Abstract
Background
The association of vitamin D status with prostate cancer is controversial; no association has been observed for overall incidence, but there is a potential link with lethal disease.
Methods
We assessed prediagnostic 25-hydroxyvitamin D [25(OH)D] levels in plasma, variation in vitamin D–related genes, and risk of lethal prostate cancer using a prospective case–control study nested within the Health Professionals Follow-up Study. We included 1260 men who were diagnosed with prostate cancer after providing a blood sample in 1993–1995 and 1331 control subjects. Men with prostate cancer were followed through March 2011 for lethal outcomes (n = 114). We selected 97 single-nucleotide polymorphisms (SNPs) in genomic regions with high linkage disequilibrium (tagSNPs) to represent common genetic variation among seven vitamin D–related genes (CYP27A1, CYP2R1, CYP27B1, GC, CYP24A1, RXRA, and VDR). We used a logistic kernel machine test to assess whether multimarker SNP sets in seven vitamin D pathway–related genes were collectively associated with prostate cancer. Tests for statistical significance were two-sided.
Results
Higher 25(OH)D levels were associated with a 57% reduction in the risk of lethal prostate cancer (highest vs lowest quartile: odds ratio = 0.43, 95% confidence interval = 0.24 to 0.76). This finding did not vary by time from blood collection to diagnosis. We found no statistically significant association of plasma 25(OH)D levels with overall prostate cancer. Pathway analyses found that the set of SNPs that included all seven genes (P = .008) as well as sets of SNPs that included VDR (P = .01) and CYP27A1 (P = .02) were associated with risk of lethal prostate cancer.
Conclusion
In this prospective study, plasma 25(OH)D levels and common variation among several vitamin D–related genes were associated with lethal prostate cancer risk, suggesting that vitamin D is relevant for lethal prostate cancer.
CONTEXT AND CAVEATS
Prior knowledge
Although vitamin D levels have not been associated with men’s overall risk of prostate cancer, it is possible that they might be associated with the risk of lethal prostate cancer.
Study design
Prediagnostic plasma vitamin D levels in 1993–1995, variation in seven genes related to vitamin D metabolism, and risk of lethal prostate cancer were analyzed for 1260 men who developed prostate cancer (through 2004) vs 1331 control subjects within the Health Professionals Follow-up Study. A multimarker global test was used to determine whether subsets of 97 single-nucleotide polymorphisms from the seven vitamin D–related genes were related to incidence of lethal prostate cancer.
Contribution
Compared with men in the lowest quartile of plasma vitamin D levels, men in the highest quartile were at 57% less risk of lethal prostate cancer, but they had no difference in risk of overall prostate cancer. Some subsets of single-nucleotide polymorphisms from vitamin D–related genes were associated with lethal prostate cancer risk.
Implication
The data suggest that variations in men’s vitamin D levels and genetic backgrounds relevant to vitamin D metabolism might contribute to their risk of lethal prostate cancer.
Limitation
Vitamin D levels were only measured one time and the study group was mostly composed of white men. The global pathway analysis did not identify individual susceptibility loci and the single nucleotide polymorphism associations may be limited by multiple testing.
From the Editors
There is abundant laboratory evidence that vitamin D may have anticancer properties. In prostate cancer cell lines, vitamin D has been reported to promote differentiation and apoptosis and to inhibit cell proliferation, cell adhesion, and angiogenesis (1–4). In contrast, results from most epidemiological studies have not supported an association of circulating plasma 25-hydroxyvitamin D [25(OH)D], the biomarker of vitamin D status, with overall prostate cancer risk (5). Fewer studies have investigated whether 25(OH)D levels are related to a lower risk of lethal prostate cancer, the most clinically relevant outcome. In a previous publication (6), we examined plasma 25(OH)D levels and prostate cancer risk in the Health Professionals Follow-up Study (HPFS). During follow-up through the year 2000, 692 men developed prostate cancer. We found no statistically significant association of plasma 25(OH)D levels with the incidence of overall prostate cancer, but we did observe a non-statistically significant association of low plasma 25(OH)D with advanced-stage and lethal prostate cancer. At the time, only 60 patients had developed advanced-stage or lethal prostate cancer, and thus, the study had insufficient power to fully examine this relationship. In addition, men with prostate cancer who had high prediagnostic circulating levels of 25(OH)D had less risk of mortality than men in the lowest quartile of 25(OH)D in both the HPFS and the Physicians’ Health Study (PHS) cohorts (6).
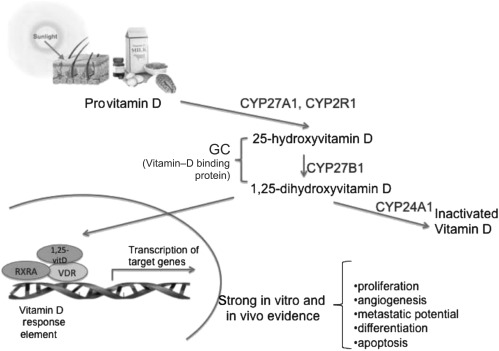
Common genetic variants of the vitamin D pathway may be associated with prostate cancer, but few studies have comprehensively assessed associations across the several genes that encode vitamin D pathway components (7,8) or with the specific endpoint of lethal prostate cancer (9). The vitamin D pathway involves several proteins (Figure 1). The 25-hydroxylases (CYP2R1 and CYP27A1) convert provitamin D that is absorbed from the diet or synthesized from the action of sunlight on the skin to the major circulating form, 25(OH)D. Next, 1-α hydroxylase (CYP27B1) converts 25(OH)D to the active metabolite 1,25-dihydroxyvitamin D [1,25(OH)2D] either in the kidney (where it is released to circulation) or in specific target organs including the prostate. Circulating 1,25(OH)2D is degraded by CYP24A1. Both vitamin D metabolites bind to vitamin D–binding protein (GC), which aids in transport to target sites. In target cells, the active form of vitamin D, 1,25(OH)2D, binds to the nuclear vitamin D receptor (VDR). This complex then forms a heterodimer with the retinoid X receptor (RXR) and attaches to vitamin D response elements on multiple target genes, including several with anticancer properties (10).
Figure 1.
Genes involved in vitamin D metabolism and signaling.
In this study, we investigated both circulating levels of 25(OH)D and genetic variants of the vitamin D pathway in participants of the prospective HPFS. With extended follow-up for incident prostate cancers through 2004, mortality data through 2011, and an increased sample size of 1260 men with prostate cancer, 114 of whom developed lethal cancers, we aimed to 1) investigate whether higher levels of plasma 25(OH)D are associated with a decreased risk of lethal prostate cancer and 2) use a pathway-based approach to assess whether common variation among seven genes related to the vitamin D pathway is associated with lethal prostate cancer.
