein kinases to small-molecule inhibitors.15,16 We mutated a single codon, that for leucine 95 in the ATP-binding pocket of Pat1, termed the “gate-keeper” residue, to a small residue (glycine or alanine). While Pat1(L95G) mutant (pat1-as1) was not fully functional (unpublished data), Pat1(L95A) (pat1-as2) appeared to be functional as judged by the nearly wild-type growth of cells on YES medium at 25°C.17 We also noticed that cells expressing Pat1(L95A) were temperature-sensitive and sporulated at 34°C in the absence of an inhibitor. Importantly, Pat1(L95A) cells grew normally at 25°C on YES medium in the absence of an inhibitor, but very poorly in the presence of the ATP-analog 1-NM-PP1.17 Thus, we conclude that at 25°C, Pat1(L95A) is functional and confers sensitivity to 1-NM-PP1. We refer to Pat1(L95A) as Pat1-as (Pat1-analog sensitive).
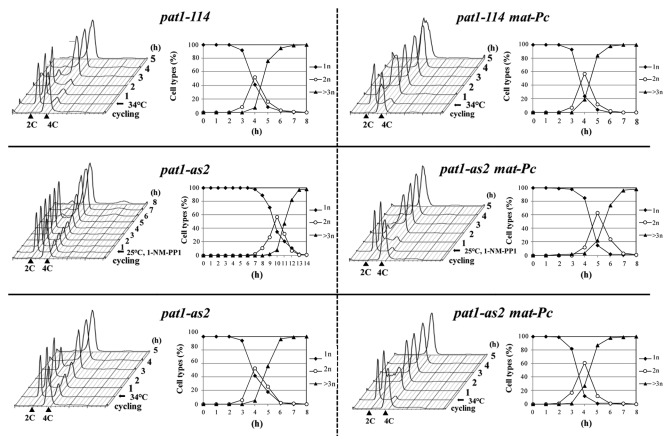
We next analyzed whether the pat1-as2 allele can be used to generate meiotic cultures that progress through meiosis with a high degree of synchrony. We arrested diploid pat1-as2/pat1-as2 cells in G1 by nitrogen starvation and subsequently inactivated the Pat1-as kinase by adding 1-NM-PP1 at 25°C. Analysis of nuclear divisions together with FACS analysis revealed that these cells underwent premeiotic S phase followed by two rounds of chromosome segregation in a very synchronous manner (Fig. 1). The level of synchrony in pat1-as2-induced meiosis was similar to that in pat1–114-induced meiosis, although the onset of S phase and meiotic divisions at 25°C were delayed by about 2–3 h in pat1-as2-induced meiosis (Fig. 1). A high level of synchrony was also observed in diploid pat1-as2/pat1-as2 cells, where the mating-pheromone signaling pathway was activated by ectopically expressing mat-Pc locus (Fig. 1). In this case, S phase was delayed by only about 1 h. We conclude that pat1-as2 can be used as a tool to generate synchronous meiotic cultures at physiological temperature 25°C.
Figure 1. Progression of diploid pat1–114/pat1–114 (JG12209), pat1-as2/pat1-as2 (JG15620), pat1–114/pat1–114 mat-Pc (JG16328) and pat1-as2/pat1-as2 mat-Pc (JG16113) cells into meiosis. Cells were cultured to mid log phase in YES-Ade medium, transferred to EMM2-NH4Cl medium for 16 h at 25°C (pat1–114 and pat1-as2) or for 7 h at 25°C (pat1–114 mat-Pc and pat1-as2 mat-Pc) to synchronize cells in G1, then shifted into EMM2 medium and incubated at 34°C or kept at 25°C with addition of 25 μM 1-NM-PP1 to inactivate the Pat1 kinase. Progression of meiosis was monitored by flow cytometry (Cytox Green staining) and by counting the nuclei (DAPI staining) of samples that were collected at the indicated time points after temperature shift or addition of 1-NM-PP1.
pat1-as2 improves fidelity of chromosome segregation and spore viability
pat1–114-induced meiosis differs from normal meiosis in several aspects, such as frequency of recombination, nuclear positioning of centromeres, segregation of sister centromeres during meiosis I, and spore viability.6,8,9 While the aberrant nuclear positioning of centromeres results from Pat1 inactivation, not the high temperature,9 some of the other defects might be due to the detrimental effects of elevated temperature, which is used to inactivate the Pat1–114 mutant kinase. To test this, we compared recombination, chromosome segregation and spore viability in wild-type cells and in cells where meiosis was induced by inactivation of Pat1 either by higher temperature (pat1–114, 34°C and pat1-as2, 34°C) or by adding inhibitor (pat1-as2, 25°C).
