Abstract
Anti-EGFR therapy is among the most promising molecular targeted therapies against cancer developed in the past decade. However, drug resistance eventually arises in most, if not all, treated patients. Emerging evidence has linked epigenetic changes, such as DNA methylation at CpG islands, to the development of resistance to multiple anticancer drugs. In addition, genes that are differentially methylated have increasingly been appreciated as a source of clinically relevant biomarker candidates. To identify genes that are specifically methylated during the evolution of resistance to anti-EGFR therapeutic agents, we performed a methylation-specific array containing a panel of 56 genes that are commonly known to be regulated through promoter methylation in two parental non-small cell lung cancer (NSCLC) and head and neck squamous cell carcinoma (HNSCC) cell lines and their resistant derivatives to either erlotinib or cetuximab. We found that death-associated protein kinase (DAPK) was hypermethylated in drug-resistant derivatives generated from both parental cell lines. Restoration of DAPK into the resistant NSCLC cells by stable transfection re-sensitized the cells to both erlotinib and cetuximab. Conversely, siRNA-mediated knockdown of DAPK induced resistance in the parental sensitive cells. These results demonstrate that DAPK plays important roles in both cetuximab and erlotinib resistance, and that gene silencing through promoter methylation is one of the key mechanisms of developed resistance to anti-EGFR therapeutic agents. In conclusion, DAPK could be a novel target to overcome resistance to anti-EGFR agents to improve the therapeutic benefit, and further evaluation of DAPK methylation as a potential biomarker of drug response is needed.
Keywords: cetuximab, DAPK, erlotinib, methylation, NSCLC
Introduction
Recent advances in molecular targeted therapies have demonstrated promising clinical activity of anti-EGFR agents in a variety of human solid malignancies. Two classes of EGFR-directed therapeutic inhibitors are currently in clinical use: anti-EGFR monoclonal antibodies, such as cetuximab, and small-molecule tyrosine kinase inhibitors (TKIs) of EGFR, such as erlotinib. Cetuximab is used in the treatment of metastatic colorectal cancer and advanced squamous cell carcinoma of the head and neck (HNSCC),1 while erlotinib is administered to patients with non-small cell lung cancer (NSCLC),2 head and neck cancer and pancreatic cancer.3 Although both classes of drugs are capable of inducing an initial therapeutic response, primary as well as acquired drug resistance is frequently observed and is associated with ultimate disease relapse and a disappointing overall response rate.4,5 Genetic analysis has revealed that several mutations in EGFR are associated with clinical response to erlotinib,4 while mutations in KRAS are associated with cetuximab resistance in colorectal cancer.6,7 However, even though these genetic alterations are firmly linked to drug response, they can only explain clinical outcome in a limited subset of patients. Thus, in order to benefit a larger population of patients, it is crucial to develop a more detailed understanding of the factors involved in the development of drug resistance and to identify additional therapeutic targets that may arise during the evolution of resistance.
Emerging evidence has linked epigenetic changes, such as DNA methylation at CpG islands, to the development of cytotoxic chemotherapeutic drug resistance as well as the initiation and progression of cancer due to the repressed transcription.8-13 Treatment-related DNA hypermethylation may play a role in creating drug-resistant phenotypes by inactivating genes that are required for cytotoxicity. For example, promoter methylation of the DNA repair enzyme O6-methylguanine-DNA methyltransferase (MGMT) contributes to resistance to carmustine (BCNU),9 while methylation of FANCF is associated with acquired cisplatin resistance in ovarian cancer.10 Both of these genes play functional roles in mediating the resistance. In addition, we have previously demonstrated that DNA methylation is a frequent event in cells that are chronically exposed to cisplatin, and that methylation of spermidine/spermine N1-acetyltransferase 1 (SAT1) and S100 calcium binding protein P (S100P) are functionally associated with cisplatin resistance.11
The rapid acquisition of drug resistance to anti-EGFR agents that is often observed clinically prompted us to explore whether epigenetic events occur during the forced evolution of resistance to cetuximab and erlotinib. To evaluate this possibility, we investigated four cell line pairs developed from two parental NSCLC and HNSCC cell lines, H226 and SCC1. For each pair, drug-resistant cells were generated via exposure of parental cells to slowly escalating doses of either erlotinib or cetuximab. DNA from each pair was extracted and screened using a methylation-specific array containing a panel of 56 genes that are commonly known to be regulated through promoter methylation. Identifying differentially methylated genes associated with acquired drug resistance, we found that death-associated protein kinase 1 (DAPK) was hypermethylated in drug-resistant cell lines generated from both parental strains, and that DAPK expression modulated the response to both cetuximab and erlotinib. .