Methods
Study Population
The HPFS cohort includes 51 529 US male health professionals who were aged 40–75 years at enrollment in 1986 when they responded to a questionnaire about their medical history and lifestyle. The men were mailed questionnaires to update this information every 2 years. Detailed dietary information was collected every 4 years. The overall follow-up rate was greater than 94% and ascertainment of deaths was more than 98% complete (11). This case–control study is nested in the HPFS blood cohort, which includes 18 018 (35%) participants who sent in chilled blood samples preserved in sodium EDTA via overnight delivery between 1993 and 1995. Samples were processed within 24 hours of blood collection; plasma, erythrocytes, and buffy coats were separated, aliquotted, and stored in liquid nitrogen freezers (12).
Case subjects in this study were 1260 men with incident prostate cancer that was diagnosed between 1993 and 2004. We excluded men with T1a tumors. Prostate cancers were first identified from self-reports on questionnaires or from death certificates and then confirmed by medical record review. Study investigators reviewed medical and pathology records to extract data on stage at diagnosis and Gleason score. Staging was classified using the TNM staging system. Histological grade was assessed using Gleason scores and summed. We used pathological stage and grade when available and clinical measures if pathological information was not available. Deaths were identified via repeated mailings, telephone calls, and searches of the National Death Index. Causes of deaths were confirmed through review of medical records and death certificates. Biennial follow-up surveys were mailed to those who reported prostate cancer to collect information on treatment and disease progression (eg, metastases). All reviews were conducted blinded to exposure information.
For each confirmed case patient who received a diagnosis of prostate cancer after having sent a blood sample, we randomly selected a control subject (n = 1331) from among those men who had not been diagnosed with prostate cancer at the time when the case patient was diagnosed. Because of the high screening rates in men who are diagnosed with prostate cancer (97% of men had received one or more prostate-specific antigen [PSA] tests before diagnosis), we required control subjects to have a PSA test within 2.5 years before the date of diagnosis of their matched case subject to provide the opportunity for occult prostate cancers to be diagnosed. Control subjects were matched to case patients on age (year of birth ± 1 year), PSA test before blood collection (yes or no), and time of blood collection—that is, time of day (midnight to before 9 AM, 9 AM to before noon, noon to before 4 PM, and 4 PM to before midnight), season (winter, spring, summer, fall), and year (exact calendar year). Follow-up for progression to prostate cancer–specific death was complete through March 31, 2011.
The racial composition of the HPFS blood cohort is 95% white men of European descent; for the plasma 25(OH)D analysis, we included all participants and adjusted for race. We also conducted a sensitivity analysis that was restricted to only white men. For the genetic analysis, we restricted the analysis to include only white men to reduce the potential for population stratification.
The main analyses focused on the risk of lethal prostate cancer, which was defined as prostate tumors with distant metastases at diagnosis, or progression to bone and/or organ metastases or prostate cancer–specific death during follow-up through March 31, 2011. We also assessed overall prostate cancer; advanced-stage prostate cancer, defined as stage T3 or T4 or M1 or N1 at diagnosis; and high Gleason grade prostate cancers, defined as those with Gleason pattern 4 + 3 or higher.
25(OH)D Assessment
Case and control subject selection occurred over four rounds of follow-up, and each batch was assayed separately: blood draw to January 1996, February 1996 to January 1998, February 1998 to January 2000, and February 2000 to January 2004. Matched case–control pairs were handled identically and assayed in the same batch in a blinded fashion along with random quality control standard aliquots. Plasma 25(OH)D was measured using a radioimmunosorbent assay in the laboratory of Dr Bruce Hollis (13). The mean intrapair coefficients of variation calculated from blinded quality control samples were 5.4%, 5.6%, 14.8%, and 5.6% for batches 1 to 4, respectively. Repeated blood samples on 144 individuals in HPFS collected approximately 3 years apart yielded a Pearson correlation coefficient of 0.7 (adjusted for age, race, and season) for 25(OH)D (14).
We accounted for batch and seasonal variation in 25(OH)D by creating separate quartile cut points for each batch and season based on the levels among the control subjects. Seasons were defined as summer (June to August), autumn (September to November), spring (March to May), and winter (December to February). We were unable to define specific cut points for deficiency and insufficiency based on clinical recommendations because of substantial batch-to-batch variation in addition to seasonal variation.
Single-Nucleotide Polymorphisms (SNP) Selection and Genotyping
Using the Tagger algorithm (15) implemented in the HaploView program (16) and dense genotyping data from the International HapMap Phase III CEU samples, we identified 97 linkage disequilibrium (LD) tagging SNPs to capture variation with R2 greater than 0.8 among the seven selected genes with additional coverage of the areas 10 kb upstream and downstream of the actual gene. Known functional SNPs and SNPs previously assessed in other studies were given priority. Otherwise, selection was restricted to those with a minor allele frequency of greater than 5% in the reference panel. One SNP in VDR and one in CYP24A1 failed genotyping.
Blood samples from matched case–control pairs were handled identically and assayed in the same batch in a blinded fashion. DNA was extracted from whole blood using a standard QIAmp kit (QIAGEN Inc, Chatsworth, CA) protocol, and genotyping was performed using the Biotrove Open Array SNP Genotyping Platform at the Harvard Medical School—Partners Health care Center for Genetics and Genomics. All SNPs had greater than 90% genotype completion, and the concordance was greater than 99% for blinded quality control samples. Participants with missing values had their genotypes imputed using the HapMap Phase III CEU data and the MACH imputation program (17). During imputation, one SNP (CYP24A1 rs6127119) was flagged because the minor allele frequency in the sample was only 3% as compared with the minor allele frequency of 22% in the phased haplotype samples from HapMap. Hardy–Weinberg equilibrium tests for each of the SNPs was performed among control subjects using the Pearson goodness-of-fit test using a predetermined cutoff of P value less than .001, as is customary. Two SNPs (CYP27A1 rs11677711 and CYP2R1 rs11023374) had evidence for departure from Hardy–Weinberg equilibrium among control subjects (P < .001). We excluded these three SNPs in the pathway analyses.
Statistical Analyses
All statistical tests were two-sided. Analyses were conducted using SAS v 9.1 (SAS Institute, Cary, NC) and R (http://www.r-project.org/) statistical packages. The participants of the study provided informed consent, and the Human Subjects Committee at the Harvard School of Public Health approved this study.
Plasma Vitamin D and Prostate Cancer.
