Abstract
Cardiac troponin I (cTnI) is the inhibitory subunit of cardiac troponin, a key myofilament regulatory protein complex located on the thin filaments of the contractile apparatus. cTnI is uniquely specific for the heart and is widely used in clinics as a serum biomarker for cardiac injury. Phosphorylation of cTnI plays a critical role in modulating cardiac function. cTnI is known to be regulated by protein kinase A and protein kinase C at five sites, Ser22/Ser23, Ser42/44, and Thr143, primarily based on results from in vitro phosphorylation assays by the specific kinase(s). However, a comprehensive characterization of phosphorylation of mouse cTnI occurring in vivo has been lacking. Herein, we have employed top-down mass spectrometry (MS) methodology with electron capture dissociation for precise mapping of in vivo phosphorylation sites of cTnI affinity purified from wild-type and transgenic mouse hearts. As demonstrated, top-down MS (analysis of intact proteins) is an extremely valuable technology for global characterization of labile phosphorylation occurring in vivo without a priori knowledge. Our top-down MS data unambiguously identified Ser22/23 as the only two sites basally phosphorylated in wild-type mouse cTnI with full sequence coverage, which was confirmed by the lack of phosphorylation in cTnI-Ala2 transgenic mice where Ser22/23 in cTnI have been rendered nonphosphorylatable by mutation to alanine.
Cardiac troponin (cTn1) along with tropomyosin (Tm) is located on the thin filaments of the contractile apparatus and represents a key myofilament regulatory protein complex. This complex plays a critical role in Ca2+-mediated regulation of skeletal and cardiac muscle contraction and relaxation (1, 2). The inhibitory subunit of cTn (cTnI) binds tightly to actin-Tm in the absence of Ca2+ and thereby inhibits the binding of myosin cross-bridges to actin (2–4). Altered structure and function of cTnI are directly linked to cardiac dysfunction in various human heart diseases. Specific proteolytic cleavages of cTnI have been correlated with myocardial stunning and ischemia/reperfusion injury (5). Mutations in the cTnI gene (TNNI3) are known to be associated with cardiomyopathies such as hypertrophic cardiomypathy (HCM) and dilated cardiomypathy (DCM) (6). cTnI is uniquely specific for the heart because of the 31 amino acid sequence on its N-terminus which is not present in TnI of skeletal muscles. Hence, cTnI has become widely used in clinics as a serum biomarker of cardiac injury given that it is released into the general circulation following necrotic death of heart muscle tissue (7).
Phosphorylation of cTnI has been extensively studied and shown to be particularly important in modulating cardiac function (8–10). cTnI is regulated by protein kinase A (PKA) (11), protein kinase C (PKC) (12–17), p21-activated kinase (PAK) (18), protein kinase D (PKD) (19), and cyclic GMP-dependent protein kinase (PKG) (8). Activated by β-adrenergic stimulation of the heart, PKA phosphorylates cTnI at residues Ser22/23 (Ser23/24 including the initiating methionine), leading to a reduction in myofilament Ca2+ sensitivity (20) and an increase in cross-bridge cycling rate (21, 22). Phosphorylation of TnI by PKC occurs at Ser22/23, Ser42/44, and Thr143 (Ser23/24, Ser43/45, and Thr144, respectively, including the initiating methionine), resulting in alterations of both calcium sensitivity and the maximum tension formed during contraction (8, 23). Ser42/44 are the common phosphorylation sites for PKC isozymes including PKC-α/δ/ε, whereas PKC-δ/β/ε also cross-phosphorylates Ser22/23 (14, 17), and Thr143 has been identified as the preferred site for PKC-βII (24). PKG and PKD also phosphorylate Ser22/23, with effects similar to those of PKA (8, 19, 25). PAK induced phosphorylation at a novel site, Ser149, located in the region forming a Ca2+-sensitive interaction with the N-terminal regulatory domain of TnC, which increases Ca2+ sensitivity of skinned fiber bundles (18). However, all of these phosphorylation sites were determined on the basis of results from in vitro phosphorylation assays using conventional strategies such as 32P-labeling, tryptic peptide mapping, and Western blotting with known phosphoserine or phosphothreonine antibodies (12, 14, 17, 24). A comprehensive and precise mapping of phosphorylation sites of mouse cTnI present in vivo is still lacking.
Top-down mass spectrometry (MS) methodology has become an extremely promising technology for the characterization of post-translational modification (PTM) including labile phosphorylation and mutations/splicing isoforms of proteins present in vivo without a priori knowledge (26–29). Conventional bottom-up MS strategies rely on analysis of small peptides generated by proteolytic digestion of the protein. This has intrinsic limitations for the characterization of protein PTMs because of partial sequence coverage and loss of connections between disparate portions of proteins containing modifications (30). In contrast, the top-down MS strategy analyzes intact proteins that reveals all possible modifications present and provides far more reliable detection and characterization of PTMs with full sequence coverage. In addition, it greatly simplifies sample preparation and reduces sample complexity as no proteolytic digestion is required. Top-down MS is considerably enhanced with a unique tandem MS (MS/MS) technique, electron capture dissociation (ECD) (27, 31). ECD cleaves NH–CHR bonds to produce mainly c and z• ions, which is complementary to traditional MS/MS techniques such as collisionally activated dissociation (CAD) (32), infrared multiphoton dissociation (IRMPD) (33), and postsource decay (PSD) which cleave CO–NH bonds to produce b and y fragment ions. ECD preserves labile modifications such as phosphorylation due to its nonergodic nature, whereas CAD/IRMPD/PSD tends to knock off such labile modifications during the MS/MS process. Therefore, top-down mass spectrometry with ECD is extremely valuable in mapping labile PTMs, most notably phosphorylation (34, 35).
Herein, we have employed top-down ECD mass spectrometry for comprehensive and precise mapping of all in vivo phosphorylation sites of cTnI affinity purified from wild-type and transgenic mouse hearts. We discovered that Ser22/23 are the only two sites basally phosphorylated in wild-type mouse cTnI, albeit the possibility of other sites with extremely low phosphorylation occupancy (< 1%) cannot be excluded.
EXPERIMENTAL PROCEDURES
Materials
All reagents were obtained from Sigma Chemical Co. (St Louis, MO, USA) unless noted otherwise. Complete protease inhibitor cocktail was from Roche Diagnostics Corporation (Indianapolis, IN, USA).
Affinity Purification of cTn
All procedures were approved by the University of Wisconsin Animal Care and Use Committee. Whole cTn complexes were affinity purified by the method developed by Marston and co-workers (36) with modifications. Briefly, hearts were surgically excised from adult FVB mice anesthetized with isoflurane and rapidly washed with ice-cold Ringer’s solution (125 mM NaCl, 5 mM KCl, 25 mM HEPES-NaOH at pH 7.4, 2 mM NaH2PO4, 1.2 mM MgSO4, 5 mM Pyruvate, and 11 mM d-glucose) to remove blood and snap-frozen in liquid nitrogen, and then stored at −80 °C. Mouse hearts (6–8 whole mouse hearts per preparation) were ground up in a ceramic mortar and homogenized in wash buffer (5 mM NaH2PO4, 5 mM Na2HPO4, 5 mM MgCl2, 0.5 mM EGTA, 1 mM ATP, 1 mM PMSF, 5 mM DTT, 2 µg/mL leupeptin, 1% Triton X-100, and 0.75 mg/mL protease inhibitor cocktail) using either a Polytron homogenizer for 1 min or a Dounce (glass) homogenizer for 6–7 min at 4 °C. The homogenate was then centrifuged at 16,100 g for 3 min, and the pellet was resuspended in 1.5 mL and incubated in extraction solution (0.7 M LiCl, 25 mM Tris pH 7.5, 5 mM EGTA, 0.1 mM CaCl2, 5 mM DTT, 1 mM PMSF, 2 µg/mL leupeptin, and 0.75 mg/mL protease inhibitor cocktail) for 30 min at 4 °C. The sample was centrifuged at 16,100 g for 3 min, the supernatant was collected, and the extraction procedure was repeated twice. After combining supernatants, cell debris was removed by centrifugation at 55,000 rpm for 45 min (80Ti rotor, Beckman L-55 ultracentrifuge). The supernatant was incubated with 0.3 mL of CNBr-activated Sepharose CL-4B conjugated with a monoclonal cTnI antibody (antitroponin I monoclonal antibody MF4 or 14G5, Hytest, Finland) for 20 min at 4 °C and then loaded into an empty disposable column. After washing with 2 mL of extraction solution, the column was eluted with 100 mM glycine at pH 2 while 0.4 mL fractions were collected and neutralized immediately with 40 µL of 1 M MOPS (pH 9). Fractions were analyzed for protein content by SDS–PAGE on 15% gels stained with Coomassie Blue.