Results
Identification of DAPK as a commonly methylated candidate gene in cetuximab- and erlotinib-resistant NSCLC and HNSCC cells
Four cell line pairs developed from two NSCLC and HNSCC cell lines (H226/H226C, H226D/H226T, SCC1/SCC1C, SCC1D/SCC1T) were used in this study. Resistance of drug-induced cells to cetuximab (C) or erlotinib (Tarceva, T) was validated by drug response assays (Fig. S1). To determine differences in promoter methylation between parental H226 and SCC1 cells and their drug-resistant progeny, we subjected DNA extracted from each cell line to a 56 gene panel methylation array containing genes that are known to be regulated by promoter methylation. This approach takes advantage of the well-defined methylation regions in the promoters of these known genes for array hybridization and offers improved sensitivity and specificity for gene discovery compared with large-scale genome-wide methylation arrays. In addition, changes in methylation are likely to be detected when any of these genes is differentially expressed in resistant cells. Using a computed threshold value to determine methylation status (genes with Cy5/Cy3 ratios < 4.0 were scored as methylated, and those with scores > 4.0 were scored as unmethylated), genes that were unmethylated in parental cells but methylated in the corresponding resistant progeny were identified (Table 1). Twenty of 56 genes were differentially methylated in at least one cell line pair. Generally, methylation pattern was unique in individual pairs, with some notable commonalities across cell lines and drugs. Three genes were differentially methylated in three cell line pairs (DAPK, CALC and GPC3). Focusing on these genes, we found that DAPK was the top-ranked gene in three of four cell line pairs. The strong and persistent change in DAPK promoter methylation observed in both NSCLC and HNSCC cells resistant to two different anti-EGFR drugs prompted us to further investigate the function of DAPK in anti-EGFR drug resistance.
Table 1. List of genes methylated in resistant daughter cells but not in parental cells.
| H226 (Cy5/Cy3) | H226C (Cy5/Cy3) | Ratio | |
|---|---|---|---|
|
DAPK |
14.5 |
1 |
14.5 |
|
MGMT |
10.3 |
1 |
10.4 |
|
PGK |
1 |
7.3 |
7.2 |
|
FAS |
1 |
7.1 |
7.1 |
|
CALC |
9.9 |
1.7 |
5.7 |
|
ERaA |
2.3 |
6.6 |
2.8 |
|
GPC3 |
5.2 |
3.3 |
1.6 |
|
P27 |
3 |
4.8 |
1.6 |
| SCC1 (Cy5/Cy3) | SCC1C (Cy5/C3) | Ratio | |
|---|---|---|---|
|
DAPK |
57.6 |
1 |
57.7 |
|
ERaB |
21.4 |
1 |
21.5 |
|
MGMT |
6.8 |
1 |
6.8 |
|
SRBC |
18 |
3.6 |
4.9 |
|
GR |
6.4 |
1.4 |
4.7 |
|
RB1 |
11 |
2.6 |
4.3 |
|
GPC3 |
4.4 |
1.7 |
2.6 |
|
SYK |
7.4 |
2.9 |
2.6 |
|
EP300 |
4.3 |
1.9 |
2.3 |
| H226D (Cy5/Cy3) | H226T (Cy5/Cy3) | Ratio | |
|---|---|---|---|
|
DAPK |
24.2 |
1 |
24 |
|
RPL15 |
7.3 |
0.4 |
20.4 |
|
VHL |
9 |
0.7 |
12.7 |
|
CALC |
9.3 |
0.8 |
11.6 |
|
GPC3 |
8.4 |
0.9 |
8.9 |
|
APAF-1 |
7 |
1 |
7 |
|
S100 |
2.1 |
14.1 |
6.8 |
|
FAS |
5.8 |
1 |
5.8 |
| SCC1 (Cy5/Cy3) | SCC1T (Cy5/Cy3) | Ratio | |
|---|---|---|---|
|
14–3-3 |
23.4 |
1 |
23.5 |