Unconditional logistic regression models estimated odds ratios (ORs) and 95% confidence intervals (CIs) of prostate cancer by quartiles (using the lowest quartile as the reference), adjusting for matching factors, age at blood draw, PSA test before blood draw, year, batch and season of blood draw, and follow-up time. We also ran the corresponding conditional logistic regression analyses with the matched pairs; however, the results were not qualitatively different, and the estimates from the unconditional models were more stable because of the inclusion of all control subjects. Thus, we present the results for the unconditional models only. To address potential confounding by PSA screening, we created a variable for PSA testing frequency. For case patients, we calculated the percentage of PSA tests a man could possibly have reported before diagnosis. For example, a case patient who was diagnosed in 2004 could have up to six recorded tests (1994, 1996, 1998, 2000, 2002, 2004), whereas a case patient who was diagnosed in 1998 could have up to three recorded tests (1994, 1996, 1998). Frequency of PSA testing in control subjects was assessed through 2004, with a maximum of six recorded tests.
We considered the following potential confounders that have been associated with prostate cancer incidence and progression: body mass index (kg/m2), race (white, nonwhite), smoking (never, past, current), vigorous physical activity (metabolic equivalent task-hours/wk), energy-adjusted calcium (mg/d) and lycopene (μg/d) intakes, height (inches), diabetes status (yes or no), total energy intake (kcal/d), coffee intake (servings per day), family history of prostate cancer (yes or no), vasectomy status (yes or no), red meat (servings per week), and fish intake (servings per week). Covariate values from the 1994 follow-up questionnaire were used as they were closest to the time of blood collection. If values in 1994 were missing, values from previous years (1986–1992) were substituted. However, after adjusting for the matching factors, there was little evidence for confounding by any of the additional covariates, so they were not included in the final model.
Tests for trends were computed using an ordinal variable with values of 1–4 corresponding to the quartile in which the individual’s plasma vitamin D metabolite fell. All statistical tests were two-sided, and P values less than .05 were considered statistically significant.
We conducted the following sensitivity analyses. Because race is known to be associated with both prostate cancer risk and vitamin D levels, we restricted to only white men (95% of the population). To assess whether the association varied by time since blood draw and to consider the possibility of reverse causation in which undiagnosed tumor could potentially change vitamin D levels, we conducted analyses excluding case patients who were diagnosed within the first 2 years after blood draw and comparing case patients who were diagnosed within 5 years of blood draw to those diagnosed 5 years or later following blood draw. We also analyzed the data using a measure of 25(OH)D that was standardized for batch and season as described by Rosner et al. (18). β coefficients from a linear regression model of 25(OH)D with batch and season indicators were averaged; for each specific batch–season combination, the difference between the corresponding β coefficient from the model and the average coefficient was subtracted from the unadjusted 25(OH)D value to create a continuous measurement that was standardized to the average batch–season.
Vitamin D, Genetic Variation, and Prostate Cancer.
Adjusted odds ratios and 95% confidence intervals of each individual SNP and prostate cancer were assessed using unconditional logistic regression, adjusting for age at blood collection. We report the nominal two-sided P values for these analyses without adjusting for multiple testing; however, we also calculated a correction for multiple comparisons using a method that takes into account the LD within the set of SNPs being tested to calculate the effective number of independent tests (19). The 95 SNPs corresponded to 89 independent tests; consequently, a P value significance threshold of .0006 controls the experiment-wide type I error rate at the .05 level.
Because genotyped SNPs may be imperfect surrogates for the true causal SNP, their individual effect estimates are likely to be modest. An advantage of considering the joint effects of several SNPs in a region or along a pathway is that if several of these SNPs are in partial LD with the casual SNP, the multimarker global test will more effectively capture the true effect. To capture the joint effect of SNPs in the vitamin D pathway, we used a logistic kernel machine model. Unlike the typical multivariable omnibus test that applies a regression using a vector of covariates corresponding to the set of SNPs and results in a χ2 distribution with degrees of freedom equal to the number of SNPs in the model, the kernel machine considers the correlation between the SNPs in the set, typically resulting in reduced degrees of freedom, improving the power of the hypothesis test. Also, in contrast to methods that create a single genetic score from a set of proposed risk alleles, a benefit of the kernel machine is that it does not require a priori knowledge of directionality for the variants. In brief, the logistic kernel machine model treats each of the included SNPs as a random effect; desired covariates can be entered as fixed effects. The null hypothesis that the variance of the SNP random effects is 0 (ie, that the SNPs individually or jointly are not associated with disease) can be tested using a score test. The kernel machine is computationally efficient compared with other methods that require permutation. The mathematical details of the logistic kernel machine model have been described elsewhere (20,21), and the method has been applied to study genetic associations in a variety of traits and diseases (22–24). We assessed global associations for SNP sets defined by all seven genes as well as each gene individually.
Results
Plasma Vitamin D and Prostate Cancer
Characteristics of the 1260 prostate cancer patients are presented in Table 1; 20% had high Gleason grade (4 + 3 or higher) tumors and 14% had advanced-stage (stage T3/T4 or N1/M1 [T1–T4]) disease, and 9% progressed to develop bone or organ metastases or died of prostate cancer during follow-up through March 31, 2011. The median time from blood collection to diagnosis was 5.2 years. In control subjects (n = 1331), higher levels of 25(OH)D were statistically significantly associated with lower body mass index and a lower prevalence of diabetes, and higher physical activity, calcium intake, vitamin D intake, and exposure to sunlight (as measured by ultraviolet-B flux based on residence). Of note, we found no statistically significant association between plasma 25(OH)D levels and history of PSA testing before blood collection or frequency of PSA screening through 2004. Overall, screening rates were very high; 97% of all case subjects had a history of PSA screening before diagnosis, and control subjects were selected based on having a history of PSA screening. The difference in the median values of batch- and season-standardized 25(OH)D levels was approximately 20 ng/mL between the highest and lowest quartiles (Supplementary Table 1, available online).
Table 1.
Characteristics of prostate cancer case patients (n = 1260)*
| Characteristic | Case patients |
| Age at blood collection (y), mean (SD) | 64.4 (7.8) |
| Age at diagnosis (y), mean (SD) | 69.5 (7.5) |
| Stage, No. (%)† | |
| T1, T2 (N0, M0) | 1010 (86) |
| T3a (N0, M0) | 92 (8) |
| T3b (N0, M0) | 43 (4) |
| T4 (N0, M0) | 1 (0) |
| N1 | 15 (1) |
| M1 | 15 (1) |
| Gleason grade, No. (%)‡ | |
| 2 to 6 | 646 (56) |
| 7: 3 + 4 or no major score defined | 275 (24) |
| 7: 4 + 3 | 110 (9) |
| 8 to 10 | 129 (11) |
| Deaths or metastases due to prostate cancer, No. (%) | 114 (9) |
| Prostate cancer deaths without recorded date of metastasis | 68 |
| Metastases to bone or organs on follow-up | 31 |
| Metastases at diagnosis | 15 |
| Time to diagnosis from blood draw (y), median (IQR) | 5.2 (2.9–7.5) |
| Time to lethal prostate cancer (death or metastases) from diagnosis (y), median (IQR) | 5.2 (3.1–8.4) |
IQR = interquartile range; SD = standard deviation.