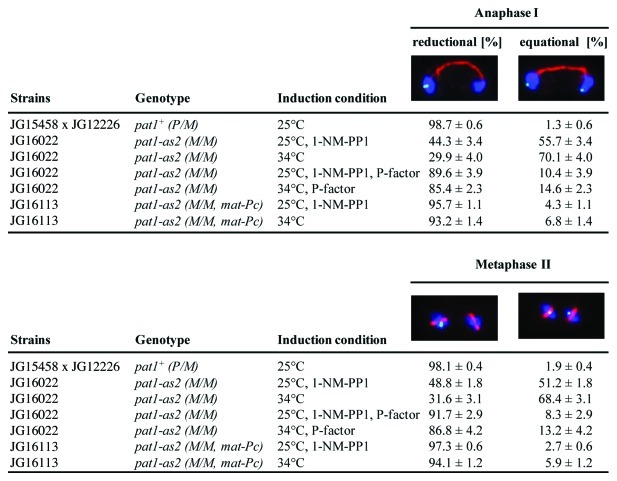
While in wild-type cells, sister centromeres segregate to the same pole in anaphase I, we observed that in cells where meiosis was induced by inactivation of Pat1-as at 34°C (note that pat1-as2 is temperature-sensitive), sister centromeres segregated to the same pole in only 30% of anaphase I cells but 45% when meiosis was induced by inhibiting Pat1-as at 25°C (Fig. 2). We confirmed, as previously reported,8 that triggering mating pheromone-signaling either by ectopically expressing mat-Pc or by adding P-factor increased the fidelity of segregation of sister centromeres during anaphase I. In cells induced into meiosis by inactivation of Pat1 by higher temperature (34°C), sister centromeres segregated to the same pole in 85% of cells treated with P-factor and in 93% of cells containing mat-Pc. The fidelity of chromosome segregation was further improved in cells where the mating pheromone-signaling pathway was activated and meiosis was induced by inhibiting Pat1-as at 25°C (Fig. 2). Proper segregation was seen in 90% of pat1-as2 cells treated with P-factor and 96% of pat1-as2 mat-Pc cells. Thus, the fidelity of chromosome segregation in synchronously induced pat1-as2 cells is nearly as high as that in wild-type cells.
Figure 2. Analysis of segregation of sister centromeres during meiosis I. Wild-type pat1+ (JG12226) cells carrying cen2-GFP (cen2-proximal loci were visualized by the LacI-GFP fusion protein bound to lac operator array inserted about 5 kb from cen2)8 were crossed to cells lacking cen2-GFP (JG15458) and sporulated in EMM2-NH4Cl medium. Diploid pat1-as2/pat1-as2 (JG16022) and pat1-as2/pat1-as2 mat-Pc (JG16113) cells carrying heterozygous cen2-GFP were cultured to mid log phase in YES-Ade medium, then transferred to EMM2-NH4Cl medium for 16 h at 25°C to synchronize cells in G1, then shifted into EMM2 medium and released into meiosis by adding 25 μM 1-NM-PP1 inhibitor at 25°C or by shifting the cells to 34°C. Mating pheromone-signaling was triggered either by addition of synthetic P-factor (70 µg/ml) or expression of the mat-Pc as indicated. Cells in anaphase I or metaphase II were fixed, stained with Hoechst 33342 and antibodies against tubulin and GFP, and examined under a fluorescence microscope. Segregation of chromosome II labeled with cen2-GFP was scored in 600 anaphase I cells and in 200 metaphase II cells.