Top-Down Mass Spectrometry
Immunoaffinity purified mouse cTn complexes were separated and desalted using an offline reverse phase protein microtrap (Michrom Bioresources, Inc., CA), with a two step reverse phase gradient elution method, first with 1% acetic acid in 50:50 methanol/water and then 1% acetic acid in 75:25 methanol/water. Samples were introduced into the mass spectrometer using an automated chip-based nanoESI source (Triversa NanoMate, Advion BioSciences, Ithaca, NY) with a spray voltage of 1.2–1.6 kV versus the inlet of the mass spectrometer, resulting in a flow of 50–200 nL/min. Protein molecular ions were analyzed using a linear trap/FTICR (LTQ FT Ultra) hybrid mass spectrometer (Thermo Scientific Inc., Bremen, Germany). Ion transmission into the linear trap and subsequently into the FTICR cell was automatically optimized for maximum ion signal. The number of accumulated ions for the full scan linear trap (LT), FTICR cell (FT), MSn FTICR cell, and ECD were 3 × 104, 8 × 106, 8 × 106, and 8 × 106 respectively. The resolving power of the FTICR mass analyzer was typically set at 200,000 resulting in an acquisition rate of 1 scan/s. Charge states of protein molecular ions were first isolated and then dissociated by ECD using 2–3% electron energy and a 45–125 ms duration with no delay. Typically, 1000 to 3000 transients were averaged to ensure high quality ECD spectra. All FTICR spectra were processed with Xtract Software (FT programs 2.0.1.0.6.1.4, Xcallibur 2.0.5, Thermo Scientific Inc., Bremen, Germany) using a S/N threshold of 1.5 and fit factor of 40%, and then validated manually. The resulting mass lists were further assigned using in-house Ion Assignment software (version 1.0) based on the protein sequence of mouse cTnI obtained from Swiss-Prot protein knowledgebase (UnitProtKB/Swiss-Prot P48787, TNNI3_mouse, http://ca.expasy.org/uniprot/P48787). Allowance was made for possible post-translational modifications such as the removal of initial methionine, acetylation of the N-terminus, and variable phosphorylation sites, using a 0.3 and 0.1 Da tolerance for precursor and fragment ions, respectively. All reported Mr values are most abundant masses. To quantitatively determine modified forms of intact cTnI, the MS signal intensity values were used to calculate the relative ratios for all protein ions observed, which is known as the protein ion relative ratio (PIRR) developed by Kelleher and co-workers (37, 38). For such quantitative analysis, the top five most abundant isotopomer peak heights were integrated to calculate the relative abundance of the intact protein or fragmentation ions.
SDS–PAGE with Pro-Q Diamond Staining
Myofilament proteins were extracted from both wild-type and cTnI-Ala2 transgenic mouse hearts as described previously (15, 16). The phosphorylation levels of myofilament proteins were analyzed by SDS–PAGE on 12% polyacrylamide gels and stained with Pro-Q diamond (Molecular Probes, Invitrogen, Eugene, OR) according to the manufacturer’s instructions.
RESULTS
High Resolution Mass Spectrometry Analysis of cTnI from Wild-Type Mouse Hearts
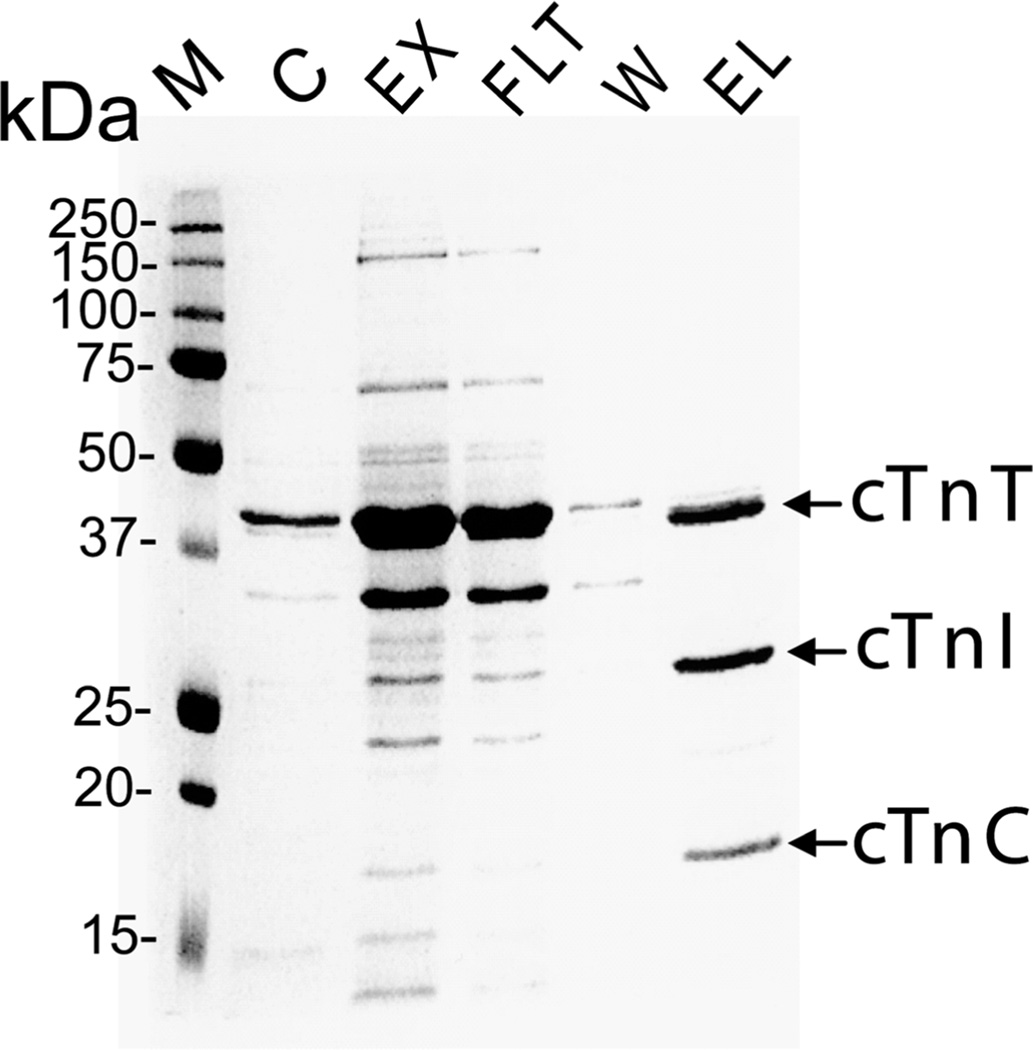
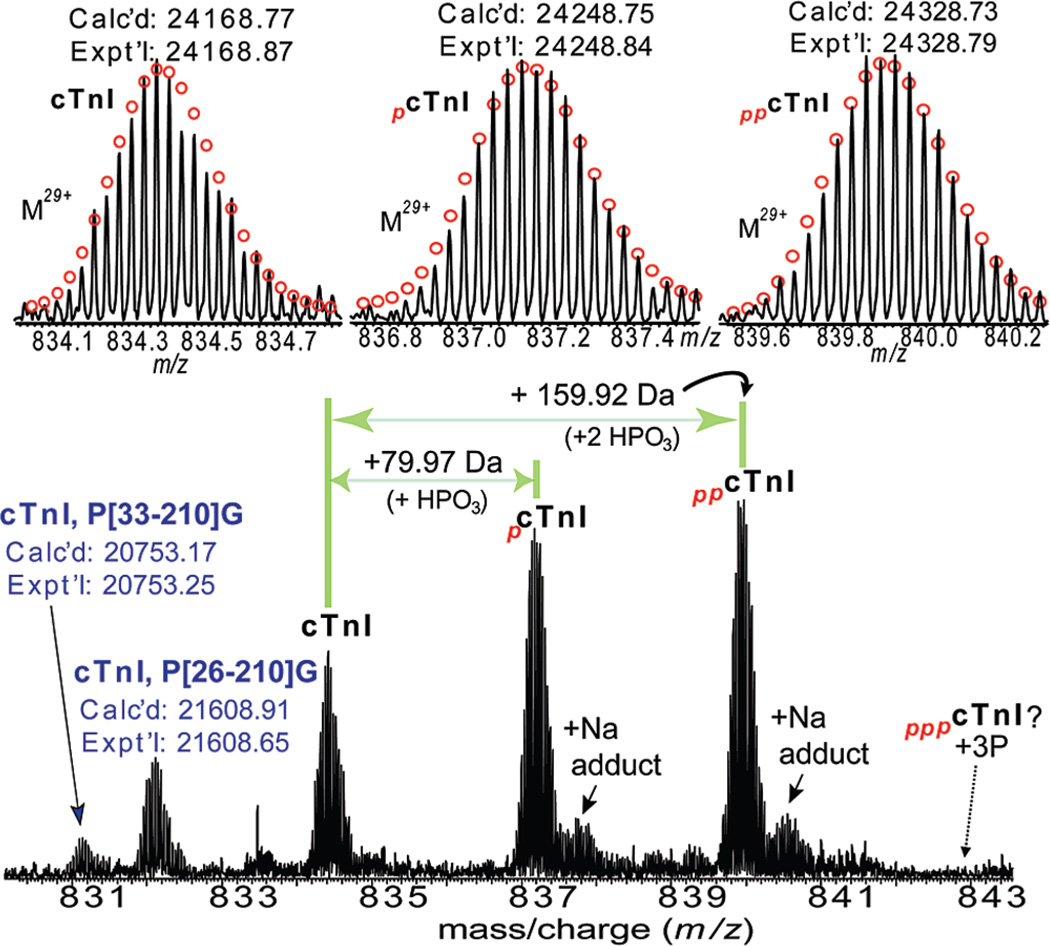
