Abstract
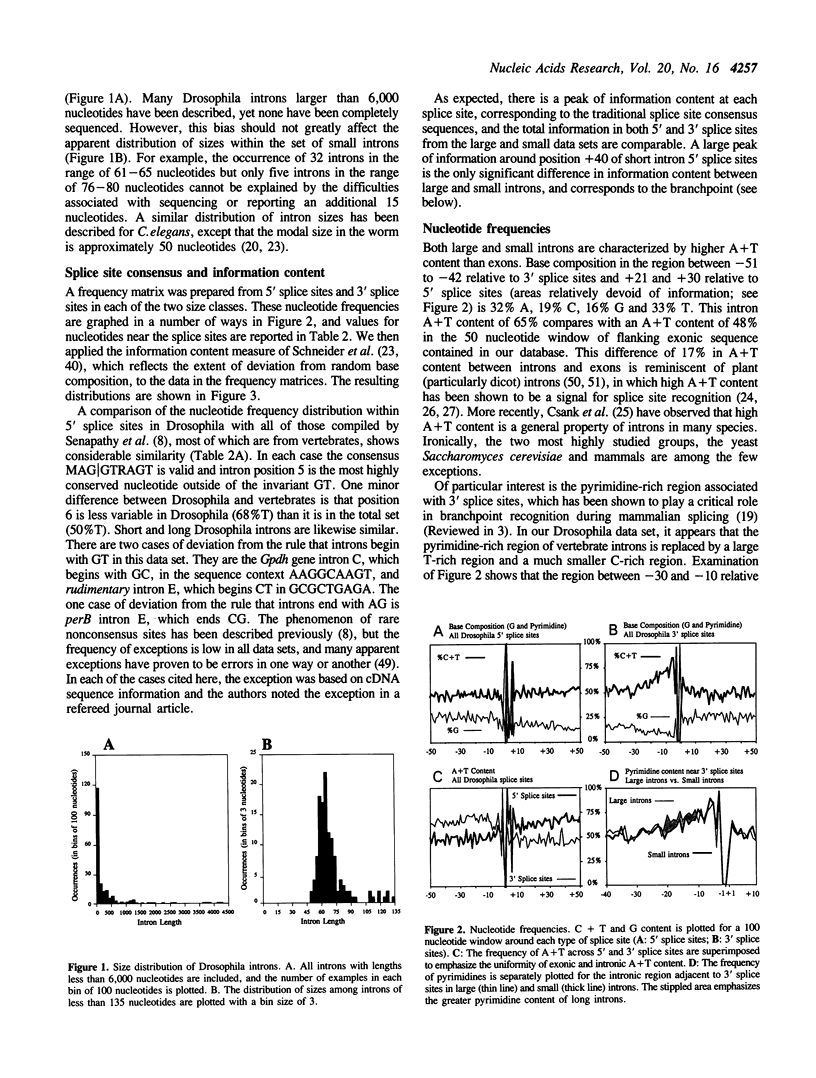
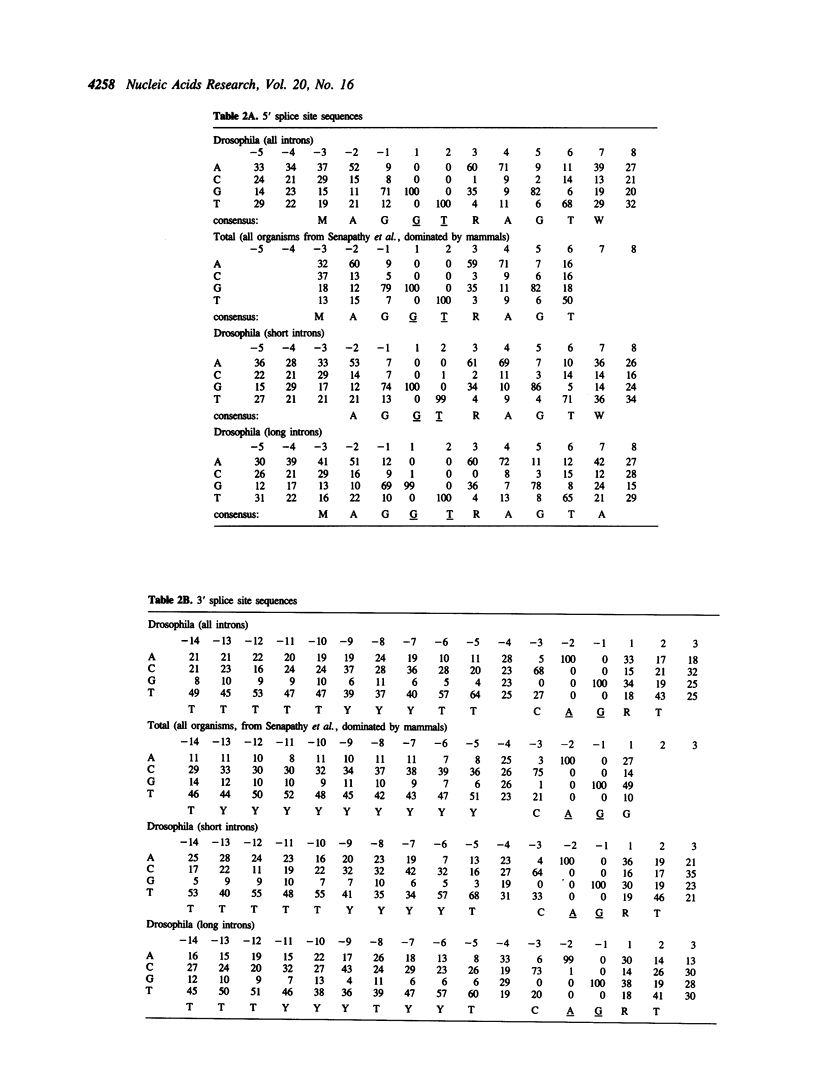
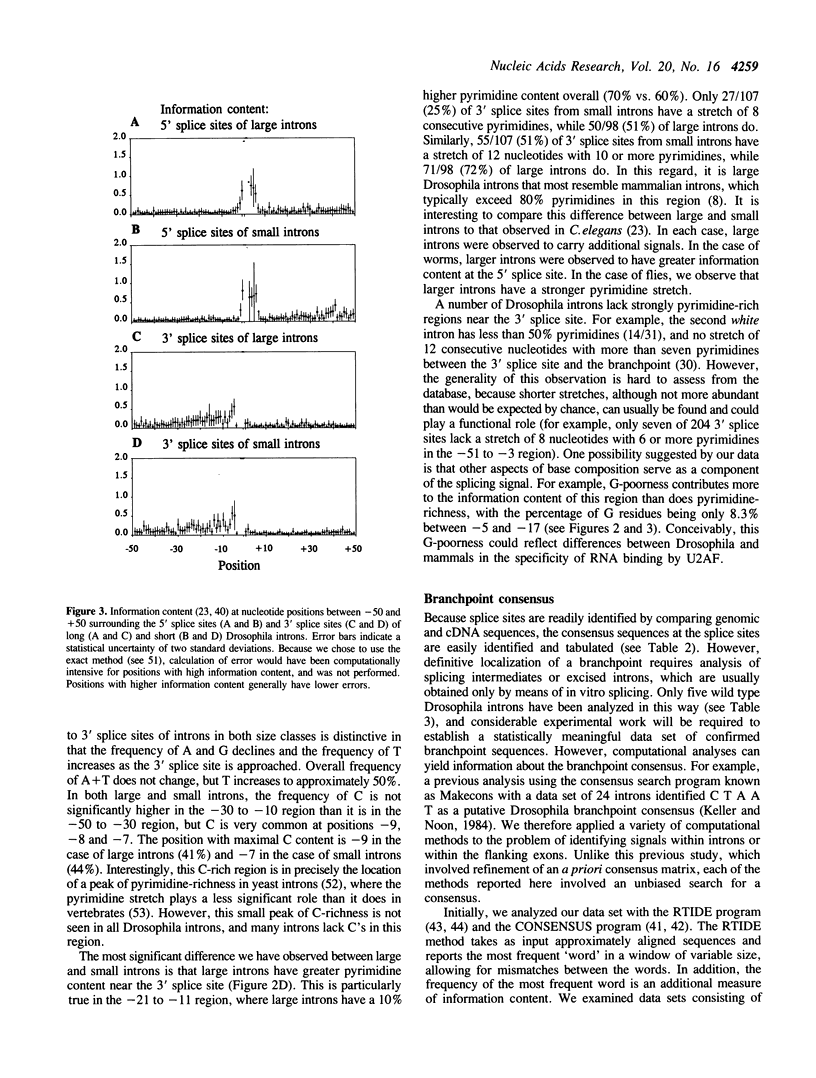
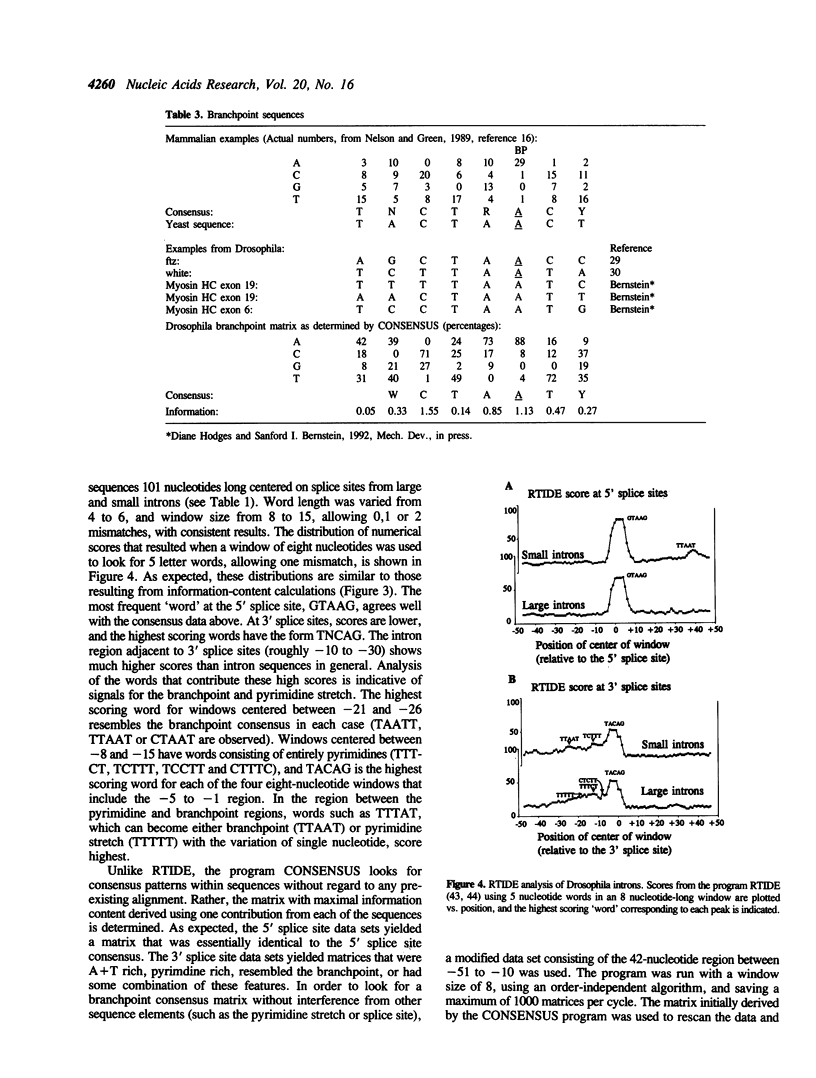
A database of 209 Drosophila introns was extracted from Genbank (release number 64.0) and examined by a number of methods in order to characterize features that might serve as signals for messenger RNA splicing. A tight distribution of sizes was observed: while the smallest introns in the database are 51 nucleotides, more than half are less than 80 nucleotides in length, and most of these have lengths in the range of 59-67 nucleotides. Drosophila splice sites found in large and small introns differ in only minor ways from each other and from those found in vertebrate introns. However, larger introns have greater pyrimidine-richness in the region between 11 and 21 nucleotides upstream of 3' splice sites. The Drosophila branchpoint consensus matrix resembles C T A A T (in which branch formation occurs at the underlined A), and differs from the corresponding mammalian signal in the absence of G at the position immediately preceding the branchpoint. The distribution of occurrences of this sequence suggests a minimum distance between 5' splice sites and branchpoints of about 38 nucleotides, and a minimum distance between 3' splice sites and branchpoints of 15 nucleotides. The methods we have used detect no information in exon sequences other than in the few nucleotides immediately adjacent to the splice sites. However, Drosophila resembles many other species in that there is a discontinuity in A + T content between exons and introns, which are A + T rich.