Data for stage missing for 84 subjects.
Data for Gleason grade missing for 100 subjects.
Table 2 presents the associations of lethal, overall, advanced-stage, and high-grade prostate cancers with plasma 25(OH)D levels, by season- and batch-specific quartile. Model 1 adjusted for matching factors, and model 2 additionally adjusted for PSA screening frequency. There was little evidence of confounding by PSA screening frequency so we refer to the results from model 1 in the text. We observed a statistically significant inverse association between plasma 25(OH)D levels and lethal prostate cancer (highest vs lowest quartile: OR = 0.43, 95% CI = 0.24 to 0.76; Ptrend = .001). Plasma 25(OH)D levels were not associated with the risk of overall prostate cancer or of prostate cancer that was high Gleason grade or advanced stage at diagnosis. We found no statistically significant association of plasma 25(OH)D levels with overall prostate cancer (highest vs lowest quartile: OR = 1.05, 95% CI = 0.84 to 1.32).
Table 2.
Odds ratios (ORs) and 95% confidence intervals (CIs) of prostate cancer by season- and batch-specific quartiles of plasma 25(OH)D
| Outcome | Quartile of plasma 25(OH)D |
Ptrend* | |||||||
| Q1 |
Q2 |
Q3 |
Q4 |
||||||
| No. of case patients/No. of control subjects | OR (95% CI) | No. of case patients/No. of control subjects | OR (95% CI) | No. of case patients/No. of control subjects | OR (95% CI) | No. of case patients/No. of control subjects | OR (95% CI) | ||
| Lethal prostate cancer | |||||||||
| Model 1† | 41/325 | 1.00 (referent) | 33/336 | 0.77 (0.47 to 1.28) | 21/334 | 0.50 (0.28 to 0.88) | 19/329 | 0.43 (0.24 to 0.76) | .001 |
| Model 2‡ | 41/325 | 1.00 (referent) | 33/336 | 0.78 (0.47 to 1.30) | 21/334 | 0.50 (0.28 to 0.88) | 19/329 | 0.44 (0.24 to 0.79) | .002 |
| Overall prostate cancer | |||||||||
| Model 1 | 310/325 | 1.00 (referent) | 298/336 | 0.93 (0.75 to 1.17) | 319/334 | 0.98 (0.78 to 1.22) | 333/329 | 1.05 (0.84 to 1.32) | .57 |
| Model 2 | 310/325 | 1.00 (referent) | 298/336 | 0.93 (0.74 to 1.17) | 319/334 | 0.99 (0.79 to 1.24) | 333/329 | 1.07 (0.86 to 1.34) | .45 |
| Advanced stage at diagnosis | |||||||||
| Model 1 | 51/325 | 1.00 (referent) | 43/336 | 0.88 (0.57 to 1.38) | 32/334 | 0.61 (0.38 to 0.97) | 40/329 | 0.78 (0.50 to 1.23) | .13 |
| Model 2 | 51/325 | 1.00 (referent) | 43/336 | 0.96 (0.61 to 1.52) | 32/334 | 0.63 (0.39 to 1.03) | 40/329 | 0.85 (0.53 to 1.35) | .22 |
| High-grade prostate cancer | |||||||||
| Model 1 | 69/325 | 1.00 (referent) | 55/336 | 0.79 (0.53 to 1.17) | 51/334 | 0.71 (0.48 to 1.05) | 64/329 | 0.92 (0.63 to 1.35) | .56 |
| Model 2 | 69/325 | 1.00 (referent) | 55/336 | 0.81 (0.54 to 1.21) | 51/334 | 0.75 (0.50 to 1.13) | 64/329 | 0.99 (0.67 to 1.46) | .87 |
P values for trend were calculated using unconditional logistic regression using an ordinal variable with values of 1–4 corresponding to the quartile of plasma 25(OH)D and were two-sided.
Model 1: adjusted for age at blood collection (year), prostate-specific antigen test before blood collection (yes or no), year of blood collection, timing of blood collection (midnight to 9 AM, 9 AM to before noon, noon to before 4 PM, 4 PM to before midnight), batch (blood collection [1993–1995]–1996, 1996–1998, 1998–2000, 2000–2004), season (winter, spring, summer, fall), and follow-up time.
Model 2: model 1 + prostate-specific antigen screening frequency (% of all possible recorded tests).
The association of lethal prostate cancer and 25(OH)D levels did not vary by time from blood collection to diagnosis (Table 3). Results were consistent when we used the standardized exposure measure instead of the batch- and season-specific quartiles and when we restricted the analysis to white participants.
Table 3.
Odds ratios (ORs) and 95% confidence intervals (CIs) of prostate cancer by season and batch-specific quartile of plasma 25(OH)D by time from blood collection to diagnosis*
| Outcome | Quartile of plasma 25(OH)D |
Ptrend† | |||||||
| Q1 |
Q2 |
Q3 |
Q4 |
||||||
| No. of case patients/No. of control subjects | OR (95% CI) | No. of case patients/No. of control subjects | OR (95% CI) | No. of case patients/No. of control subjects | OR (95% CI) | No. of case patients/No. of control subjects | OR (95% CI) | ||
| Lethal prostate cancer | |||||||||
| ≥2 y since blood collection | 29/325 | 1.00 (referent) | 24/338 | 0.82 (0.46 to 1.46) | 17/333 | 0.57 (0.30 to 1.07) | 13/335 | 0.41 (0.21 to 0.82) | .006 |
| <5 y since blood collection | 24/325 | 1.00 (referent) | 27/338 | 1.08 (0.59 to 1.99) | 14/333 | 0.59 (0.29 to 1.20) | 14/335 | 0.53 (0.26 to 1.08) | .03 |
| ≥5 y since blood collection | 15/325 | 1.00 (referent) | 8/338 | 0.50 (0.20 to 1.22) | 7/333 | 0.43 (0.17 to 1.09) | 5/335 | 0.32 (0.11 to 0.92) | .02 |
| Overall prostate cancer | |||||||||
| ≥2 y since blood collection | 258/325 | 1.00 (referent) | 245/338 | 0.91 (0.72 to 1.16) | 280/333 | 1.03 (0.82 to 1.30) | 269/335 | 1.02 (0.81 to 1.30) | .61 |
| <5 y since blood collection | 140/325 | 1.00 (referent) | 142/338 | 0.97 (0.71 to 1.33) | 139/333 | 1.04 (0.76 to 1.42) | 183/335 | 1.23 (0.91 to 1.67) | .14 |
| ≥5 y since blood collection | 167/325 | 1.00 (referent) | 158/338 | 0.87 (0.65 to 1.17) | 176/333 | 1.00 (0.74 to 1.33) | 155/335 | 0.92 (0.69 to 1.25) | .83 |
| Advanced stage at diagnosis | |||||||||
| ≥2 y since blood collection | 38/325 | 1.00 (referent) | 32/338 | 0.87 (0.53 to 1.44) | 24/333 | 0.61 (0.36 to 1.05) | 31/335 | 0.83 (0.50 to 1.38) | .27 |
| <5 y since blood collection | 27/325 | 1.00 (referent) | 27/338 | 1.05 (0.58 to 1.90) | 14/333 | 0.51 (0.25 to 1.02) | 25/335 | 0.85 (0.46 to 1.56) | .27 |
| ≥5 y since blood collection | 23/325 | 1.00 (referent) | 17/338 | 0.72 (0.37 to 1.42) | 18/333 | 0.81 (0.42 to 1.56) | 15/335 | 0.72 (0.36 to 1.45) | .42 |
| High-grade prostate cancer | |||||||||
| ≥2 y since blood collection | 54/325 | 1.00 (referent) | 47/338 | 0.83 (0.54 to 1.26) | 41/333 | 0.72 (0.46 to 1.12) | 51/335 | 0.91 (0.60 to 1.39) | .55 |
| <5 y since blood collection | 41/325 | 1.00 (referent) | 24/338 | 0.58 (0.33 to 1.01) | 24/333 | 0.57 (0.33 to 0.99) | 33/335 | 0.73 (0.44 to 1.22) | .23 |
| ≥5 y since blood collection | 28/325 | 1.00 (referent) | 31/338 | 1.01 (0.58 to 1.77) | 26/333 | 0.87 (0.48 to 1.55) | 32/335 | 1.21 (0.69 to 2.11) | .64 |
Adjusted for matching factors as in model 1 from previous table.