The cTn complex was successfully affinity purified from wild-type mouse heart tissue as shown in Figure 1. SDS–PAGE analysis with Coomassie blue staining of the elution from the affinity chromatography revealed three major bands at approximate molecular weights of 18, 26, and 37 kDa corresponding to the three subunits of the cTn complex (cTnC, cTnI, and cTnT), suggesting that this elution contained a high purity cTn. The cTn subunits were further separated and desalted via an offline reverse phase HPLC with a two step reverse phase gradient elution method before loading onto an LTQ/FT mass spectrometer. The cTnI subunit was detected in the elution of 75:25 methanol/water with 1% acetic acid. High resolution ESI/FTMS analysis revealed that the cTnI subunit is present as un-, mono-, and bis-phosphorylated forms (Figure 2). The high accuracy molecular weight of unphosphorylated cTnI matched the mouse cTn with the removal of N-terminal methionine and addition of acetylation (Expt’l, 24168.87; Calc’d, 24168.77, 4.1 ppm) based on DNA-predicted protein sequence for mouse cTnI (UnitProtKB/Swiss-Prot P48787, TNNI3_mouse, http://ca.expasy.org/uniprot/P48787). The other two highly abundant peaks of molecular weight 24248.84 and 24328.79 were assigned as mono- and bis-phosphorylated cTnI containing either one or two covalently bound phosphoryl moieties (HPO3, 79.98 Da) by the highly accurate molecular weight measurements (Calc’d, 24248.75, and 24328.73, 3.7, and 2.5 ppm, respectively). No tris-phosphorylated cTnI was detected (estimated to be <0.5% of the total cTnI population based on the signal intensity at the expected position of tris-phosphorylated cTnI over the summed intensity of all cTnI protein forms). The intensity ratio for cTnI in its unphosphorylated (cTnI), monophosphorylated (pcTnI), and bis-phosphorylated forms (ppcTnI) was approximately 24%, 36%, and 40%, respectively. Two minor peaks of N-terminal proteolytically processed products were also observed at molecular weights of 21608.65 and 20753.25, which matched with Y[26–210]G and P[33–210]G, respectively. These two proteolytic products are likely relevant in vivo rather than artifactual in nature since our experimental procedures including tissue collection and sample purification are well controlled to minimize the influence of in vitro proteolysis.
Figure 1.
Immunoaffinity purification of cTn complexes from mouse hearts. SDS–PAGE analysis of affinity purified cardiac troponin complex stained with Coomassie Blue. M, molecular weight markers, C, chicken cTn (obtained from Sigma-Aldrich), EX, extracted cardiac myofilament proteins; FLT, column flow through; W, wash; EL, elution of troponin complex.
Figure 2.
High resolution MS analysis of intact cTnI purified from wild-type mouse hearts. Bottom panel: ESI/FTMS spectrum of cTnI showing it is mono- and bis-phosphorylated. N-terminally proteolytic degraded products of cTnI were also observed. Top panel: isotopically resolved molecular ions of un-, mono-, and bis-phosphorylated cTnI (M29+) with highly accurate molecular weights measured. pcTnI and ppcTnI represent mono- (+79.97 Da, HPO3), and bis-phosphorylated (+159.92 Da, 2 HPO3) cTnI. +Na indicates cTnI ion with sodium adduct peaks. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. Dashed arrow indicates the expected position of tris-phosphorylated cTnI (pppcTnI), which is not observed here. Calc’d, calculated most abundant molecular weight; Expt’l, experimental most abundant molecular weight.