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Bernstein S. I., Hansen C. J., Becker K. D., Wassenberg D. R., 2nd, Roche E. S., Donady J. J., Emerson C. P., Jr Alternative RNA splicing generates transcripts encoding a thorax-specific isoform of Drosophila melanogaster myosin heavy chain. Mol Cell Biol. 1986 Jul;6(7):2511–2519. doi: 10.1128/mcb.6.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Thomas J. Cis and trans mRNA splicing in C. elegans. Trends Genet. 1988 Nov;4(11):305–308. doi: 10.1016/0168-9525(88)90107-2. [DOI] [PubMed] [Google Scholar]
- Bruzik J. P., Steitz J. A. Spliced leader RNA sequences can substitute for the essential 5' end of U1 RNA during splicing in a mammalian in vitro system. Cell. 1990 Sep 7;62(5):889–899. doi: 10.1016/0092-8674(90)90264-f. [DOI] [PubMed] [Google Scholar]
- Csank C., Taylor F. M., Martindale D. W. Nuclear pre-mRNA introns: analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res. 1990 Sep 11;18(17):5133–5141. doi: 10.1093/nar/18.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveleth D. D., Gietz R. D., Spencer C. A., Nargang F. E., Hodgetts R. B., Marsh J. L. Sequence and structure of the dopa decarboxylase gene of Drosophila: evidence for novel RNA splicing variants. EMBO J. 1986 Oct;5(10):2663–2672. doi: 10.1002/j.1460-2075.1986.tb04549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenthal S., Parker V. P., Davidson N. Developmental variations in the splicing pattern of transcripts from the Drosophila gene encoding myosin alkali light chain result in different carboxyl-terminal amino acid sequences. Proc Natl Acad Sci U S A. 1985 Jan;82(2):449–453. doi: 10.1073/pnas.82.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields C. Information content of Caenorhabditis elegans splice site sequences varies with intron length. Nucleic Acids Res. 1990 Mar 25;18(6):1509–1512. doi: 10.1093/nar/18.