P values for trend were calculated using unconditional logistic regression using an ordinal variable with values of 1–4 corresponding to the quartile of plasma 25(OH)D and were two-sided.
Variation of Vitamin D Pathway Genes and Prostate Cancer
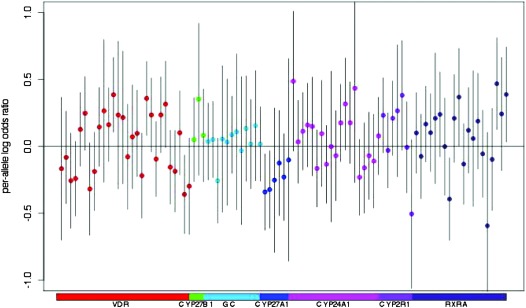
A total of 1193 case patients and 1244 control subjects were included in the genetic analyses. In our global pathway analysis, we observed a statistically significant association for the total pathway SNP set and lethal prostate cancer (P = .008). We also observed nominally statistically significant associations for SNP sets defined by the specific genes VDR (P = .01) and CYP27A1 (P = .02) with lethal prostate cancer (Table 4). Adjusting for plasma 25(OH)D levels did not change the results of the pathway analyses. Figure 2 shows the per-allele log odds ratio for each individual SNP and lethal prostate cancer. Although none of the SNPs was statistically significant at the more stringent multiple testing threshold, the overall pathway effect was statistically significant, illustrating the concept that variance in lethal prostate cancer may be explained by the combined effect of SNPs with more modest effects. Several individual SNPs had moderate point estimates and nine SNPs in VDR, RXRA, and CYP27A1 were nominally associated with lethal prostate cancer (P < .05). Under the null distribution (calculated from 10 000 permutations where we randomly shuffled case–control status), the probability of observing at least nine nominally statistically significant SNPs from the 95 individual tests was 0.09. Supplementary Table 2 (available online) lists the results for each individual SNP and lethal prostate cancer. Supplementary Table 3 (available online) presents the results for commonly studied VDR variants and two SNPs that were associated with circulating 25(OH)D levels in a large Genome-Wide Association Study (GWAS) (GC: rs2282679 and CYP2R1: rs2060793) (25). Bsm1 (rs1544410) was nominally associated with lethal cancer (per-allele OR = 1.47, 95% CI = 1.11 to 1.95, Ptrend = .007). Neither of the SNPs that predicted circulating 25(OH)D levels in the GWAS were statistically significantly associated with lethal prostate cancer.
Table 4.
Pathway analysis results for lethal prostate cancer and gene sets in the vitamin D pathway
| Gene | Chromosome | Size, kb | No. of SNPs included* | Global P from kernel machine analysis† | Global P from kernel machine analysis adjusted for 25(OH)D† |
| VDR | 12 | 63.49 | 28 | .01 | .01 |
| CYP27B1 | 12 | 4.86 | 3 | .75 | .70 |
| GC | 4 | 42.48 | 12 | .80 | .65 |
| CYP27A1 | 20 | 20.54 | 5 | .02 | .02 |
| CYP2R1 | 2 | 33.31 | 6 | .14 | .10 |
| CYP24A1 | 11 | 14.2 | 18 | .45 | .45 |
| RXRA | 9 | 114 | 20 | .09 | .10 |
| Total Pathway | 92 | .008 | .006 |
Ninety-seven SNPs originally selected; two SNPs failed genotyping; two were out of Hardy–Weinberg equilibrium, at P < .001; and one was eliminated because frequency in sample did not match that of phased haplotypes for imputation; therefore, a total of 92 SNPs were included in pathway analysis.
P values were calculated using a logistic kernel machine model and were two-sided.
Figure 2.
Per-allele log odds ratio for each single-nucleotide polymorphism (SNP) and lethal prostate cancer. Each dot represents a SNP, color coded by gene.
We did not observe global pathway associations for the vitamin D pathway–related SNPs with overall prostate cancer (P = .28) or with prostate cancers that were advanced stage at diagnosis (P = .89), or high Gleason grade (P = .35). No individual SNPs were statistically significantly associated with overall, advanced-stage, or high Gleason grade prostate cancer after considering multiple testing (Supplementary Table 4, available online).
Discussion
In this large prospective study of lethal prostate cancer, we comprehensively examined its relationship with vitamin D, focusing on circulating 25(OH)D levels and genetic variation in the vitamin D pathway. We observed that plasma 25(OH)D levels in prospectively collected blood samples and common genetic variation across the vitamin D pathway were associated with the risk of lethal prostate cancer. Men with the highest quartile of plasma 25(OH)D levels had less than half the risk of lethal prostate cancer compared with men who were in the lowest quartile of plasma 25(OH)D levels, regardless of time from blood collection to diagnosis. Furthermore, we performed the first pathway-based analyses, to our knowledge, of lethal prostate cancer risk in relation to common genetic variation across vitamin D pathway. We found a statistically significant association in the total pathway analysis consisting of seven vitamin D pathway genes and the risk of lethal prostate cancer. In addition, gene-specific analyses provided suggestive evidence for associations of lethal prostate cancer and CYP27A1 and VDR.