Localization of Basal Phosphorylation Sites in the Wild-Type Mouse cTnI
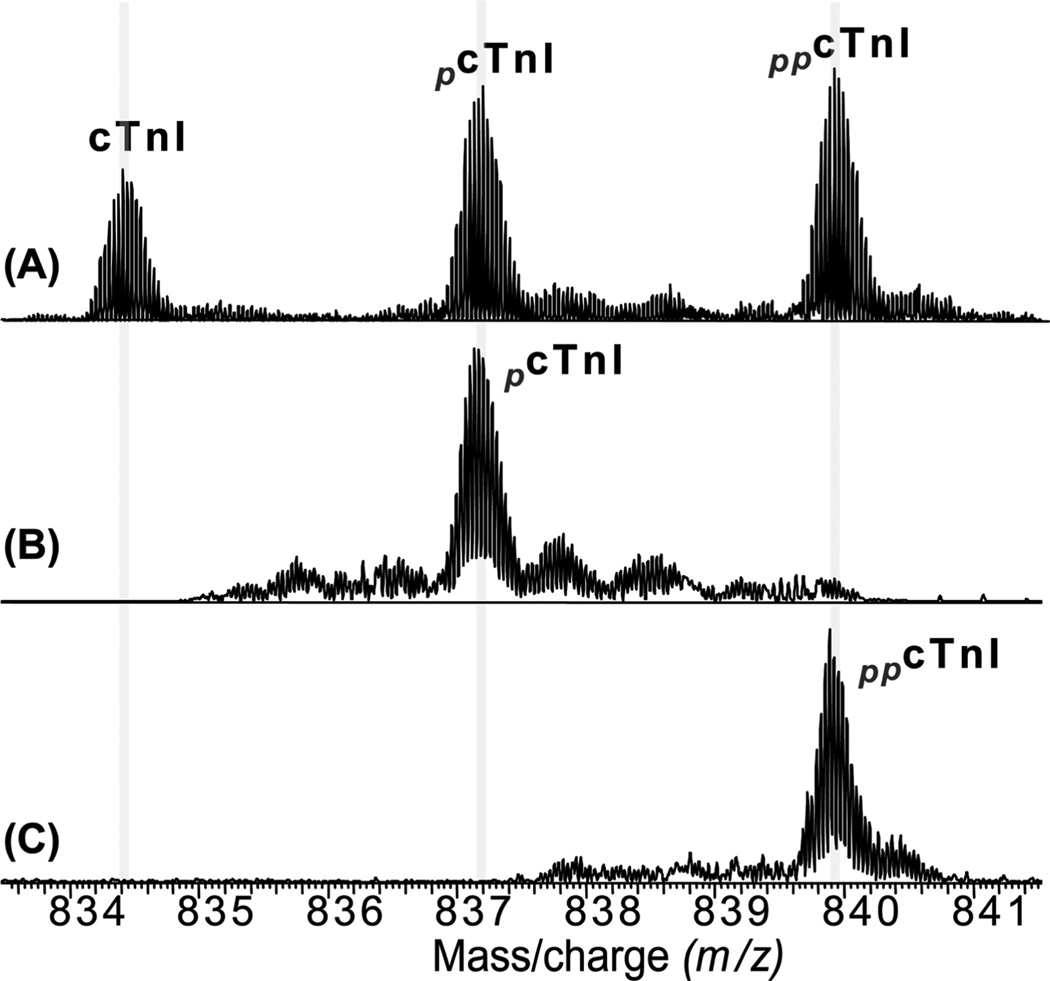
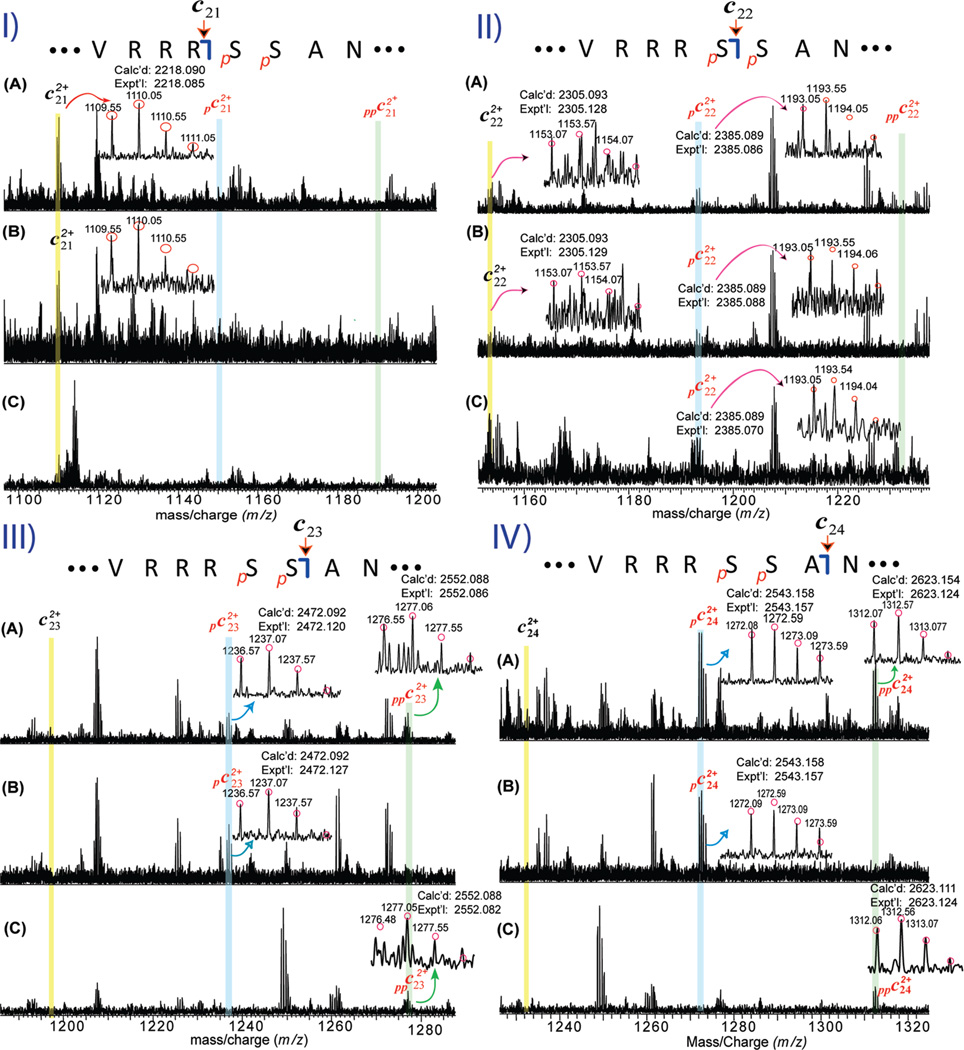
To map the basal phosphorylation sites, individual charge states of a mixture of un-, mono-, and bis-phosphorylated cTnI (Figure 3A) or monophosphorylated cTnI (pcTnI) alone (Figure 3B), or bis-phosphorylated cTnI (ppcTnI) alone (Figure 3C) were isolated in the LTQ/FT mass spectrometer, which could be considered as gas phase separation or purification of cTnI ions. These isolated (or purified) ions were then fragmented by ECD. The key fragmentation ions are shown in Figure 4, and the complete fragmentation maps are shown in Figure 5. No phosphorylated c21 ions (pc21) were detected (Figure 4-I) suggesting that the first 21 amino acids from the N-terminus (Ala[1–21]Arg) do not contain a phosphorylation site. In contrast, phosphorylated c22 ions (pc22) were detected, which unambiguously localized a phosphorylation site to Ser22 (Figure 4-II), similar to that in previous reports for human and rat cTnI (35, 39). Lower abundant unphosphorylated c22 ions were also present from ECD of the monophosphorylated cTnI ions alone with the ratio for unphosphorylated c22 and phosphorylated c22 of approximately 2/3 (Figure 4-II-B). This differs in the case of c23 ions that were detected only in the monophosphorylated state (pc23) and never in the unphosphorylated state, clearly revealing Ser23 as the other phosphorylation site in pcTnI. Considering the potential ion suppression of phosphorylated ions in the positive ion mode ESI/MS from the negatively charged phosphoryl group, this suggests that more than 60% of pcTnI are phosphorylated at Ser22 and that less than 40% are phosphorylated at Ser23.
Figure 3.
Gas-phase separation (purification) of cTnI phosphorylated ions. Isolation of a single charge state (M29+) of a mixture of un-, mono-, and bis-phosphorylated cTnI (A), monophosphorylated cTnI alone (B), and bis-phosphorylated cTnI alone (C). pcTnI and ppcTnI represent mono-, and bis-phosphorylated cTnI, respectively.
Figure 4.
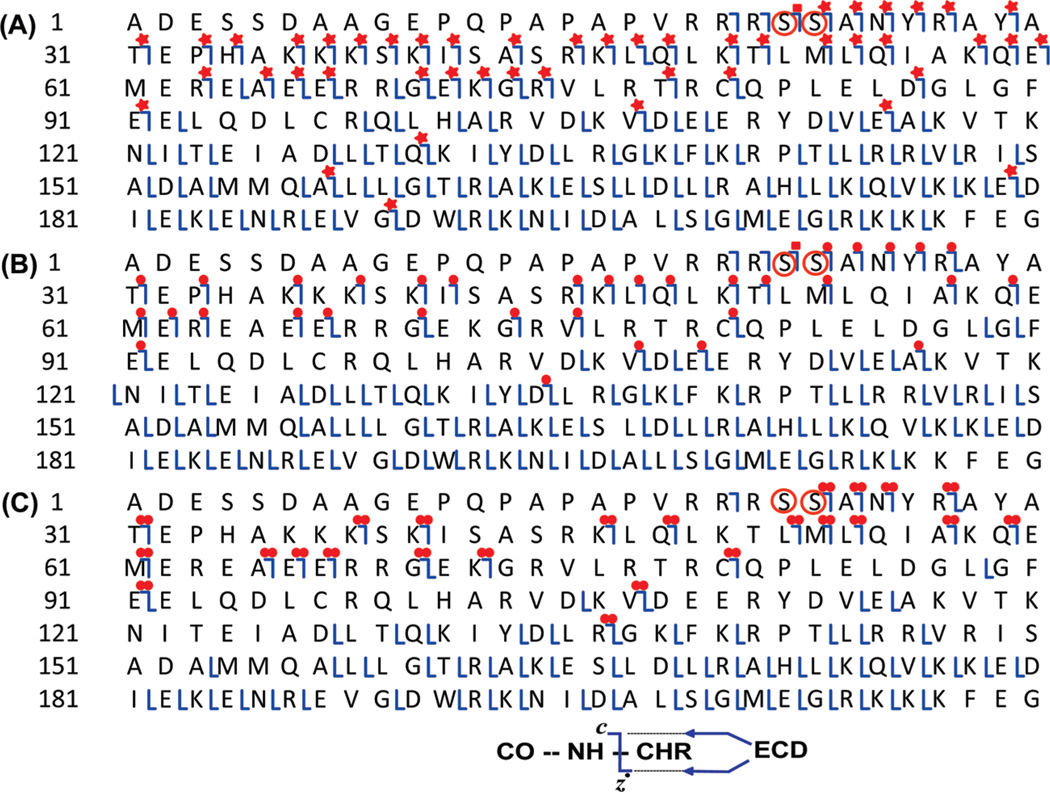
Localization of basal phosphorylation sites to Ser22/23 in wild-type mouse cTnI by ECD mass spectrometry. (I–IV) Representative spectra of doubly charged (2+) c21–c24 fragment ions, respectively, from ECD of a single charge state (M29+) of a mixture of un-, mono-, and bis-phosphorylated cTnI (A), monophosphorylated cTnI alone (B), and bis-phosphorylated cTnI alone (C). pc and ppc represent mono- and bis-phosphorylated c fragmentation ions. The abbreviated amino acid sequences for ions c21–c24 were accordingly denoted on top of each panel. The positively identified isotopomer peak profiles of un-, mono-, and bis-phosphorylated c21–c24 (2+) ions were correspondingly expanded in each spectrum. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned molecular weight. Calc’d, calculated most abundant molecular weight; Expt’l, experimental most abundant molecular weight.