|
CALC |
4.3 |
0.9 |
4.8 |
| PGK | 2.1 | 6 | 2.9 |
Validation of DAPK promoter methylation and downregulation in drug-resistant cells
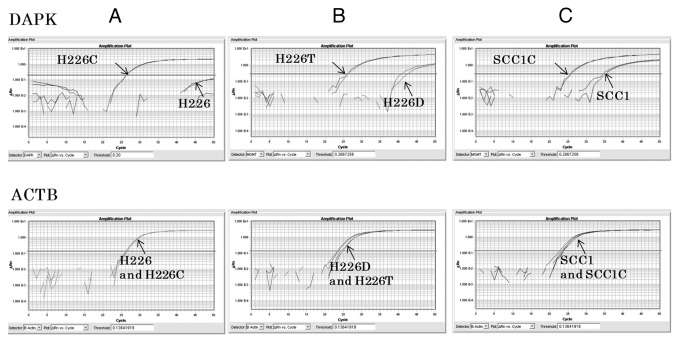
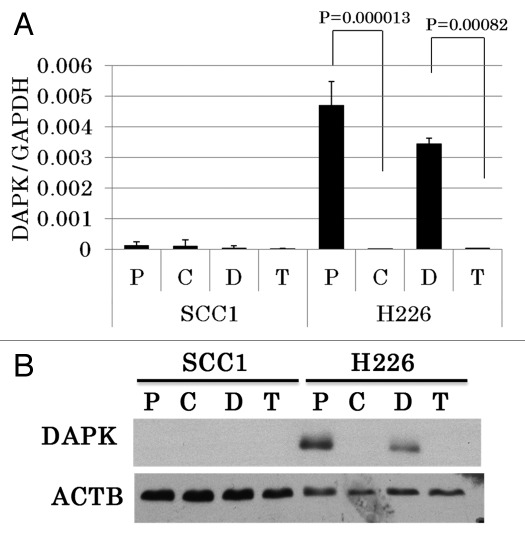
To analyze promoter methylation of DAPK in resistant cells, we used quantitative methylation-specific PCR (qMSP) with primers and probes specifically designed to amplify the methylated promoter region. Amplification of β-actin was used to control for DNA input. We found that the DAPK promoter region of H226 parental cells was minimally amplified, suggesting it was largely unmethylated. In contrast, DNA extracted from drug-resistant H226C cells produced strong amplification, indicative of heavy promoter methylation (Fig. 1A). The methylation is determined to be 100% when using in vitro methylated human normal lymphocyte DNA as standard reference. H226 cells exposed to long-term DMSO treatment (H226D), which served as a control for erlotinib-resistant H226T cells, manifested low-level (1%) methylation of DAPK, while H226T cells demonstrated complete methylation of the DAPK promoter region (100%, Fig. 1B). Unlike H226 cells, SCC1 parental cells displayed a medium level of methylation (35%) in the promoter of DAPK. However, the degree of methylation was further increased in cetuximab-resistant SCC1C cells (100%, Fig. 1C). We next performed real-time RT-PCR to examine whether methylation of the DAPK promoter leads to transcriptional repression, and found that while DAPK was present in H226 and H226D cells, it was almost absent in both resistant cell lines, consistent with methylation-induced gene silencing (Fig. 2A). In parental SCC1 cells, which displayed a moderate level of methylation, however, the expression of DAPK was markedly lower, indicating that the promoter methylation effectively suppressed its expression. Increased promoter methylation of DAPK was further observed in SCC1C cells without further transcription suppression (Fig. 2A). The expected change in DAPK level suggested by our RT-PCR data was further confirmed by western blot analysis using an antibody against DAPK (Fig. 2B). Taken together, these results demonstrated that DAPK was silenced during the process of acquiring cetuximab and erlotinib resistance in H226 cells via promoter hypermethylation.
Figure 1. The promoter of DAPK is methylated in acquired resistant cells to erlotinib and cetuximab. qMSP results of DAPK on H226 and H226C (A), H226D and H226T(B), and SCC1 and SCC1C (C). 100% of DNA methylations were observed in H226C, H226T and SCC1C. The results of qMSP of ACTB as internal control are displayed in the bottom panel.
Figure 2. The expression of DAPK is silenced in acquired resistant cells to erlotinib and cetuximab. (A) RT-PCR results of normalized DAPK mRNA expression in H226, H226C, H226D, H226T, SCC1, SCC1C, SCC1D and SCC1T cells. P, parental; C, cetuximab; D, DMSO; T, tarceva (erlotinib). (B) Immunoblotting of DAPK in the indicated cell lines. β-actin (ACTB) was used as internal control.