Although our findings for the individual genes should be interpreted cautiously, there is evidence that CYP27A1 and VDR are expressed in prostate tissue. VDR is part of the main receptor complex that mediates many of the biological actions of vitamin D. In men who were treated with prostatectomy, we previously found that those who had higher VDR expression had increased survival compared with men who had lower VDR expression (26). Finally, expression of CYP27A1 in the prostate could be important for regulating tissue levels of 25(OH)D, and one study has observed that CYP27A1 may also function to convert 25(OH)D to 1,25(OH)2D (27). Laboratory studies provide evidence that vitamin D can act to reduce cell adhesion and angiogenesis, which may be particularly relevant to decreasing metastatic potential (1,28).
We cannot rule out the possibility that our findings are due to chance, but we did consider other potential explanations for bias or confounding, in particular that by PSA screening. Whereas men with higher plasma 25(OH)D levels had a slighter higher frequency of PSA screening, overall screening rates were high in our cohort; greater than 97% of case patients had at least one PSA screening test. Furthermore, when we adjusted directly for PSA screening frequency or for correlates of healthy behavior that could be associated with PSA testing and 25(OH)D levels in our multivariable model such as body mass index and physical activity, we found no evidence for confounding. Thus, we believe that this bias is unlikely to explain the entire association of 25(OH)D with lethal prostate cancer. We considered several other potential confounders for prostate cancer incidence and progression and did not observe evidence of strong confounding. We believe it is unlikely that residual confounding alone could explain the full magnitude of the observed associations; however, we cannot completely rule out some bias by unmeasured confounding. Our observation that common genetic variants in the vitamin D pathway were associated with lethal prostate cancer also supports that vitamin D may be the causal factor rather than confounding by factors correlated with 25(OH)D levels.
Because of the long natural history of prostate cancer and the possibility that undiagnosed tumors could affect plasma 25(OH)D levels, we conducted sensitivity analyses in which we excluded tumors that were diagnosed within 2 and 5 years of blood collection. The association with lethal prostate cancer remained consistent in case subjects who were diagnosed 5 or more years after blood collection or approximately 10 or more years before the occurrence of a lethal outcome. Thus, it is unlikely that reverse causation could explain our findings.
These results are consistent with growing evidence that risk factors for lethal prostate cancer are different than those for overall prostate cancer (29). There is scant observational data on vitamin D and lethal prostate cancer. In a case-only analysis, we previously assessed the association of prediagnostic 25(OH)D and cancer-specific survival among a subset of prostate cancer patients in the HPFS and the PHS cohorts; higher 25(OH)D levels were associated with better outcomes (6).
Other studies (30–32) did not find beneficial associations between higher levels of 25(OH)D and prostate cancer, which was defined as “aggressive” based on Gleason grade or stage at diagnosis. Similarly, we did not observe associations with tumors characterized as advanced stage at diagnosis or high grade. One potential explanation for our observation that plasma 25(OH)D levels were associated with lethal prostate cancer but not high-grade cancers might be that tumor grade, as an indicator of differentiation, is an important predictor of lethal outcome, but far from a perfect surrogate. Most high-grade cancers are diagnosed at an early stage and do not progress to a lethal outcome, and the overall positive predictive value of Gleason score for prostate cancer mortality is relatively low (33). In our study, only 16% of men diagnosed with a Gleason score of 7 (4 + 3) or higher progressed to lethal prostate cancer. In addition, known inter- and intraobserver variability in the evaluation of Gleason grade and systematic scoring changes have occurred over time, which could result in further misclassification (34). A low stage at diagnosis also does not guarantee that a man will not develop lethal prostate cancer, especially in the context of high PSA screening rates. In our cohort, 86% of the prostate cancer case patients were diagnosed at stage T1 or T2, and more than half of the lethal prostate cancers in our study were diagnosed at a localized stage. Furthermore, our findings for 25(OH)D and lethal cancer were similar when we restricted to men diagnosed with earlier stage disease (T3a or less) that progressed to lethal cancer (OR comparing extreme quartiles = 0.40; 95% CI = 0.19 to 0.83; Ptrend = .02). It is possible that the relevant action of 25(OH)D exposure on lethal prostate cancer may be through influencing other characteristics of aggressive behavior (eg, apoptosis, angiogenesis, and/or cell adhesion) independent of grade and stage (28,35,36).
No previous studies have assessed the joint effects of SNPs in the vitamin D pathway and lethal prostate cancer risk. A small case-only study found suggestive evidence of associations for some individual variants in VDR, CYP24A1, and CYP27B1 and progression to prostate cancer–specific mortality (9). One nominally statistically significant SNP in VDR from that study (rs3782905) was in partial LD with rs10875693 from our study; the minor alleles of both SNPs were associated with an increased risk of prostate cancer mortality. We did not find gene-specific associations with CYP24A1 and CYP27B1, and we did not replicate the case-only study's findings for the corresponding individual SNPs within those genes.
A case-only GWAS of prostate cancer mortality did not note any susceptibility loci in the selected vitamin D pathway genes. However, only individual SNP analyses were conducted, and the study was powered to detect relatively strong associations (OR > 2 for a minor allele frequency of 0.20 using the genome-wide threshold α of 1 × 10-7) and thus would not capture smaller effect sizes that are likely to be the norm for most common variants (37). In contrast to the GWAS, we had an a priori defined set of SNPs in the vitamin D pathway. Thus, we were able to apply a more powerful pathway-based kernel machine approach, which allowed assessment of the joint association of variants in the predefined pathway.
A GWAS study found that polymorphisms in the GC and CYP2R1 genes were associated with circulating 25(OH)D levels (25). We conducted a pathway analysis for 25(OH)D levels and found a statistically significant association with the SNP sets defined by GC and CYP2R1 (Supplementary Tables 5 and 6, available online). However, we did not find statistically significant associations with the GC or CYP2R1 genes or the individual SNPs in GC or CYP2R1 with lethal prostate cancer. One possible explanation is that because only a small amount of variation (<3%) in 25(OH)D levels can be explained by the SNPs, we are not powered to detect the expected small associations in disease risk. It is also possible that the changes in circulating 25(OH)D levels observed with the alleles may not reflect the levels in the local prostate environment. Additionally, the SNPs may have pleiotropic effects. For example, the GC SNP may also affect the vitamin D–binding protein's affinity, and thus, circulating 25(OH)D levels may not reflect true bioavailability (38).