Figure 5.
MS/MS product map from the ECD spectra for assignments to mouse cTnI. Fragment assignments were made to the DNA-predicted sequence of mouse cTnI (UnitProtKB/Swiss-Prot P48787, TNNI3_mouse) with the removal of N-terminal methionine and acetylation at the new terminus. (A) ECD of a single charge state (M29+) of a mixture of un-, mono-, and bis-phosphorylated cTnI; (B) ECD of one charge state (M29+) of monophosphorylated cTnI alone; (C) ECD of a single charge state (M29+) of bis-phosphorylated cTnI alone. Square, both un- and monophosphorylated fragment ions were observed. Star, un-, mono-, and bis-phosphorylated ions were observed. Single dot, only monophosphorylated fragment ions observed. Double dots, only bis-phosphorylated fragment ions were observed. Ser22 and Ser23 are highlighted in circles.
To determine whether Ser22/23 are the only two sites for phosphorylation, the bis-phosphorylated form of cTnI (ppcTnI) alone was isolated and fragmented by ECD (Figures 3C and 4C). Only bis-phosphorylated forms of c23 and c24 (ppc23 and ppc24) were detected, but neither unphosphorylated c23 and c24 nor monophosphorylated c23 and c24 (pc23 and pc24) were observed (estimated to be <2–3% of the bis-phosphorylated c23 and c24 ions, respectively) (Figure 4-III-C and IV-C). These observations suggested that Ser22 and Ser23 are the only two sites basally phosphorylated in wild-type mouse cTnI. Conversely, if other sites were phosphorylated (in the case of positional isomers), one would expect to observe un- and monophosphorylated c23 and c24 ions represented in the ECD spectrum of bis-phosphorylated cTnI, but such fragments were not observed.
In summary, one ECD spectrum from the mixture of un-, mono-, and bis-phosphorylated cTnI (Supporting Information Figure 1) generated 50 c and 92 z• fragment ions representing 130 cleavages of the total 210 NH–CH available backbone bonds (Figure 5A). Consistently, all the c ions prior to c21 (i.e., c20 and c21) were present as unphosphorylated forms, c22 was present as both un- and monophosphorylated forms, and all c ions after c22 (i.e., c23–c189) were present as un-, mono-, and bis-phorphorylated forms. One ECD spectrum from isolated monophosphorylated cTnI (pcTnI) alone (Figure 4B) generated 37 c and 84 z• fragment ions representing 112 cleavages of the total 210 NH–CH available backbone bonds (Figure 5B). All of the c ions prior to c21 (i.e., c20 and c21) were present as unphosphorylated forms, c22 was present as both un- and monophosphorylated forms, and all c ions after c22 (i.e., c22–c134) were present solely as monophosphorylated forms. One ECD spectrum from isolated bis-phosphorylated cTnI (ppcTnI) alone (Figure 4C) generated 25 c and 61 z• fragment ions representing 81 cleavages of the total 210 NH–CH available backbone bonds (Figure 5C). All of the c ions prior to c21 (i.e., c20) were present as the unphosphorylated form, whereas all c ions after c22 (i.e., c23–c136) were present solely as bis-phosphorylated forms. Complementary c and z• fragmentation pairs (e.g., c107 and z•103, and c80 and z•130) were observed for all three ECD spectra, which ensures 100%sequence coverage for mouse cTnI. Three replicates have been performed for each experiment, and a total of 45 healthy wild-type mouse hearts have been used for all of the MS experiments described here. All of the data are in good agreement with the assignment that Ser22/23 are the only basally phosphorylated sites.
High Resolution Mass Spectrometry Analysis of cTnI from cTnI-Ala2 Transgenic Mouse Hearts
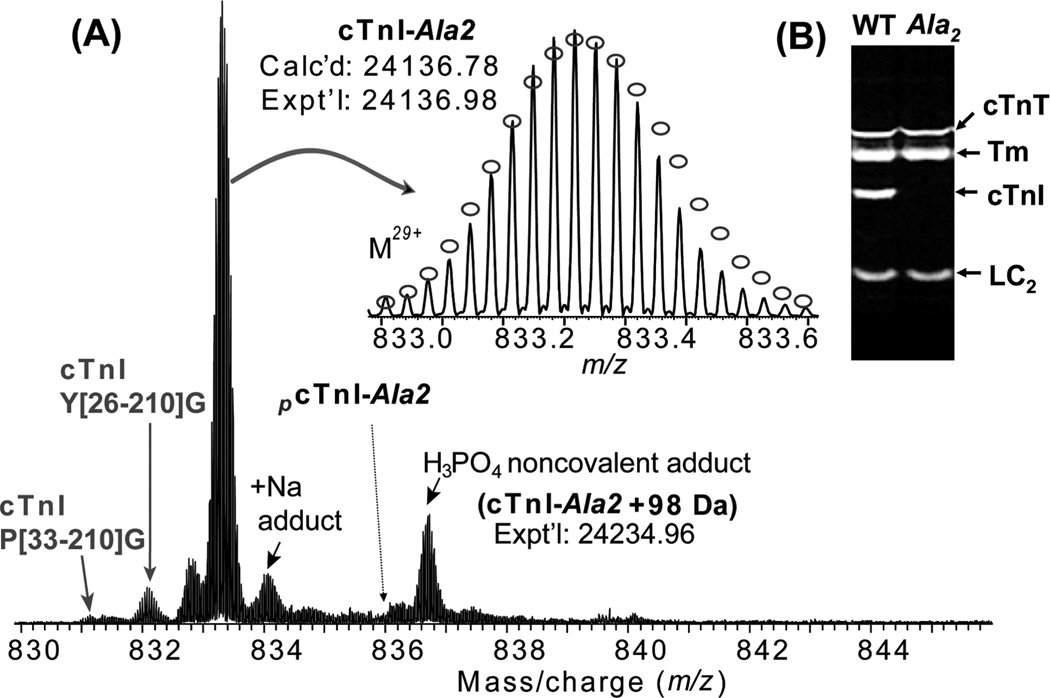
To verify whether Ser22/23 are the only basal phosphorylation sites, we applied high resolution ESI/FTMS to analyze cTnI affinity purified from cTnI-Ala2 transgenic mice where Ser22/23 (Ser23/24 including the N-terminal methionine) of cTnI have been rendered nonphosphorylatable by mutation to alanine (15), later referred to as cTnI-Ala2. The cTnI-Ala2 transgenic mice show no hypertrophy, exercise deficits, or reduced lifespan, indicating that cTnI with Ser22/23 mutated to alanine still supports cardiac function well (10, 15, 16). The ESI/FTMS spectrum of cTnI-Ala2 shows that it is present solely as an unphosphorylated form (Figure 6A). The high accuracy molecular weight of unphosphorylated cTnI (Expt’l, 24136.98; Calc’d, 24136.78, 8.3 ppm) matched with the removal of N-terminal methionine and addition of acetylation at the N-terminus. Minor proteolytic degradation products of cTnI-Ala2 were detected similar to cTnI from wild-type mouse. In addition, noncovalent adducts of sodium (+22 Da) and phosphoric acids (+98 Da) were observed. However, no discernible phosphorylated cTnI (+80 Da) ions were found (estimated to be <1% of the total cTnI population). ECD of unphosphorylated cTnI-Ala2 generated 26 c and 28 z• fragment ions including two complementary pairs (c82 and z•128, and c102 and z•108), which unequivocally confirmed the sequence of cTnI-Ala2 (Supporting Information Figure 2).
Figure 6.
(A) High resolution MS analysis of cTnI affinity purified from cTnI-Ala2 transgenic mouse hearts. No phosphorylated ions were detected, confirming Ser22 and Ser23 are the only basally phosphorylated sites in wild-type mouse cTnI. N-terminally proteolytic degraded products of cTnI were also observed. Inset, isotopically resolved molecular ions of unphosphorylated cTnI (M29+) with highly accurate molecular weight measured. Dashed arrow indicates the expected position of monophosphorylated cTnI from cTnT-Ala2 transgenic mice (pcTnI-Ala2), which is not observed here. +Na indicates cTnI ion with sodium adduct peaks. Circles represent the theoretical abundance isotopic distribution of the isotopomer peaks corresponding to the assigned mass. Calc’d, calculated most abundant molecular weight; Expt’l, experimental most abundant molecular weight. (B) SDS–PAGE analysis of myofilament proteins extracted from both wild-type and cTnI-Ala2 transgenic mouse hearts stained with Pro-Q diamond. Only lower molecular weight regions are shown. Lane 1, WT, wild-type mouse; lane 2, Ala2, cTnI-Ala2 transgenicmouse. cTnT, cardiac troponin T; Tm, tropomyosin; LC2, myosin regulatory light chain 2.