Restoration of cetuximab and erlotinib sensitivity in resistant NSCLC cells upon reconstitution of DAPK expression
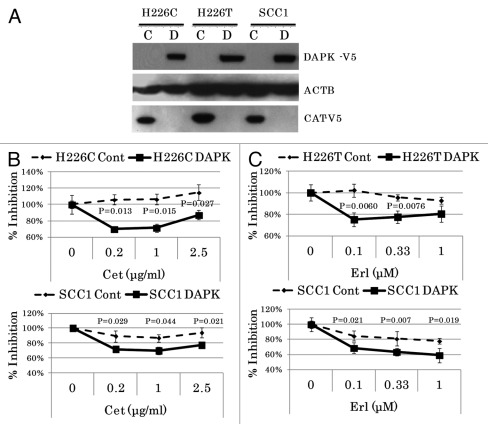
To further investigate the role of DAPK in mediating drug response in H226 cells, we cloned the full-length coding region of DAPK into the pcDNA™6.2/V5-DEST expression vector and transfected either control or DAPK vector into H226C and H226T cells. Pooled cells with stable expression were generated by blasticidine selection. Immunoblot analysis demonstrated either specific expression of the exogeneous DAPK-V5 fusion protein or the CAT-V5 fusion protein in the control vector-transfected cells (Fig. 3A). Importantly, we observed a marked re-sensitization of H226C cells to cetuximab and H226T to erlotinib after DAPK restoration (Fig. 3B and C, upper panels). On the other hand, although restoration of DAPK expression in parental SCC1 cells further increased the cellular response to both drugs (Fig. 3B and C, lower panels), ectopic expression of DAPK-V5 in SCC1C and SCC1T cells failed to restore sensitivity of these cells to the drugs (data not shown), suggesting that the evolution of anti-EGFR resistance in these cell lines is likely to be independent of DAPK.
Figure 3. Ectopic expression of DAPK increased sensitivity of H226 cells to erlotinib and cetuxiamb. (A) Immunoblotting analysis demonstrated exogeneous expression of fusion proteins. The pools of stable clones of DAPK using pcDNA6.2/V5-DEST-DAPK were compared with that of control empty vector using pcDNA6.2/V5/GW/CAT. Expression of β-actin (ACTB) was used as the internal control. Significantly decreased cell viability after cetuximab treatment in DAPK-expressed H226C and SCC1 cells (B) or after erlotinib treatment in DAPK-expressed H226T and SCC1 cells (C) was observed.
Acquisition of resistance to cetuximab and erlotinib after siRNA-mediated knockdown of DAPK in the NSCLC cells
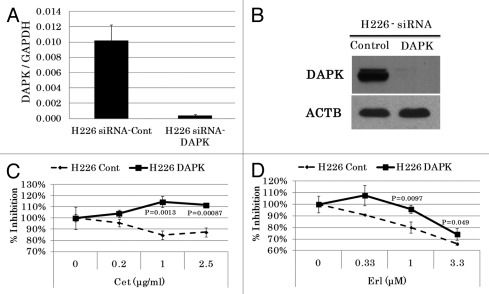
Since ectopic overexpression of genes does not always replicate physiologically relevant conditions, we next used siRNA to knockdown the endogenous expression of DAPK in H226 and H226D cells. Knockdown of DAPK was confirmed by both real-time RT-PCR and western blot for up to 5 d (Fig. 4A and B). As expected, DAPK knockdown in both cell lines resulted in the development of resistance to both drugs (Fig. 4C and D). Taken together, these results clearly demonstrated that loss of DAPK expression played a key role in the acquired resistance of H226 cells to anti-EGFR agents.
Figure 4. siRNA-mediated DAPK knockdown rendered cells more resistant to cetuximab and erlotinib. Scramble siRNA or siRNA against DAPK were transfected into H226 cells. DAPK knockdown was confirmed by either real-time RT-PCR (A), or immunoblotting with antibody against DAPK (B). (C) Transfected cells were exposed to either erlotinib or cetuximab and increased resistance was observed after DAPK knockdown.
Discussion
The impact of gene methylation on drug response has broad clinical implications. Methylation frequently leads to silencing of genes that may play a causal role in drug resistance. Thus, detection of promoter methylation can be a rapid and efficient approach to identifying transcriptional regulation of key resistance genes. In addition, genes that are differentially methylated have increasingly been appreciated as a source of clinically relevant biomarker candidates.14,15 Cell-free circulating tumor DNA found in the bloodstream of cancer patients as well as tumor DNA obtained non-invasively from other body fluids, such as sputum, saliva or urine, can provide sufficient quantities of DNA for the purpose of gene methylation analysis.16-18 We have found that bisulfite treatment of 100 ng of tumor DNA is often sufficient for subsequent methylation analysis at a confident level, and that this quantity can often be obtained from around 1 ml of patient serum. We have also successfully performed 56 gene panel methylation arrays, such as those used in this study, with only 5 ng of plasma DNA.19 Thus, non-invasive identification of specific gene methylation patterns associated with both patient disease and drug response is increasingly technically feasible and can provide important information in order to optimize treatment and improve patient outcomes.