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Colgan J. D., Manley J. L. Multiple cis-acting sequence elements are required for efficient splicing of simian virus 40 small-t antigen pre-mRNA. Mol Cell Biol. 1988 Sep;8(9):3582–3590. doi: 10.1128/mcb.8.9.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Eggert M., Waterman M. S. Rigorous pattern-recognition methods for DNA sequences. Analysis of promoter sequences from Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):117–128. doi: 10.1016/0022-2836(85)90262-1. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 1991 Sep;10(9):2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hanley B. A., Schuler M. A. Plant intron sequences: evidence for distinct groups of introns. Nucleic Acids Res. 1988 Jul 25;16(14B):7159–7176. doi: 10.1093/nar/16.14.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G. Z., Hartzell G. W., 3rd, Stormo G. D. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput Appl Biosci. 1990 Apr;6(2):81–92. doi: 10.1093/bioinformatics/6.2.81. [DOI] [PubMed] [Google Scholar]
- Himmelspach M., Gattoni R., Gerst C., Chebli K., Stévenin J. Differential block of U small nuclear ribonucleoprotein particle interactions during in vitro splicing of adenovirus E1A transcripts containing abnormally short introns. Mol Cell Biol. 1991 Mar;11(3):1258–1269. doi: 10.1128/mcb.11.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson I. J. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991 Jul 25;19(14):3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob M., Gallinaro H. The 5' splice site: phylogenetic evolution and variable geometry of association with U1RNA. Nucleic Acids Res. 1989 Mar 25;17(6):2159–2180. doi: 10.1093/nar/17.