We have also directly employed FTMS analysis of the entire cTn mixture without chromatography separation, which revealed 12 major and 63 minor protein forms including cTnI-Ala2 and cTnT (Supporting Information Figure 3A). In this spectrum, cTnI is present as the unphosphorylated form again with no phosphorylated cTnI-Ala2 observed (estimated to be <2% of the total cTnI protein forms) (Supporting Information Figure 3B). cTnT is present as un- and monophosphorylated forms (Supporting Information Figure 3C), similar to that in a previous report for rat cTnT (39).
Furthermore, we have utilized an orthogonal phosphorylation detection method, Pro-Q diamond labeling, to examine the phosphorylation level in cTnI and other prominent myofilament proteins. Myofilament proteins extracted from wild-type and cTnI-Ala2 transgenic mouse hearts were analyzed by SDS–PAGE using Pro-Q Diamond staining (40) (Figure 6B). cTnI from the wild-type mouse hearts is highly phosphorylated, whereas no phosphorylation was detected (estimated to be <1% of wild-type cTnI phosphoprotein) for cTnI-Ala2 purified from transgenic mouse hearts. In contrast, the phosphorylation levels on cTnT, Tm, and myosin regulatory light chain 2 (LC2) remained the same in wild-type and transgenic hearts. Hence, the lack of phosphorylation in cTnI-Ala2 purified from cTnI-Ala2 transgenic mouse hearts confirms that Ser22 and Ser23 are the only basally phosphorylated sites in cTnI from wild-type mice.
DISCUSSION
Ser22/23 as the Only Basally Phosphorylated Sites in Mouse cTnI
Our high resolution high accuracy MS analysis of wild-type mouse cTnI revealed that it is mono- and bis-phosphorylated, suggesting that at least two basal phosphorylation sites were present in mouse cTnI. The lack of tris-phosphorylated cTnI (Figure 2) significantly decreases the probability of three or more phosphorylation sites as both mono- and bis-phosphorylated cTnI were highly abundant. Furthermore, we employed top-down MS for a comprehensive and precise localization of the phosphorylation sites, and our data unambiguously localized two phosphorylation sites to Ser22/23. To determine whether Ser22/23 are the only sites phosphorylated in mouse cTnI, we isolated the bis-phosphorylated cTnI alone in the mass spectrometer (which could be considered as gas-phase purification) and subjected it to fragmentation by ECD. Our data clearly indicates that there are no other discernible basally phosphorylated sites in cTnI. In addition, no phosphorylation was observed for cTnI-Ala2 purified from transgenic mouse heart where Ser22/23 were replaced with alanine (15) on the basis of both MS data and Pro-Q diamond staining, strongly supporting the conclusion that Ser22/23 are the only sites basally phosphorylated in wild-type mouse cTnI.
One concern is whether other sites in cTnI apart from Ser22/23 with extremely low phosphorylation occupancy might be lost in the noise if they are less than a few percent of the total population. As every analytical method has its own detection limit, the observation that only Ser22/23 are detected to be phosphorylated does not completely exclude the possibility of other sites basally phosphorylated at low occupancy. Combining the MS and Pro-Q diamond data, we are confident that we identified all phosphospecies representing > 1–3% of the total cTnI population. In other words, we cannot exclude the possibility of other sites of very low (< 1%) phosphorylation occupancy.
Another concern is whether affinity purification may select for complexes with overall lower phosphorylation. We have used an orthogonal method, Pro-Q diamond staining, for analysis of the entire myofilament extract before affinity purification. The Pro-Q diamond data of cTnI before affinity purification is consistent with the MS data of cTnI after the affinity purification. In addition, MS data acquired using two different affinity resins that target distinct epitope regions (N-terminal [residues 1–23] versus C-terminal [residues 190–197] of cTnI) showed highly comparable phosphorylation levels for cTnI (data not shown). Hence, our data suggest that the overall impact of affinity chromatography on phosphoprotein selection was minimal.
Our findings for mouse cTn differ to some extent from what we have previously reported for human cTnI where three basal phosphorylation sites were localized including Ser22/23 and a new site at Ser76 (35). Additionally, our analysis of monophosphorylated human cTnI showed an almost exclusive preference for basal phosphorylation of Ser22 over Ser23, which was taken as evidence of ordered or sequential phosphorylation of these two sites. ECD data of monophosphorylated mouse cTnI presented here indicates that both Ser22 and Ser23 are basally phosphorylated as positional isomers, although monophosphorylated cTnI molecules with phosphorylated Ser22 were slightly more abundant than phosphorylated Ser23. This apparent difference could be explained by species differences and tissue sources of cTnI. One is commercially available human cTnI (Calbiochem) presumably purified from healthy human hearts, and the other is mouse cTnI affinity purified in-house from mouse hearts. Although cTnI is highly conserved in rodents and human myocardium, distinctions have been reported between rodents and humans. For example, mouse cTnI lacks Ser76, which is present in human cTnI. Degradation and modification of cTn have been observed in the stunned myocardium of mice (41) and rats (42, 43) but not in large mammalian models (44). Myofilament Ca2+ sensitivity is increased in the human end stage failing myocardium (23, 36, 45), yet a decrease in myofilament Ca2+ sensitivity was observed in rodent models of heart failure resulting from chronic pressure overload and myocardial infarction (46, 47). Furthermore, the assumption here is made that no changes in phosphorylation occur after death prior to harvest of the heart so that sample preparation methods could also contribute to the observed differences for human and mouse cTnI. The limitation of our previous human cTnI study (35) is that the source of the human cTnI is not well defined, although the company’s technical staff believed it was purified from healthy human heart tissue. The extensive proteolytic degradations and oxidations observed for human cTnI (35) indicate that the heart tissue samples have been collected some time after the death of the subject possibly without proper tissue preservation. Hence, one would argue that actual phosphorylation in a pumping healthy human heart might be different. In contrast, the mouse cTnI data described here were freshly purified in-house from well-preserved healthy mouse heart tissue sources. Postmortum proteolysis and other modifications have been minimized as only minor proteolytic fragments, and no artifactual modifications such as oxidation were detected for mouse cTnI.
Physio-Pathological Implication of Ser22/23 Basal Phosphorylation in Mouse cTnI
Ser22/23 are the bona fide phosphorylation sites for PKA upon β-adrenergic stimulation of the heart, and ample evidence indicates that PKA phosphorylation at Ser22/23 can result in a reduction in myofilament Ca2+ sensitivity (20) and an increase in cross-bridge cycling rate (21, 22). A decreased PKA phosphorylation was reported in the human myocardium with dilated cardiomypathy (48), and dephosphorylation at PKA sites could contribute to the Ca2+ sensitization of the myofilaments in heart failure (36, 49, 50). PKC can also cross-phosphorylate the PKA sites of cTnI at Ser22/23 and induce similar functional effects as PKA phosphorylation and reduced Ca2+ sensitivity of Ca2+ stimulated MgATPase of reconstituted actomyosin S-1 (14, 17). Phosphorylation specificity of PKC isozymes has been studied by Jideama et al. who demonstrated that PKC-δ uniquely phosphorylates Ser22/23 of cTnI, whereas PKC-δ and PKC-α phosphorylated Ser42/44 (Ser43/45 including the initiation methionine) (14). Ser22/23 have also been identified by Kobayashi et al. (14, 17) and Wang et al. (24) as substrates for PKC-β and PKC-ε. PKC also phosphorylates cTnI on Thr143 (Thr144 including the initiation Met), which may sensitize cardiac myofilaments to Ca2+ when Ser22/23 are unphosphorylated (24) or desensitize when Ser22/23 are pseudobis-phosphorylated (51). Additionally, PKG and PKD phosphorylate Ser22 and Ser23, with effects similar to those of PKA (8, 19, 25). Moreover, Vahebi et al. showed that Rho-A-dependent kinase (ROCK-II) phosphorylated cTnI at Ser22/23 and Thr143 (52).