Next, we determined spore viability by both tetrad and random spore analyses. Cells induced into meiosis by inactivation of Pat1 by higher temperature (34°C) produced spores with low viability (about 30%). Notably, spore viability was higher (about 57%) when cells were induced into meiosis by inactivation of Pat1-as at 25°C (Table 1). Triggering mating pheromone-signaling, either by addition of P-factor or addition of the mat-Pc, increased spore viability to about 73% and 80%, respectively, in cells induced into meiosis by inactivation of Pat1 by higher temperature (34°C). Spore viability was further improved in cells where the mating pheromone-signaling pathway was activated and meiosis was induced by inhibiting Pat1-as at 25°C: spore viability was increased from about 57% to about 80% or 86% by addition of P-factor or the mat-Pc gene (Table 1).
Table 1. Spore viability of S. pombe strains.
| Strain |
Alleles |
|
P-factor addition |
Mutation |
Temperature |
1-NM-PP1 |
Viability (%) |
|
|---|---|---|---|---|---|---|---|---|
| mat1 | Ectopic | - | TAa | RSAb | ||||
| JG11315 |
P/M |
- |
- |
none |
25°C |
- |
91.5 |
92.5 |
| JG12209 |
M/M |
- |
- |
pat1–114 |
34°C |
- |
31.5 |
29.2 |
| JG16022 |
M/M |
- |
- |
pat1-as2 |
25°C |
+ |
56.0 |
55.8 |
| JG16022 |
M/M |
- |
- |
pat1-as2 |
34°C |
- |
30.5 |
32.1 |
| JG15912 |
P/P |
- |
- |
pat1-as2 |
25°C |
+ |
58.5 |
57.4 |
| JG15912 |
P/P |
- |
- |
pat1-as2 |
34°C |
- |
31.5 |
27.8 |
| JG16022 |
M/M |
- |
+ |
pat1-as2 |
25°C |
+ |
79.5 |
81.3 |
| JG16022 |
M/M |
- |
+ |
pat1-as2 |
34°C |
- |
72.0 |
73.6 |
| JG16113 |
M/M |
mat-Pc |
- |
pat1-as2 |
25°C |
+ |
87.5 |
85.4 |
| JG16113 | M/M | mat-Pc | - | pat1-as2 | 34°C | - | 81.0 | 77.8 |
a TA – spore viability examined by tetrad analysis (tetrads examined ≥ 75)
b RSA – spore viability examined by random spore analysis (spores examined ≥ 6000)
Data are the means of 3 independent experiments. SEM was ˂ 10% of the mean.
In summary, precocious segregation of sister centromeres and low spore viability in cells induced into meiosis by inactivation of Pat1 by higher temperature (34°C) is partly due to high temperature (34°C), and using pat1-as2 to induce meiosis at 25°C improves fidelity of segregation of sister centromeres as well as spore viability. The fidelity of chromosome segregation and spore viability are almost at the wild-type level when induction of meiosis by inhibiting Pat1-as at 25°C is combined with the activation of mating pheromone-signaling pathway.
Meiotic recombination is slightly reduced in the pat1-as2 strain
The frequency of recombination in pat1-as2 strains in the presence of inhibitor was measured both for gene conversion (intragenic recombination) and for crossovers (intergenic recombination); the results were compared with those in pat1+ and pat1–114 strains (Table 2). The recombinant frequency was reduced about 3-fold for intragenic ade6 recombination and about 2-fold in the intergenic ade6 – arg1 and lys3 – ura1 intervals in pat1-as2 strains compared with those in pat1+ strains. This reduction of recombination was also seen in the pat1–114 strain, suggesting that this deficiency is a result of the loss of the active Pat1 kinase, regardless of the higher temperature.
Table 2. Meiotic recombination in pat1 strains.
| Straina | Mutation | ade6-M26 x 52b | lys3-ura1c | ade6-arg1 |
|---|---|---|---|---|
| GP1943 |
none |
3900, 5600 20000* |
33/280 = 12% 14cM |
NDd 38% = 68cMe |
| GP7382 |
pat1-as2 |
1300, 1600 7600* |
69/876 = 8% 9 cM |
146/743 = 20% 27 cM |
| GP1973 | pat1–114 | 2000 5000, 5200* |
290/420 = 7% 8 cM |
NDd |
a Recombination data are from sporulated diploid cultures: two meiotically induced cultures for pat1+ at 25°C, three for pat1-as2 at 25°C with 1-NM-PP1, and three for pat1–114 at 34°C.
b Measured as Ade+ colonies per 106 total colonies.