A single measurement of plasma 25(OH)D level has reasonable validity over a 5-year period (14,39). Nevertheless, multiple measurements would result in a better estimate of longer-term exposure and reduce the degree of non-differential measurement error. We are limited in our ability to determine the relevant etiologic period with only a single measurement. Prostate cancer cells may be present decades before diagnosis, and the median follow-up time from blood collection to diagnosis in our study was 5.4 years. We found that the relationship of lethal prostate cancer incidence to plasma 25(OH)D levels remained consistent when we compared case patients diagnosed within 5 years of blood collection to those diagnosed 5 years or more after blood collection. Conversely, because 25(OH)D levels, as repeatedly measured over time in a subsample of our population, had a Pearson correlation coefficient of 0.7, our single measure of 25(OH)D could be interpreted to be correlated with vitamin D status at or after diagnosis. We did not assign clinical cut points for vitamin D insufficiency and deficiency because blood samples were assayed at different times, which introduced extraneous laboratory batch variation. However, the rankings within each batch remain valid, the range of season and batch standardized values was within that of other studies, and plasma 25(OH)D concentrations varied by other known predictors such as vitamin D intake, sunlight exposure, and physical activity.
Our results may not be generalizable to other racial groups due to differences in genetic variation, vitamin D levels, and prostate cancer risk. A limitation of our pathway kernel machine model is that the method does not identify individual susceptibility loci and therefore does not directly translate to improved risk prediction. Nevertheless, the pathway result does contribute to the growing evidence that vitamin D plays a role in preventing prostate cancer progression. Our study only focused on common variation in the vitamin D genes; future studies that also consider rare variants, which may be in LD with causal SNP(s), may yield additional associations. We restricted all genetic analyses to white men to reduce the potential for population stratification. For a large number of individual markers (>500), the kernel machine method may be sensitive to detecting associations due to latent population structure; however, given the smaller number of SNPs in our study and the homogeneity of our study population, we do not believe this bias to be a major concern. Finally, it is possible that our findings could be due to chance, and it is important to consider the multiple testing, especially given the several genes, SNPs, and prostate cancer subtypes that were assessed when interpreting the findings. Larger studies with more lethal cancers would have more power to identify individual SNPs with similar point estimates; for lethal prostate cancer, our study was powered to detect an odds ratio greater than 1.6 (using α = .05) or greater than 2.0 (using the more stringent threshold α = .0005) for a minor allele frequency of 0.20. Should our results be replicated, studies are needed to explore potential functionality of the SNPs in the pathway.
There are several strengths of our study, including the detailed covariate information to assess confounding, prediagnostic blood samples, and the availability of medical record review, allowing for assessment of tumor characteristics. We used a comprehensive LD strategy for selection of tagSNPs across seven genes involved in vitamin D metabolism and signaling. We also used a multimarker kernel machine approach to detect the collective effect of SNPs across this pathway, even when power was limited for individual SNP analyses. Moreover, our study had substantial longitudinal follow-up required to assess lethal prostate cancer.
In summary, the findings from this large prospective study support the hypothesis that higher plasma 25(OH)D levels are associated with lower risk of lethal prostate cancer. We also observed complementary evidence suggesting that common variation across several genes involved in vitamin D metabolism and signaling is associated with the risk of lethal prostate cancer. Further research focusing on lethal prostate cancer and vitamin D is needed to confirm or refute these findings and advance our understanding of the underlying mechanisms behind these relationships.
Funding
National Institutes of Health (R01CA133891, R01CA141298-02, P01CA055075, and T32CA09001 to IMS and KLP) and the Prostate Cancer Foundation (to LAM).
Supplementary Material
Footnotes
We thank Hardeep Ranu and Patrice Soule of the DF/HCC High Throughput Polymorphism Detection Laboratory and Carolyn Guo for her assistance in preparing the data and performing quality control analysis.
In addition, we would like to thank the participants and staff of the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: Alabama, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Idaho, Illinois, Indiana, Iowa, Kentucky, Louisiana, Maine, Maryland, Massachusetts, Michigan, Nebraska, New Hampshire, New Jersey, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Rhode Island, South Carolina, Tennessee, Texas, Virginia, Washington, Wyoming.
The design of the study, collection, analysis and interpretation of the data, and the article preparation and submission were not influenced by the study sponsor.
References
- 1.Hsu JW, Yasmin-Karim S, King MR, et al. Suppression of prostate cancer cell rolling and adhesion to endothelium by 1alpha,25-dihydroxyvitamin D3. Am J Pathol. 2011;178(2):872–880. doi: 10.1016/j.ajpath.2010.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen TC, Schwartz GG, Burnstein KL, et al. The in vitro evaluation of 25-hydroxyvitamin D3 and 19-nor-1alpha,25-dihydroxyvitamin D2 as therapeutic agents for prostate cancer. Clin Cancer Res. 2000;6(3):901–908. [PubMed] [Google Scholar]
- 3.Kizildag S, Ates H. Treatment of K562 cells with 1,25-dihydroxyvitamin D(3) induces distinct alterations in the expression of apoptosis-related genes BCL2, BAX, BCL(XL), and p21. Ann Hematol. 2010;89(1):1–7. doi: 10.1007/s00277-009-0766-y. [DOI] [PubMed] [Google Scholar]
- 4.Krishnan AV, Peehl DM, Feldman D. Inhibition of prostate cancer growth by vitamin D: regulation of target gene expression. J Cell Biochem. 2003;88(2):363–371. doi: 10.1002/jcb.10334. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert R, Martin RM, Beynon R, et al. Associations of circulating and dietary vitamin D with prostate cancer risk: a systematic review and dose-response meta-analysis. Cancer Causes Control. 2011;22(3):319–340. doi: 10.1007/s10552-010-9706-3. [DOI] [PubMed] [Google Scholar]