The observation here that Ser22 and Ser23 were identified as the only basal phosphorylation sites of mouse cTnI does not diminish the significance of other sites regulated by PKC and other kinases. In fact, the cTnI analyzed here is purified only from healthy mouse heart, and a heart in the end stage failure is known to be markedly different from a healthy heart at the molecular and cellular level. Therefore, it is reasonable to suspect that other sites in cTnI might be related to cardiac dysfunction. Vlahos et al. showed that PKC is upregulated in the human heart in end stage failure particularly the Ca2+-dependent isoforms PKC-α and PKC-β (53). PKC phosphorylation might contribute to slowed filament sliding rates in failing human hearts as demonstrated by Knott et al. via an in vitro motility assay (54). Studies with transgenic mouse models generally support the concept that excess phosphorylation of cTnI by PKC may result in impaired relaxation and even systolic dysfunction (15, 16, 55–58). For example, Solaro and co-workers developed transgenic mouse lines that overexpress PKC in the myocardium which exhibit a steady progression to heart failure (59, 60). The results from a transgenic mouse line by crossing PKC-ε with cTnI alanine 42/44 mice suggest that phosphorylation of cTnI at Ser42/44 is associated with contractile dysfunction when PKC is up-regulated in heart failure (61). Hence, PKC sites at Ser42/44 and Thr 143 could be considered to be pathological or maladaptive as they are connected with disorders of the heart (9). To verify whether these PKC sites are pathological, one key subject of our future work will be to precisely locate the specific phosphorylation sites of cTnI in disease animal models.
Top-Down ECD MS for the Characterization of Basal Phosphorylation Occurring in Vivo
Here, we present a comprehensive characterization of cTnI directly purified from mouse heart tissue and precisely mapped all its basal phosphorylation sites occurring in vivo by top-down ECD mass spectrometry. Mapping in vivo phosphorylation sites in proteins have been difficult with traditional technologies such as 32P labeling and tryptic peptide mapping (12, 14, 25). Western blotting with known phosphoserine or phosphothreonine antibodies enables the identification of specific types of phosphorylation in vivo; however, it is limited by the availability of antibodies, and prior knowledge regarding the phosphorylation is needed (61). Most of the antibodies are specific toward one epitope (one phosphorylation site), and the few available generic antiphosphoserine/threonine antibodies are of extremely low specificity (62). The introduction of phosphor-protein specific gel stain, Pro-Q diamond, allows measurements of protein phosphorylation in situ (36, 52), yet it does not provide information on the sites of phosphorylation.
MS is the only detection method that can universally provide information about specific protein modifications without a priori knowledge of the modification (63). Recently, MS has been utilized to identify the phosphorylation sites from the enzymatic digest of cTnI after in vitro phosphorylation by a specific kinase as reported in several studies (17, 18, 24, 52). However, the information on the phosphorylation status obtained through this peptide-based bottom-up MS strategy is typically limited because of the partial sequence coverage and loss of connections between disparate portions of proteins containing modifications. In contrast, a top-down MS strategy allows an unbiased analysis of a whole protein globally to characterize any protein modifications (PTMs, mutations, etc.) providing a bird’s eye view to observe all possible modifications and to precisely locate the sites of modifications of proteins present in vivo (35, 64). As demonstrated, top-down MS with ECD is an extremely valuable method for the characterization of labile PTMs such as phosphorylation, as ECD preserves the labile phosphoryls during its fragmentation process. In contrast, the conventional MS/MS techniques such as CAD (also known as CID) and PSD (often used in MALDI-TOF instruments) tend to knock off labile PTMs, making it very difficult for site mapping. Here, we were able to examine all of the possible phosphorylation sites for the intact mouse cTnI with 100% sequence coverage and unambiguously identified the Ser22/23 as the only basal phosphorylation sites. More importantly, tandem MS allows a direct separation of intermediate and end product of phosphorylation (gas phase purification), which allows one to explore the potential interdependence of two phosphorylation sites. We have been able to isolate the un-, mono-, and bis-phosphorylated cTnI molecules and locate the phosphorylation sites for each cTnI form. No clear interdependency was indicated for Ser22 and Ser23 in mouse cTnI as both Ser22 and Ser23 have been phosphorylated in mono- and bis-phosphorylated cTnI, although phosphorylation on Ser22 is slightly more abundant than phosphorylation on Ser23. On the contrary, an ordered or sequential phosphorylation was suggested for human cTnI since Ser22 was phosphorylated in both mono- and bis-phosphorylated cTnI, whereas Ser23 was only phosphorylated in bis-phosphorylated cTnI (35).
To recapitulate, we have comprehensively characterized mouse cTnI with a precise mapping of all its phosphorylation sites using top-down ECD mass spectrometry. Ser22/23 was identified as the only sites phosphorylated basally in mouse cTnI affinity purified directly from wild-type and transgenic mouse hearts, although we cannot exclude the possibility of other sites with extremely low phosphorylation occupancy (<1 %). As demonstrated, top-down MS with ECD provides unique opportunities to identify the phosphorylation sites globally with 100% sequence coverage and explore the interdependency between multiple phosphorylation sites.
Supplementary Material
ACKNOWLEDGMENT
We thank Chris Doede, Huseyin Guner, Lisa Xu, Raquel Sancho Solis, and Matt Lawrence for technical assistance and helpful discussions.
Footnotes
Financial support was kindly provided by the Wisconsin Partnership Fund for a Healthy Future, NIH grant RO1 HL081386 (to J.W.W.), and American Heart Association Scientist Development Grant (to Y.G.).
Abbreviations: cTn, cardiac troponin; cTnI, cardiac troponin I; cTnT, cardiac troponin T; Tm, tropomyosin; LC2, myosin regulatory light chain 2; ECD, electron capture dissociation; MS, mass spectrometry; MS/MS, tandem mass spectrometry; CAD, collisionally activated dissociation; IRMPD, infrared multiphoton dissociation; PSD, postsource decay; LTQ, linear ion trap; FT, Fourier transform; PTM, post-translational modification; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; PKA, protein kinase A; PKC protein kinase C; PAK, p21-activated kinase; PKG, cyclic GMP-dependent protein kinase; PKD, protein kinase D; S/N, signal-to-noise ratio; m/z, mass to charge ratio.
SUPPORTING INFORMATION AVAILABLE
Representative ECD spectrum of cTnI; MS/MS product map from the ECD spectra for assignments to cTnI-Ala2; high resolution FTMS analysis of cTn affinity purified cTnI-Ala2 transgenic mouse hearts. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Solaro RJ, Rarick HM. Troponin and tropomyosin: Proteins that switch on and tune in the activity of cardiac myofilaments. Circ. Res. 1998;83:471–480. doi: 10.1161/01.res.83.5.471. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi T, Solaro RJ. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 3.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 4.Huang XP, Pi YQ, Lee KJ, Henkel AS, Gregg RG, Powers PA, Walker JW. Cardiac troponin I gene knockout: A mouse model of myocardial troponin I deficiency. Circ. Res. 1999;84:1–8. doi: 10.1161/01.res.84.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Murphy AM, Kogler H, Georgakopoulos D, McDonough JL, Kass DA, Van Eyk JE, Marban E. Transgenic mouse model of stunned myocardium. Science. 2000;287:488–491. doi: 10.1126/science.287.5452.488. [DOI] [PubMed] [Google Scholar]
- 6.Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. J. Clin. Invest. 2005;115:518–526. doi: 10.1172/JCI200524351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babuin L, Jaffe AS. Troponin: the biomarker of choice for the detection of cardiac injury. Can. Med. Assoc. J. 2005;173:1191–1202. doi: 10.1503/cmaj.050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Layland J, Solaro RJ, Shah AM. Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc. Res. 2005;66:12–21. doi: 10.1016/j.cardiores.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Solaro RJ. Multiplex kinase signaling modifies cardiac function at the level of sarcomeric proteins. J. Biol. Chem. 2008;283:26829–26833. doi: 10.1074/jbc.R800037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker JW. Protein kinase C, troponin I and heart failure: overexpressed, hyperphosphorylated and underappreciated? J. Mol. Cell. Cardiol. 2006;40:446–450. doi: 10.1016/j.yjmcc.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Solaro RJ, Moir AJG, Perry SV. Phosphorylation of troponin-I and inotropic effect of adrenaline in perfused rabbit heart. Nature. 1976;262:615–617. doi: 10.1038/262615a0. [DOI] [PubMed] [Google Scholar]