Indicates meiotic inductions done concurrently with higher Ade+ frequency in all strains; other experiments were done on a different day.
c Data are the number of recombinants/number of spore colonies tested. Genetic distance in cM was calculated using Haldane’s equation.
d ND, not determined.
e From Davis et al. (2008).18
Recombination data are from sporulated diploid cultures: two meiotically induced cultures for pat1+, three for pat1-as2, and three for pat1–114. The reduction is based on the mean frequencies from all colonies scored.
Discussion
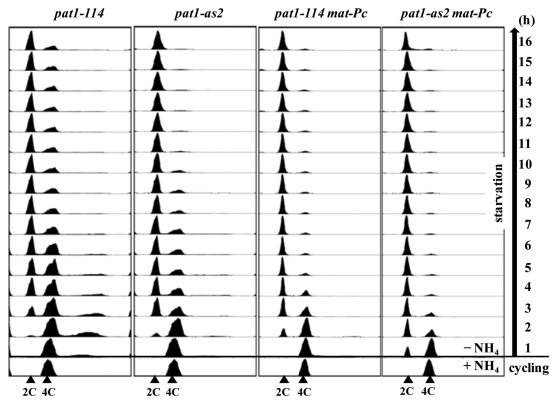
Development of the pat1–114 as a tool to induce synchronous meiotic cultures has been crucial for progress in the field of S. pombe meiosis. However, a major disadvantage of using pat1–114 is the higher temperature (34°C) needed to inactivate the Pat1–114 mutant kinase. Here, we show that this disadvantage can be overcome by using the pat1-as2 allele, which allows induction of synchronous meiosis at physiological temperature (25°C). Importantly, pat1-as2-induced meiosis eliminates some, but not all, abnormalities observed in pat1–114-induced meiosis. Ectopic expression of genes from both mat loci, which activates mating pheromone-signaling,8 eliminates further abnormalities, such as improving the fidelity of chromosome segregation and spore viability of pat1-as2-induced meiosis (Table 1 and Fig. 2). Induction of pat1-as2 mat-Pc with inhibitor at 25°C results in chromosome segregation at MI and spore viability only slightly different from those of wild type. In addition, cells arrest in G1 and upon induction replicate their DNA more quickly than do pat1–114 cells (Figs. 1 and 3). Although recombination in pat1-as2 at 25°C is reduced by a factor of 2–3 compared with that in pat1+ (Table 2), the addition of mating-pheromone or mat-Pc may increase recombination to the wild-type level. Thus, pat1-as2 mat-Pc cells offer the most synchronous meiosis with characteristics close to that of wild-type meiosis.
Figure 3.mat-Pc shortens the time for nitrogen starvation-induced G1 arrest and DNA replication. Diploid pat1–114/pat1–114 (JG12209), pat1-as2/pat1-as2 (JG15620), pat1–114/pat1–114 mat-Pc (JG16328) and pat1-as2/pat1-as2 mat-Pc (JG16113) cells were cultured to mid-log phase in YES-Ade medium and transferred to EMM2-NH4Cl medium. Cells were harvested at the indicated times and analyzed by flow cytometry (Cytox Green staining).
Another advantage of using pat1-as2 is that it can be combined with temperature-sensitive alleles of other essential proteins. For example, meiosis can be induced by inhibiting pat1-as2 at 25°C, and at the desired time, a temperature-sensitive form of a different protein can be inactivated by shifting the culture to non-permissive temperature (e.g., 34°C). This will, of course, introduce all the negative effects of higher temperature. Nevertheless, some of the meiotic processes occur normally at 34°C; therefore, this may still be a useful experimental approach. In addition to being useful for generating synchronous meiotic cultures, pat1-as2 provides a valuable tool for elucidating Pat1 function itself.
We anticipate that the pat1-as2 allele will be useful for additional studies of meiosis in which synchrony is important. For example, determination of the timing of events, such as the localization of proteins to chromosomes by microscopy, is simplest when cells are all at the same stage. Biochemical analyses, such as assaying protein abundance or modification and determining the frequency and structure of DNA intermediates of recombination, are most powerful with synchronous cultures, which can give maximal signals. In addition, novel DNA species, such as single-end invasion intermediates19 not yet reported in S. pombe, may be detectable at the lower temperature because of a possible longer life-time during meiosis that progresses more slowly.