In this study, we probed for alterations in gene methylation associated with acquired resistance to EGFR-targeted therapeutic agents, and discovered that in a number of cell line pairs, DAPK becomes methylated during the evolution of drug resistance. DAPK (also known as DAPK1) belongs to the calcium/calmodulin (CaM)-regulated serine/threonine protein kinase family. DAPK exhibits pro-apoptotic function and mediates cell death triggered by a variety of death-inducers, including interferon-γ,20 TGF,21 TNFα and Fas ligand.22 The expression of DAPK mRNA and protein are often lost in many major types of human cancers, at times as a result of silencing by DNA methylation.23 Similarly, we found here that when resistance to anti-EGFR agents was induced, DAPK-expressing cancer cells silenced expression through promoter methylation. Ectopic expression of DAPK restored sensitivity to these drugs in the resistant H226C and H226T cell lines, while knockdown resulted in acquisition of drug resistance in the parental cells, suggesting the functionality of this gene in mediating drug resistance. In the drug-sensitive HNSCC cell line SCC1, however, DAPK was already methylated and silenced prior to exposure to anti-EGFR agents, and restoration of DAPK in the induced drug-resistant clones failed to re-sensitize cells to either erlotinib or cetuximab. Interestingly, restoration of DAPK expression in parental SCC1 cells further increased the cellular response to the drug. These results suggest that acquisition of resistance to either erlotinib or cetuximab in SCC1 is most likely independent of DAPK, and that other unknown molecular determinants of resistance may counteract the effect of DAPK in inducing drug sensitivity specifically in these resistant clones.
Although the precise mechanism of DAPK-mediated resistance to anti-EGFR agents is unknown, we speculate that it may involve DAPK’s role in mediating apoptosis. Enhanced anti-apoptotic and pro-survival signaling is commonly seen in cancer cells that display primary or acquired resistance to conventional chemotherapeutic drugs and molecular targeted agents. For example, elevations in Bcl-2 expression cause resistance to multiple conventional chemotherapeutic drugs, while decreases in Bcl-2 expression promote apoptotic responses to anticancer drugs.24-28 Patients with ovarian cancer who have p53 mutations in their tumors are generally more resistant to platinum-based drugs such as cisplatin as compared with patients with wild-type p53.29 In addition, elevated activity of pro-survival PI3K-Akt signaling has also been observed to mediate resistance of NSCLC cells to both erlotinib and gefitinib, another anti-EGFR TKI. Interestingly, a reciprocal regulation between DAPK and ERK, one of the major downstream signal transducers of EGFR, has been established that promotes the apoptotic activity of DAPK. DAPK interacts with ERK through a docking sequence within its death domain and is a substrate of ERK. Conversely, DAPK promotes the cytoplasmic retention of ERK, thereby inhibiting ERK signaling in the nucleus.30 Thus, it is likely that the loss of expression of DAPK through methylation weakens pro-apoptotic signaling, compromising the effectiveness of anti-EGFR agents.
In summary, this study compared the 56 gene methylation profiles of parental H226 and SCC-1 cells to their erlotinib- and cetuximab-resistant daughter cell lines. We found that DAPK was methylated in the resistant derivatives of both type of drugs, and that its silencing was a strong mediator of acquired drug resistance in NSCLC but not HNSCC cells used in this study. While DAPK expression has previously been demonstrated to be associated with resistance to other cytotoxic chemotherapeutic agents used to treat gastric cancer31 and acute lymphocytic leukemia,32 its relationship to anti-EGFR therapeutic agents has never before been reported. Finally, while our results support a strong predictive value of DAPK methylation to drug response in NSCLC, further studies are required to clinically confirm these findings, including evaluation of the methylation status of clinical tumor specimens obtained from patients with a known drug response.
Materials and methods
Cell lines and chemicals
Eight cell lines were used in this study: the non-small lung cancer cell (NSCLC) cell line H226 and its cetuximab-resistant progeny H226C; the Dimethyl sulfoxide (DMSO)-treated control cell line H226D and the erlotinib-resistant H226T; the head and neck squamous cell carcinoma (HNSCC) cell line SCC1 and its cetuximab-resistant progeny SCC1C; the DMSO-treated control cell line SCC1D and the erlotinib-resistant derivative SCC1T. These cell lines were generous gifts from Dr. P. M. Harari. Drug resistance arose during a process of slowly escalating drug exposure over a period of 6 mo as reported previously.33 H226 and its derivatives were cultured in RPMI1640 (Mediatech Inc.), and SCC1 and its resistant derivatives were cultured in DMEM (Mediatech Inc.), supplemented with 10% FBS (Invitrogen). Cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C. All cells were routinely monitored for mycoplasma. Both cetuximab and erlotinib were ordered from the Johns Hokpins Pharmacy.