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay R. J., Russnak R. H., Jones D., Mathias C., Candido E. P. Expression of intron-containing C. elegans heat shock genes in mouse cells demonstrates divergence of 3' splice site recognition sequences between nematodes and vertebrates, and an inhibitory effect of heat shock on the mammalian splicing apparatus. Nucleic Acids Res. 1987 May 11;15(9):3723–3741. doi: 10.1093/nar/15.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo R., Lo P. C., Mount S. M. Structure and expression of the Drosophila melanogaster gene for the U1 small nuclear ribonucleoprotein particle 70K protein. Mol Cell Biol. 1990 Jun;10(6):2492–2502. doi: 10.1128/mcb.10.6.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Steitz J. A. Sequence of U1 RNA from Drosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucleic Acids Res. 1981 Dec 11;9(23):6351–6368. doi: 10.1093/nar/9.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989 Oct;3(10):1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Ogg S. C., Anderson P., Wickens M. P. Splicing of a C. elegans myosin pre-mRNA in a human nuclear extract. Nucleic Acids Res. 1990 Jan 11;18(1):143–149. doi: 10.1093/nar/18.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson T., Beggs J. D., Finnegan D. J., Lührmann R. Polypeptide components of Drosophila small nuclear ribonucleoprotein particles. Nucleic Acids Res. 1991 Nov 11;19(21):5877–5882. doi: 10.1093/nar/19.21.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. A U-rich tract enhances usage of an alternative 3' splice site in yeast. Cell. 1991 Jan 11;64(1):181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- Rio D. C. Accurate and efficient pre-mRNA splicing in Drosophila cell-free extracts. Proc Natl Acad Sci U S A. 1988 May;85(9):2904–2908. doi: 10.1073/pnas.85.9.