Materials and Methods
S. pombe strains and general methods
The strains used and sources of alleles are listed in Table 3. Standard molecular and genetic procedures and media for growth have been described by Forsburg et al.20 and Moreno et al.21 Transformation of S. pombe with plasmids for deletion and integration was performed using a lithium acetate method as previously described by Gregan et al.22-26
Table 3.S. pombe strains used in this study.
| Strain | Genotypea |
|---|---|
| JG11315 |
h-/h+ leu1–32/leu1–32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 |
| JG15101 |
h-/h+ leu1–32/leu1–32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 pat1::NatR/pat1+ |
| JG12209 |
h-/h- ade6-M210/ade6-M216 pat1–114/pat1–114 |
| JG12226 |
h- leu1–32 lys1–131 ura4-D18 cen2(D107)::KanR ura4+-lacO his7+::lacI-GFP |
| JG15404 |
h- leu1–32 ade6-M216 ura4-D18 pat1::NatR pat1-as(L95G)::HygR |
| JG15458 |
h+ |
| JG15466 |
h- ade6-M216 pat1::NatR pat1-as(L95A)::HygR |
| JG15912 |
h+/h+ ade6-M210/ade6-M216 pat1::NatR/pat1::NatR pat1-as(L95A)::HygR/pat1-as(L95A)::HygR cen2(D107)::KanR-ura4+-lacO his7+::lacI-GFP |
| JG15978 |
h- ade6-M210 pat1::NatR pat1-as(L95A)::HygR cen2(D107)::KanR-ura4+-lacO his7+::lacI-GFP |
| JG16022 |
h-/h- ade6-M210/ade6-M216 pat1::NatR/pat1::NatR pat1-as(L95A)::HygR/pat1-as(L95A)::HygR cen2(D107)::KanR-ura4+-lacO his7+::lacI-GFP |
| JG16113 |
h-/h- ade6-M210/ade6-M216 pat1::NatR/pat1::NatR pat1-as(L95A)::HygR/pat1-as(L95A)::HygR cen2(D107)::KanR-ura4+-lacO his7+::lacI-GFP lys1::BleoMX-mat-Pc |
| JG16328 |
h-/h- lys1/lys1 ade6-M210/ade6-M216 pat1–114/pat1–114 lys1::BleoMX-mat-Pc |
| GP1943 |
h-/h+ ade6-M26/ade6–52 +/lys3–37 ura1–171/+ +/pro1–1 |
| GP1973 |
h-/h- ade6-M26/ade6–52 +/lys3–37 ura1–171/+ +/pro1–1 pat1–114 |
| GP7382 | h-/h- ade6-M26/ade6–52 pat1::NatR/pat1::NatR pat1-as(L95A)::HygR/pat1-as(L95A)::HygR arg1–114/+ +/lys3–37 +/ura1–171 |
a Alleles other than commonly used autoxtrophies are described in the following references: pat1::NatR17, pat1-as(L95G)::HygR17, pat1–1144, cen2(D107)::KanR-ura4+-lacO his7+::lacI-GFP8
Deletion of the pat1 gene
DNA flanking the pat1 gene (SPBC19C2.05) was PCR-amplified from genomic DNA using primers 5′-AAAATCTAGAcgcaagcgttgattgtcgat-3′ and 5′-AAAACTCGAGgtcccaattgatggcgaaaa-3′ for the upstream region and primers 5′-AAAATCTAGAttcgtattccaaaagcttagtttgc-3′ and 5′-AAAAAGATCTtcgctaccgcacgttgtttt-3′ for the downstream region (upper case letters indicate sequences used for the cloning, and lower case letters indicate S. pombe sequences). The products were digested with XbaI, ligated to each other and cloned into a pCloneNat1 vector (EF101285)22,27 carrying drug resistance markers for E. coli (ampicillin) and S. pombe (nourseothricin) using XhoI and BglII enzymes. The resulting pCloneNat1-△pat1 plasmid (p132) was amplified in E. coli, linearized by cutting with XbaI and used to delete one copy of the pat1 gene in diploid JG11315 strain. The deletion of the pat1 gene, which is essential for viability1,5 was confirmed by colony PCR and by tetrad analysis.