Cell viability and drug sensitivity assay
Cells were plated at a density of 3000/well in 96-well plates. The following day, cells were treated with 0, 0.01, 0.033, 0.1, 0.33, 1 or 3.3 μM erlotinib or 0, 0.2, 1, 2.5 or 5 μg/ml cetuximab for an additional 72 h. Cell viability was subsequently assayed using the MTT colorimetric assay. Briefly, MTT was dissolved in PBS at 5 mg/mL and then diluted in serum-free culture medium at a 1:10 ratio. Cells were incubated for 2–3 h at 37°C, followed by medium removal and color extraction by DMSO. The plates were read using a SpectraMax plate reader (Molecular Devices Corp.) at 570 nm with a reference wavelength of 650 nm. A minimum of 6 wells were tested for each erlotinib dose.
DNA extraction and methylation array analysis
Genomic DNA was extracted using standard phenol/chloroform technique followed by ethanol precipitation. Methylation detection in a 56-gene (MethDet-56) test was performed as described before.19,34 Briefly, equal amounts of DNA were treated with Hin6I (Fermentas Inc.) or mock digested, and each was amplified by PCR using gene-specific primers as described in reference 35. The Hin6- treated PCR product was labeled with Cy3, and the undigested PCR product was labeled with Cy5 (both from GE HealthCare). The labeled DNA was combined and hybridized to custom microarrays (Microarrays, Inc.), washed and scanned using a GenePix 4000B scanner (Molecular Devices). Signal intensity for each spot was determined using GenePix Pro 6 software after subtraction of background.
Each MethDet-56 microarray was printed in triplicate as 8 x 8 subarrays with 56 promoter-specific, five control and three empty spots. Signal intensities for each promoter spot were compared with the average intensities of five control spots, and values less than 2.5 times the control were considered non-informative (NA). The Cy5/Cy3 ratio was determined for each remaining spot; spots with ratios < 4.0 were considered methylated, while spots with ratios > 4.0 were considered unmethylated.34 The final methylation call for each promoter was determined by the majority call in at least two of the subarrays; if no majority call could be determined (e.g., two different calls and an NA call), the promoter was excluded from analysis (NA).
Quantitative methylation-specific PCR (qMSP)
Up to 2 μg of DNA was bisulfate converted and subsequently cleaned using the EpiTect kit (Qiagen) according to the manufacturer's instructions. Taqman quantitative methylation-specific PCR (qMSP) was performed as described previously.36 In vitro-methylated human lymphocyte DNA (recognized to be 100% methylated) from normal donors was used to generate standard curves and served as reference to estimate methylation levels in samples.36 The primer sequences of DAPK were: 5′-GGATAGTCGGATCGAGTTAACGTC-3′ (forward) and 5′-CCCTCCCAAACGCCGA-3′ (reverse), and the probe sequence was: 5′-TTCGGTAATTCGTAGCGGTAGGGTTTGG-3′.
RNA extraction, reverse transcription and real-time PCR
RNA was extracted using RNeasy mini kit (Qiagen) following manufacturer’s instructions. 400 ng of total RNA was reverse transcribed to single stranded cDNA using qScript cDNA SuperMix (Quanta BioSciences), which was then used as template for real-time PCR. Real-time PCR was performed using Fast SYBR® Green Master Mix (Applied Biosystems) with the following fast amplification program for 40 cycles: 95°C for 1 sec and 60°C for 20 sec on an ABI PRISM7000 Sequence Detection System (Applied Biosystems). All reactions were performed in duplicate or triplicate with water controls. The expression of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was monitored as an internal control. Specific PCR primer sequences were designed using Primer Express software (Applied Biosystems). The primer sequences of DAPK were 5′-ACCAGTGCCCTAGCCAAAGA-3′ (forward) and 5′-ATCGCTTCTGGCAACACTCA-3′ (reverse), and primer sequences for GAPDH were: 5′-CAACTACATGGTTTACATGTTC-3′ (forward) and 5′-GCCAGTGGACTCCACGAC (reverse). Expression of DAPK mRNA relative to GAPDH was calculated based on the threshold cycle (Ct) as 2-Δ(ΔCt), where Δ(ΔCt) = ΔCtDAPK – ΔCtGAPDH.