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Séraphin B. Who's on first? The U1 snRNP-5' splice site interaction and splicing. Trends Biochem Sci. 1991 May;16(5):187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Stormo G. D. Computer methods for analyzing sequence recognition of nucleic acids. Annu Rev Biophys Biophys Chem. 1988;17:241–263. doi: 10.1146/annurev.bb.17.060188.001325. [DOI] [PubMed] [Google Scholar]
- Stormo G. D. Consensus patterns in DNA. Methods Enzymol. 1990;183:211–221. doi: 10.1016/0076-6879(90)83015-2. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Hartzell G. W., 3rd Identifying protein-binding sites from unaligned DNA fragments. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Exon mutations uncouple 5' splice site selection from U1 snRNA pairing. Cell. 1990 Nov 2;63(3):619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991 May;10(5):1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman M. S., Jones R. Consensus methods for DNA and protein sequence alignment. Methods Enzymol. 1990;183:221–237. doi: 10.1016/0076-6879(90)83016-3. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Herrero J. J., Filipowicz W. Nuclear pre-mRNA processing in plants: distinct modes of 3'-splice-site selection in plants and animals. Mol Cell Biol. 1988 May;8(5):2042–2051. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Zapp M. L., Berget S. M. Evidence for nuclear factors involved in recognition of 5' splice sites. Nucleic Acids Res. 1989 Apr 11;17(7):2655–2674. doi: 10.1093/nar/17.7.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]