Creation of pat1 analog-sensitive (as) mutants and testing their sensitivity to ATP analogs
The pat1 gene together with its promoter and terminator regions was PCR-amplified from genomic DNA using primers 5′-ATATCTCGAGcgattgtgtttccttctcatcc-3′ and 5′-ATATGGATCCggtgatacaatatgactgcatgc-3′. The sequence was cloned into a pCloneHyg1 vector (EF101286)22 carrying drug resistance markers for E. coli (ampicillin) and S. pombe (hygromycin B) using XhoI and BamHI restriction sites, resulting in pCloneHyg1-pat1 plasmid (p133). Site-directed mutagenesis (QuikChangeII Site Directed Mutagenesis Kit, Agilent Technologies, Inc.) of this plasmid was used to change leucine 95 of Pat1, predicted to be the “gate-keeper” residue by a protein kinase sequence database (sequoia.ucsf.edu/ksd/sequences/family178/family178.html), to glycine or alanine. Oligonucleotides used for mutagenesis were as follows: 5′-aagacgccatttatgtcgttggccagtattgtccgaatgg-3′ (sense) and 5′-ccattcggacaatactggccaacgacataaatggcgtctt-3′ (anti-sense) for the Leu95Gly mutant and 5′-aagacgccatttatgtcgttgcccagtattgtccgaatgg-3′ (sense) and 5′-ccattcggacaatactgggcaacgacataaatggcgtctt-3′ (anti-sense) for the Leu95Ala mutant (mutant codons are italicized). The mutations were confirmed by sequencing. The resulting plasmids pCloneHyg1-pat1-as(L95G) (p134) and pCloneHyg1-pat1-as(L95A) (p135) were linearized with Psp5II and transformed into diploid △pat1/pat1+ strain JG15101. The transformants were sporulated on EMM2-NH4Cl plates (3.0 g/l potassium hydrogen phthalate, 2.2 g/l Na2HPO4, 1.0% (w/v) glucose, 2.0% agar, supplemented with amino acids, salts, vitamins and minerals) at 25°C for 36 h, and the haploids carrying mutant pat1-as(L95G) (further referred to as pat1-as1) or pat1-as(L95A) (further referred to as pat1-as2) alleles were selected based on resistance to Nourseothricin and Hygromycin. Correct integration of mutant alleles was confirmed by colony PCR. The pat1-as alleles were further PCR-amplified from genomic DNA and sequenced to finally prove the presence of the expected pat1-as alleles.
Sensitivity of pat1-as mutants to ATP analogs was tested as described by Cipak et al.17 Briefly, serial dilutions of exponentially growing pat1+ (JG15458) and pat1-as mutants (JG15404, JG15466) were plated on YES plates (5.0 g/l yeast extract, 3.0% glucose, 0.1 g/l l-leucine, 0.1 g/l l-lysine hydrochloride, 0.1 g/l l-histidine, 0.1 g/l uracil, 0.15 g/l adenine sulfate, 2.0% agar) supplemented with 25 μM ATP analogs, namely 1-NM-PP1 (4-amino-1-tert-butyl-3-(1′-naphtylmethyl)pyrazolo[3,4-d]pyrimidine) or 2-NA-PP1 (4-amino-1-tert-butyl-3-(2′-naphthyl)pyrazolo[3,4-d]pyrimidine). The plates were incubated at 25°C for 3 d and examined for visible colonies.
To activate mating pheromone signaling, we constructed diploid h−/h− strains, in which the h+-specific mating pheromone P-factor was artificially expressed from the mat-Pc gene.8 To create diploid h−/h− cells expressing mat-Pc gene, pCloneBleoMX-mat-Pc plasmid (p165) was used. To create the pCloneBleoMX-mat-Pc plasmid, lys1-mat-Pc was PCR-amplified from pYC36 vector8 using primers 5′-ATATTTAATTAAttttttgaacgctaaactttctaag-3′ and 5′-CCCCCTCGAGaaatgattctatcgtatcc-3′. The amplified fragment containing lys1-mat-Pc was digested with PacI and XhoI enzymes and cloned into pCloneBle1 vector (GQ354685). The resulting pCloneBleoMX-mat-Pc plasmid (p165) was linearized with PpuMI and integrated into the lys1+ locus of pat1–114 and pat1-as2 diploid strains.