Antibodies and immunoblot analysis
Antibody against DAPK was obtained from Cell Signaling Technology, Inc. Monoclonal anti-β-actin antibody was obtained from Sigma. Monoclonal V5 antibody was obtained from Invitrogen. EGFR and phosphor-EGFR antibodies (pY1068) were obtained from Cell Signaling Technology and Invitrogen, respectively. Cells were lysed in RIPA lysis buffer containing protease inhibitors (Roche Diagnostic Systems) and phosphatase inhibitor cocktails (Sigma-Aldrich). Samples were homogenized in RIPA buffer followed by sonication. Protein concentrations were determined by the Lowry protein assay (Bio-Rad Laboratories). Equal amounts of protein were mixed with Laemmli sample buffer (62.5 mM TRIS-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1 M DTT and 0.01% bromophenol blue), run on 4–12% NuPAGE gels and transferred to nitrocellulose membrane (Bio-Rad Laboratories). The membrane was blocked with PBS supplemented with 0.1% Tween 20 and 5% nonfat milk for 1 h at room temperature and probed with primary antibody for overnight at 4°C, followed by HRP-conjugated appropriate secondary antibodies (Santa Cruz Biotech). Signals from immunoreactive bands were detected by enhanced chemiluminescence (ECL, GE Health Care according to the manufacturer's instructions).
Cloning, transfection and establishment of stable cell lines
pDONR223-DAPK, which carries the full-length coding regions of DAPK, excluding stop codon, was purchased from Addgene. The coding region of the DAPK was subcloned into pcDNATM6.2/V5-DEST vector (Invitrogen) using Gateway LR Clonase II enzyme Mix (Invitrogen). The expression vector, here named pcDNA6.2-DAPK-V5, was then used for transfection to establish stable cell lines. FuGENE® HD or X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostic Systems) were used to transfect H226C, H226T, SCC1, SCC1C and SCC1T cells according to the manufacturer’s instructions. pcDNA™6.2/V5/GW/CAT (Invitrogen) was used as the experimental control to establish the stable control cell lines. 48 h after transfection, cells were exposed to 5 µg/ml blasticidine for 10 to 14 d to establish pooled stable cell lines. DAPK expression was confirmed by both RT-PCR and western blotting.
Short interference RNA transfection
The expression of DAPK was knocked down by ON-TARGETplus SMARTpool short interfering RNA (Thermo Scientific) against DAPK. The siRNA was dissolved into Dharmacon siRNA buffer (Thermo Scientific) to a final concentration of 20 μmol/L. H226 and H226D cells were plated in either 6-well or 96-well plates and were transfected with siRNA against DAPK using RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. Twenty-four hrs after transfection, cells were treated with either cetuxiamb or erlotinib for additional 72 h. ON-TARGETplus Non-Targeting Pool siRNA (Thermo Scientific) was used as the experimental control.
Statistical analyses
All analyses were performed in duplicate or triplicate. Two-sided t-tests were performed to compare the mean of two samples with Sigmastat 3.0 software. The statistical significance was established at p < 0.05.
Supplementary Material
Supplementary PDF file supplied by authors.
Glossary
Abbreviations:
- DAPK
death-associated protein kinase
- NSCLC
Non-small cell lung cancer
- HNSCC
head and neck squamous cell carcinoma
- EGFR
epidermal growth factor receptor
- TKIs
small-molecule tyrosine kinase inhibitors
- qMSP
quantitative methylation-specific PCR
- ERK
extracellular signal-regulated kinases
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NCI grant EDRN (grant number 5U01 CA084986–12), NIDCR grant R01 (grant number R01 DE013152–11), NIH R21 (grant number RR024420) and FAMRI-funded 072017_YCSA.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20120
References
- 1.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 2.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. National Cancer Institute of Canada Clinical Trials Group Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–32. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, et al. National Cancer Institute of Canada Clinical Trials Group Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–6. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–44. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 6.Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 7.Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–92. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–74. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 11.Chang X, Monitto CL, Demokan S, Kim MS, Chang SS, Zhong X, et al. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 2010;70:2870–9. doi: 10.1158/0008-5472.CAN-09-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10:4420–6. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 13.Segura-Pacheco B, Perez-Cardenas E, Taja-Chayeb L, Chavez-Blanco A, Revilla-Vazquez A, Benitez-Bribiesca L, et al. Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J Transl Med. 2006;4:32. doi: 10.1186/1479-5876-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levenson VV. DNA methylation as a universal biomarker. Expert Rev Mol Diagn. 2010;10:481–8. doi: 10.1586/erm.10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10:123–32. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 17.Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, Van Criekinge W, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 18.Ostrow KL, Hoque MO, Loyo M, Brait M, Greenberg A, Siegfried JM, et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin Cancer Res. 2010;16:3463–72. doi: 10.1158/1078-0432.CCR-09-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liggett T, Melnikov A, Tilwalli S, Yi Q, Chen H, Replogle C, et al. Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis. J Neurol Sci. 2010;290:16–21. doi: 10.1016/j.jns.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 21.Jang CW, Chen CH, Chen CC, Chen JY, Su YH, Chen RH. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat Cell Biol. 2002;4:51–8. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 22.Cohen O, Inbal B, Kissil JL, Raveh T, Berissi H, Spivak-Kroizaman T, et al. DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J Cell Biol. 1999;146:141–8. doi: 10.1083/jcb.146.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy. 2006;2:74–9. doi: 10.4161/auto.2.2.2459. [DOI] [PubMed] [Google Scholar]
- 24.van de Donk NW, Kamphuis MM, van Dijk M, Borst HP, Bloem AC, Lokhorst HM. Chemosensitization of myeloma plasma cells by an antisense-mediated downregulation of Bcl-2 protein. Leukemia. 2003;17:211–9. doi: 10.1038/sj.leu.2402768. [DOI] [PubMed] [Google Scholar]
- 25.De Cesare M, Perego P, Righetti SC, Pratesi G, Carenini N, Rivoltini L, et al. Enhanced antitumour efficacy of gimatecan in combination with Bcl-2 antisense oligonucleotide in human melanoma xenografts. Eur J Cancer. 2005;41:1213–22. doi: 10.1016/j.ejca.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Jansen B, Schlagbauer-Wadl H, Brown BD, Bryan RN, van Elsas A, Müller M, et al. bcl-2 antisense therapy chemosensitizes human melanoma in SCID mice. Nat Med. 1998;4:232–4. doi: 10.1038/nm0298-232. [DOI] [PubMed] [Google Scholar]
- 27.Leung S, Miyake H, Zellweger T, Tolcher A, Gleave ME. Synergistic chemosensitization and inhibition of progression to androgen independence by antisense Bcl-2 oligodeoxynucleotide and paclitaxel in the LNCaP prostate tumor model. Int J Cancer. 2001;91:846–50. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1131>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Miayake H, Tolcher A, Gleave ME. Chemosensitization and delayed androgen-independent recurrence of prostate cancer with the use of antisense Bcl-2 oligodeoxynucleotides. J Natl Cancer Inst. 2000;92:34–41. doi: 10.1093/jnci/92.1.34. [DOI] [PubMed] [Google Scholar]
- 29.Reles A, Wen WH, Schmider A, Gee C, Runnebaum IB, Kilian U, et al. Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer. Clin Cancer Res. 2001;7:2984–97. [PubMed] [Google Scholar]
- 30.Chen CH, Wang WJ, Kuo JC, Tsai HC, Lin JR, Chang ZF, et al. Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. EMBO J. 2005;24:294–304. doi: 10.1038/sj.emboj.7600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Yashiro M, Qiu H, Nishii T, Matsuzaki T, Hirakawa K. Establishment and characterization of multidrug-resistant gastric cancer cell lines. Anticancer Res. 2010;30:915–21. [PubMed] [Google Scholar]
- 32.Holleman A, den Boer ML, de Menezes RX, Cheok MH, Cheng C, Kazemier KM, et al. The expression of 70 apoptosis genes in relation to lineage, genetic subtype, cellular drug resistance, and outcome in childhood acute lymphoblastic leukemia. Blood. 2006;107:769–76. doi: 10.1182/blood-2005-07-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benavente S, Huang S, Armstrong EA, Chi A, Hsu KT, Wheeler DL, et al. Establishment and characterization of a model of acquired resistance to epidermal growth factor receptor targeting agents in human cancer cells. Clin Cancer Res. 2009;15:1585–92. doi: 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melnikov AA, Scholtens DM, Wiley EL, Khan SA, Levenson VV. Array-based multiplex analysis of DNA methylation in breast cancer tissues. J Mol Diagn. 2008;10:93–101. doi: 10.2353/jmoldx.2008.070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melnikov AA, Gartenhaus RB, Levenson AS, Motchoulskaia NA, Levenson Chernokhvostov VV. MSRE-PCR for analysis of gene-specific DNA methylation. Nucleic Acids Res. 2005;33:e93. doi: 10.1093/nar/gni092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF file supplied by authors.