- 6.Fang F, Kasperzyk JL, Shui I, et al. Prediagnostic plasma vitamin D metabolites and mortality among patients with prostate cancer. PLoS One. 2011;6(4):e18625. doi: 10.1371/journal.pone.0018625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt SK, Kwon EM, Peters U, et al. Vitamin D pathway gene variants and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1929–1933. doi: 10.1158/1055-9965.EPI-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn J, Albanes D, Berndt SI, et al. Vitamin D-related genes, serum vitamin D concentrations and prostate cancer risk. Carcinogenesis. 2009;30(5):769–776. doi: 10.1093/carcin/bgp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt SK, Kwon EM, Koopmeiners JS, et al. Vitamin D pathway gene variants and prostate cancer prognosis. Prostate. 2010;70(13):1448–1460. doi: 10.1002/pros.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovannucci E. The epidemiology of vitamin D and cancer incidence and mortality: a review (United States) Cancer Causes Control. 2005;16(2):83–95. doi: 10.1007/s10552-004-1661-4. [DOI] [PubMed] [Google Scholar]
- 11.Rimm EB, Stampfer MJ, Colditz GA, et al. Effectiveness of various mailing strategies among nonrespondents in a prospective cohort study. Am J Epidemiol. 1990;131(6):1068–1071. doi: 10.1093/oxfordjournals.aje.a115598. [DOI] [PubMed] [Google Scholar]
- 12.Verhoef P, Rimm EB, Hunter DJ, et al. A common mutation in the methylenetetrahydrofolate reductase gene and risk of coronary heart disease: results among U.S. men. J AmColl Cardiol. 1998;32(2):353–359. doi: 10.1016/s0735-1097(98)00244-7. [DOI] [PubMed] [Google Scholar]
- 13.Hollis BW, Kamerud JQ, Selvaag SR, et al. Determination of vitamin D status by radioimmunoassay with an 125I-labeled tracer. Clin Chem. 1993;39(3):529–533. [PubMed] [Google Scholar]
- 14.Platz EA, Leitzmann MF, Hollis BW, et al. Plasma 1,25-dihydroxy- and 25-hydroxyvitamin D and subsequent risk of prostate cancer. Cancer Causes Control. 2004;15(3):255–265. doi: 10.1023/B:CACO.0000024245.24880.8a. [DOI] [PubMed] [Google Scholar]
- 15.de Bakker PI, Yelensky R, Pe’er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Willer C, Sanna S, et al. Genotype imputation. Ann Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosner B, Cook N, Portman R, et al. Determination of blood pressure percentiles in normal-weight children: some methodological issues. Am J Epidemiol. 2008;167(6):653–666. doi: 10.1093/aje/kwm348. [DOI] [PubMed] [Google Scholar]
- 19.Gao X. Multiple testing corrections for imputed SNPs. Genet Epidemiol. 2011;35(3):154–158. doi: 10.1002/gepi.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu MC, Kraft P, Epstein MP, et al. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, Ghosh D, Lin X. Estimation and testing for the effect of a genetic pathway on a disease outcome using logistic kernel machine regression via logistic mixed models. BMC Bioinformatics. 2008;9:292. doi: 10.1186/1471-2105-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor KC, Small CM, Epstein MP, et al. Associations of progesterone receptor polymorphisms with age at menarche and menstrual cycle length. Horm Res Paediatr. 2010;74(6):421–427. doi: 10.1159/000316961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locke AE, Dooley KJ, Tinker SW, et al. Variation in folate pathway genes contributes to risk of congenital heart defects among individuals with Down syndrome. Genet Epidemiol. 2010;34(6):613–623. doi: 10.1002/gepi.20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu CY, Wu MC, Chen F, et al. A large-scale genetic association study of esophageal adenocarcinoma risk. Carcinogenesis. 2010;31(7):1259–1263. doi: 10.1093/carcin/bgq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn J, Yu K, Stolzenberg-Solomon R, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19(13):2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendrickson WK, Flavin R, Kasperzyk JL, et al. Vitamin D receptor protein expression in tumor tissue and prostate cancer progression. J Clin Oncol. 2011;29(17):2378–2385. doi: 10.1200/JCO.2010.30.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokar EJ, Webber MM. Cholecalciferol (vitamin D3) inhibits growth and invasion by up-regulating nuclear receptors and 25-hydroxylase (CYP27A1) in human prostate cancer cells. Clin Exp Metastasis. 2005;22(3):275–284. doi: 10.1007/s10585-005-8393-z. [DOI] [PubMed] [Google Scholar]
- 28.Chung I, Han G, Seshadri M, et al. Role of vitamin D receptor in the antiproliferative effects of calcitriol in tumor-derived endothelial cells and tumor angiogenesis in vivo. Cancer Res. 2009;69(3):967–975. doi: 10.1158/0008-5472.CAN-08-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giovannucci E, Liu Y, Platz EA, et al. Risk factors for prostate cancer incidence and progression in the health professionals follow-up study. Int J Cancer. 2007;121(7):1571–1578. doi: 10.1002/ijc.22788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn J, Peters U, Albanes D, et al. Serum vitamin D concentration and prostate cancer risk: a nested case-control study. J Natl Cancer Inst. 2008;100(11):796–804. doi: 10.1093/jnci/djn152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travis RC, Crowe FL, Allen NE, et al. Serum vitamin D and risk of prostate cancer in a case-control analysis nested within the European Prospective Investigation into Cancer and Nutrition (EPIC) Am J Epidemiol. 2009;169(10):1223–1232. doi: 10.1093/aje/kwp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Stampfer MJ, Hollis JB, et al. A prospective study of plasma vitamin D metabolites, vitamin D receptor polymorphisms, and prostate cancer. PLoS Med. 2007;4(3):e103. doi: 10.1371/journal.pmed.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andren O, Fall K, Franzen L, et al. How well does the Gleason score predict prostate cancer death? A 20-year followup of a population based cohort in Sweden. J Urol. 2006;175(4):1337–1340. doi: 10.1016/S0022-5347(05)00734-2. [DOI] [PubMed] [Google Scholar]
- 34.Stark JR, Perner S, Stampfer MJ, et al. Gleason score and lethal prostate cancer: does 3 + 4 = 4 + 3? J Clin Oncol. 2009;27(21):3459–3464. doi: 10.1200/JCO.2008.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mucci LA, Powolny A, Giovannucci E, et al. Prospective study of prostate tumor angiogenesis and cancer-specific mortality in the health professionals follow-up study. J Clin Oncol. 2009;27(33):5627–5633. doi: 10.1200/JCO.2008.20.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu JY, Feldman D, McNeal JE, et al. Reduced 1alpha-hydroxylase activity in human prostate cancer cells correlates with decreased susceptibility to 25-hydroxyvitamin D3-induced growth inhibition. Cancer Res. 2001;61(7):2852–2856. [PubMed] [Google Scholar]
- 37.Penney KL, Pyne S, Schumacher FR, et al. Genome-wide association study of prostate cancer mortality. Cancer Epidemiol Biomarkers Prev. 2010;19(11):2869–2876. doi: 10.1158/1055-9965.EPI-10-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun RF, Lauridsen AL, Suon L, et al. Vitamin D-binding protein directs monocyte responses to 25-hydroxy- and 1,25-dihydroxyvitamin D. J Clin Endocrinol Metab. 2010;95(7):3368–3376. doi: 10.1210/jc.2010-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmann JN, Yu K, Horst RL, et al. Long-term variation in serum 25-hydroxyvitamin D concentration among participants in the prostate, lung, colorectal, and ovarian cancer screening trial. Cancer Epidemiol Biomarkers Prev. 2010;19(4):927–931. doi: 10.1158/1055-9965.EPI-09-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.