Induction of meiosis, monitoring of progression of meiosis and analysis of chromosome segregation
Diploid pat1–114/pat1–114 and pat1-as2/pat1-as2 strains were grown in YES-Ade liquid medium to an OD600 = 0.5 at 25°C. The cells were collected by centrifugation, resuspended in EMM2-NH4Cl medium (3.0 g/l potassium hydrogen phthalate, 2.2 g/l Na2HPO4, 1.0% (w/v) glucose, supplemented with salts, vitamins and minerals) and incubated at 25°C for 16 h [pat1–114 (JG12209) and pat1-as2 (JG16022)] or for 7 h [pat1–114 mat-Pc (JG16328) and pat1-as2 mat-Pc (JG16113)] to arrest cells in G1 (Fig. 3). The cells were resuspended in fresh EMM2 medium [3.0 g/l potassium hydrogen phthalate, 2.2 g/l Na2HPO4, 5.0 g NH4Cl, 1.0% (w/v) glucose, agar, supplemented with salts, vitamins and minerals] and induced into meiosis by shifting to 34°C or by adding 25 μM 1-NM-PP1 at 25°C. To induce the mating pheromone response in pat1-as2 cells (JG16022), synthetic P-factor (Peptide 2.0 Inc.) was added to the EMM2 medium at a concentration of 70 μg/ml. To monitor the progression of meiosis, 0.5 ml of culture was fixed in 70% ethanol. Nuclear divisions were monitored by counting the number of nuclei after DAPI staining. Pre-meiotic S phase was monitored by flow cytometry. To follow the segregation of sister centromeres, the cells in various stages of meiosis (anaphase I and metaphase II) were collected, fixed in 2.0% paraformaldehyde and treated with 0.5 mg/ml Zymolase T-100 (cat# 8062H, MP Biomedicals, Inc.) to digest the cell walls. Primary antibodies TAT1 mouse monoclonal anti-tubulin (1:200, kindly provided by Dr. Gull) and rabbit polyclonal anti-GFP (1:800, cat# A11122, Molecular Probes) were used, followed by secondary antibodies Alexa Fluor 568 goat anti-mouse IgG (H+L), (1:500, cat# A11031, Molecular Probes) and Alexa Fluor 488 goat anti-rabbit IgG (H+L), (1:500, cat# A11034, Molecular Probes). DNA was visualized by staining with Hoechst 33342 (cat# H-3570, Molecular Probes).
Assay for meiotic recombination
Cultures were grown to saturation in EMM2*28 at 25°C, diluted to OD600 = 0.1 in EMM2*, and grown overnight at 25°C. When the OD600 reached 0.3 – 0.4, the cells were collected by centrifugation, washed once with water, suspended in EMM2* - NH4Cl, and incubated at 25°C for 18 h. NH4Cl was added to 0.5%; 20 μM 1-NM-PP1 was added to pat1-as2 cultures, or the temperature was raised to 34°C for pat1–114 cultures to initiate meiosis. At 24 h, the cells from 10 ml of culture were washed three times with water, suspended in 0.5 ml of water with 5 μl of glusulase, and incubated for 6 h at 25°C. 0.5 ml of 60% ethanol was added, and the mixture incubated at RT for 15 min. The spores were washed three times with water and suspended in 1 ml of water. The spores were plated and recombination frequencies determined as described.28
Acknowledgments
We thank K. Gull for the TAT1 antibody, G. Stengl for help with FACS analysis, S. Westermann and J.M. Peters for allowing us to use the tetrad dissection microscope, S. Polakova and I. Kovacikova for help with experiments, and K. Shokat and C. Zhang for help with construction of analog-sensitive alleles. We also thank A. Yamamoto and K. Tanaka for helpful discussions. This work was supported by Austrian Science Fund grants P23609 and F3403 to J.G. and United States of America National Institutes of Health grant GM032194 to G.R.S. L.C. was supported by the (European Community’s) Seventh Framework Programme (FP7/2007–2013) under grant agreement number PERG07-GA-2010–268167.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20052
References
- 1.Nurse P. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet. 1985;198:497–502. doi: 10.1007/BF00332946. [DOI] [Google Scholar]
- 2.Watanabe Y, Yamamoto MS. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–98. doi: 10.1016/0092-8674(94)90426-X. [DOI] [PubMed] [Google Scholar]
- 3.Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 4.Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet. 1985;198:416–21. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 5.Iino Y, Yamamoto M. Negative control for the initiation of meiosis in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1985;82:2447–51. doi: 10.1073/pnas.82.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bähler J, Schuchert P, Grimm C, Kohli J. Synchronized meiosis and recombination in fission yeast: observations with pat1-114 diploid cells. Curr Genet. 1991;19:445–51. doi: 10.1007/BF00312735. [DOI] [PubMed] [Google Scholar]
- 7.Kakui Y, Sato M, Tanaka K, Yamamoto M. A novel fission yeast mei4 mutant that allows efficient synchronization of telomere dispersal and the first meiotic division. Yeast. 2011;28:467–79. doi: 10.1002/yea.1851. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 2003;22:2284–96. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikashige Y, Kurokawa R, Haraguchi T, Hiraoka Y. Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2004;9:671–84. doi: 10.1111/j.1356-9597.2004.00760.x. [DOI] [PubMed] [Google Scholar]
- 10.Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell. 2005;16:2325–38. doi: 10.1091/mbc.E04-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu BC. Dark dependence of meiosis at elevated temperatures in the basidiomycete Coprinus lagopus. J Bacteriol. 1972;111:833–4. doi: 10.1128/jb.111.3.833-834.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malik CP. Effect of variations in temperature on meiosis in Gagea reticulata schultes. Nature. 1960;187:805–6. doi: 10.1038/187805b0. [DOI] [PubMed] [Google Scholar]
- 13.Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:3913–8. doi: 10.1073/pnas.0608936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loidl J. Effects of elevated temperature on meiotic chromosome synapsis in Allium ursinum. Chromosoma. 1989;97:449–58. doi: 10.1007/BF00295029. [DOI] [Google Scholar]
- 15.Gregan J, Zhang C, Rumpf C, Cipak L, Li Z, Uluocak P, et al. Construction of conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe. Nat Protoc. 2007;2:2996–3000. doi: 10.1038/nprot.2007.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 17.Cipak L, Zhang C, Kovacikova I, Rumpf C, Miadokova E, Shokat KM, et al. Generation of a set of conditional analog-sensitive alleles of essential protein kinases in the fission yeast Schizosaccharomyces pombe. Cell Cycle. 2011;10:3527–32. doi: 10.4161/cc.10.20.17792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis L, Rozalén AE, Moreno S, Smith GR, Martín-Castellanos C. Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr Biol. 2008;18:849–54. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/S0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 20.Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010;470:759–95. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- 21.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-L. [DOI] [PubMed] [Google Scholar]
- 22.Gregan J, Rabitsch PK, Rumpf C, Novatchkova M, Schleiffer A, Nasmyth K. High-throughput knockout screen in fission yeast. Nat Protoc. 2006;1:2457–64. doi: 10.1038/nprot.2006.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, et al. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol. 2005;15:1663–9. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 24.Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, et al. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle. 2010;9:2657–62. doi: 10.4161/cc.9.13.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rumpf C, Cipak L, Schleiffer A, Pidoux A, Mechtler K, Tolić-Nørrelykke IM, et al. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 2010;9:3997–4004. doi: 10.4161/cc.9.19.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rumpf C, Cipak L, Novatchkova M, Li Z, Polakova S, Dudas A, et al. High-throughput knockout screen in Schizosaccharomyces pombe identifies a novel gene required for efficient homolog disjunction during meiosis I. Cell Cycle. 2010;9:1802–8. doi: 10.4161/cc.9.9.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spirek M, Benko Z, Carnecka M, Rumpf C, Cipak L, Batova M, et al. S. pombe genome deletion project: an update. Cell Cycle. 2010;9:2399–402. doi: 10.4161/cc.9.12.11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Methods Mol Biol. 2009;557:235–52. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith GR. Genetic analysis of meiotic recombination in Schizosaccharomyces pombe. Methods Mol Biol. 2009;557:65–76. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]