- 12.Noland TA, Raynor RL, Kuo JF. Identification of sites phosphorylated in bovine cardiac troponin-I and troponin-T by protein kinase-C and comparative substrate activity of synthetic peptides containing the phosphorylation sites. J. Biol. Chem. 1989;264:20778–20785. [PubMed] [Google Scholar]
- 13.Noland TA, Guo XD, Raynor RL, Jideama NM, Averyhartfullard V, Solaro RJ, Kuo JF. Cardiac troponin-I mutants: Phosphorylation by protein-kinase-C and protein-kinase-a and regulation of Ca2+-stimulated mgatpase of reconstituted actomyosin S-1. J. Biol. Chem. 1995;270:25445–25454. doi: 10.1074/jbc.270.43.25445. [DOI] [PubMed] [Google Scholar]
- 14.Jideama NM, Noland TA, Raynor RL, Blobe GC, Fabbro D, Kazanietz MG, Blumberg PM, Hannun YA, Kuo JF. Phosphorylation specificities of protein kinase C isozymes for bovine cardiac troponin I and troponin T and sites within these proteins and regulation of myofilament properties. J. Biol. Chem. 1996;271:23277–23283. doi: 10.1074/jbc.271.38.23277. [DOI] [PubMed] [Google Scholar]
- 15.Pi YQ, Kemnitz KR, Zhang DH, Kranias EG, Walker JW. Phosphorylation of troponin I controls cardiac twitch dynamics: Evidence from phosphorylation site mutants expressed on a troponin I-null background in mice. Circ. Res. 2002;90:649–656. doi: 10.1161/01.res.0000014080.82861.5f. [DOI] [PubMed] [Google Scholar]
- 16.Pi YQ, Zhang DH, Kemnitz KR, Wang H, Walker JW. Protein kinase C and A sites on troponin I regulate myofilament Ca2+ sensitivity and ATPase activity in the mouse myocardium. J. Physiol. 2003;552:845–857. doi: 10.1113/jphysiol.2003.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi T, Yang XF, Walker LA, Van Breemen RB, Solaro RJ. A non-equilibrium isoelectric focusing method to determine states of phosphorylation of cardiac troponin I: Identification of Ser-23 and Ser-24 as significant sites of phosphorylation by protein kinase C. J. Mol. Cell. Cardiol. 2005;38:213–218. doi: 10.1016/j.yjmcc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Buscemi N, Foster DB, Neverova I, Van Eyk JE. p21-activated kinase increases the calcium sensitivity of rat triton-skinned cardiac muscle fiber bundles via a mechanism potentially involving novel phosphorylation of troponin I. Circ. Res. 2002;91:509–516. doi: 10.1161/01.res.0000035246.27856.53. [DOI] [PubMed] [Google Scholar]
- 19.Haworth RS, Cuello F, Herron TJ, Franzen G, Kentish JC, Gautel M, Avkiran M. Protein kinase D is a novel mediator of cardiac troponin I phosphorylation and regulates myofilament function. Circ. Res. 2004;95:1091–1099. doi: 10.1161/01.RES.0000149299.34793.3c. [DOI] [PubMed] [Google Scholar]
- 20.Kranias EG, Solaro RJ. Phosphorylation of troponin-I and phospholamban during catecholamine stimulation of rabbit heart. Nature. 1982;298:182–184. doi: 10.1038/298182a0. [DOI] [PubMed] [Google Scholar]
- 21.Kentish JC, McCloskey DT, Layland J, Palmer S, Leiden JM, Martin AF, Solaro RJ. Phosphorylation of troponin I by protein kinase A accelerates relaxation and crossbridge cycle kinetics in mouse ventricular muscle. Circ. Res. 2001;88:1059–1065. doi: 10.1161/hh1001.091640. [DOI] [PubMed] [Google Scholar]
- 22.Strang KT, Sweitzer NK, Greaser ML, Moss RL. Beta-adrenergic-receptor stimulation increases unloaded shortening velocity of skinned single ventricular myocytes from rats. Circ. Res. 1994;74:542–549. doi: 10.1161/01.res.74.3.542. [DOI] [PubMed] [Google Scholar]
- 23.van der Velden J, Papp Z, Zaremba R, Boontje NM, de Jong JW, Owen VJ, Burton PBJ, Goldmann P, Jaquet K, Stienen GJM. Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins. Cardiovasc. Res. 2003;57:37–47. doi: 10.1016/s0008-6363(02)00606-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Grant JE, Doede CM, Sadayappan S, Robbins J, Walker JW. PKC-beta II sensitizes cardiac myofilaments to Ca2+ by phosphorylating troponin I on threonine-144. J. Mol. Cell. Cardiol. 2006;41:823–833. doi: 10.1016/j.yjmcc.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Murphy AM. Another new kinase targets troponin I. Circ. Res. 2004;95:1043–1045. doi: 10.1161/01.RES.0000150051.81155.88. [DOI] [PubMed] [Google Scholar]
- 26.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- 27.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, Park JH, Begley TP, McLafferty FW. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 28.Ge Y, Rybakova I, Xu Q, Moss RL. Top-down high resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc. Natl. Acad. Sci. U.S.A. 2009 doi: 10.1073/pnas.0813369106. [Online early access] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn P, Xu Q, Cline E, Zhang D, Ge Y, Xu W. Delineating Anopheles gambiae coactivator associated arginine methyltransferase 1 (AgCARM1) by top-down high resolution tandem mass spectrometry. Protein Sci. 2009;18:1272–1280. doi: 10.1002/pro.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chait BT. Mass spectrometry: Bottom-up or top-down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 31.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 32.Senko MW, Speir JP, McLafferty FW. Collisional activation of large multiply-charged ions using Fourier-transform mass-spectrometry. Anal. Chem. 1994;66:2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 33.Little DP, Speir JP, Senko MW, Oconnor PB, McLafferty FW. Infrared multiphoton dissociation of large multiply-charged ions for biomolecule sequencing. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 34.Shi SDH, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal. Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 35.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Unraveling molecular complexity of phosphorylated human cardiac troponin I by top down electron capture dissociation/electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics. 2008:1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Messer AE, Jacques AM, Marston SB. Troponin phosphorylation and regulatory function in human heart muscle: Dephosphorylation of Ser23/24 on troponin I could account for the contractile defect in end-stage heart failure. J. Mol. Cell. Cardiol. 2007;42:247–259. doi: 10.1016/j.yjmcc.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: Human histone H4. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 38.Pesavento JJ, Bullock CR, Leduc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sancho Solis R, Ge Y, Walker JW. Single amino acid sequence polymorphisms in rat cardiac troponin revealed by top-down tandem mass spectrometry. J. Muscle Res. Cell Motil. 2008;29:203–212. doi: 10.1007/s10974-009-9168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, Schulenberg B, Hendrickson J, Beechem JM, Haugland RP, Patton WF. Global quantitative phosphoprotein analysis using multiplexed proteomics technology. Proteomics. 2003;3:1128–1144. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- 41.Murphy AM, Kogler H, Georgakopoulos D, McDonough JL, Kass DA, Van Eyk JE, Marban E. Transgenic mouse model of stunned myocardium. Science. 2000;287:488–491. doi: 10.1126/science.287.5452.488. [DOI] [PubMed] [Google Scholar]
- 42.McDonough JL, Arrell DK, Van Eyk JE. Troponin I degradation and covalent complex formation accompanies myocardial ischemia/reperfusion injury. Circ. Res. 1999;84:9–20. doi: 10.1161/01.res.84.1.9. [DOI] [PubMed] [Google Scholar]
- 43.Van Eyk JE, Powers F, Law W, Larue C, Hedges RS, Solaro RJ. Breakdown and release of myofilament proteins during ischemia and ischemia/reperfusion in rat hearts: Identification of degradation products and effects on the pCa-force relation. Circ. Res. 1998;82:261–271. doi: 10.1161/01.res.82.2.261. [DOI] [PubMed] [Google Scholar]
- 44.Colantonio DA, Van Eyk JE, Przyklenk K. Stunned peri-infarct canine myocardium is characterized by degradation of troponin T, not troponin I. Cardiovasc. Res. 2004;63:217–225. doi: 10.1016/j.cardiores.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Hamdani N, Kooij V, van Dijk S, Merkus D, Paulus WJ, dos Remedios C, Duncker DJ, Stienen GJM, van der Velden J. Sarcomeric dysfunction in heart failure. Cardiovasc. Res. 2008;77:649–658. doi: 10.1093/cvr/cvm079. [DOI] [PubMed] [Google Scholar]
- 46.Belin RJ, Sumandea MP, Allen EJ, Schoenfelt K, Wang H, Solaro RJ, de Tombe PP. Augmented protein kinase C-alpha-induced myofilament protein phosphorylation contributes to myofilament dysfunction in experimental congestive heart failure. Circ. Res. 2007;101:195–204. doi: 10.1161/CIRCRESAHA.107.148288. [DOI] [PubMed] [Google Scholar]
- 47.Belin RJ, Sumandea MP, Kobayashi T, Walker LA, Rundell VL, Urboniene D, Yuzhakova M, Ruch SH, Geenen DL, Solaro RJ, de Tombe PP. Left ventricular myofilament dysfunction in rat experimental hypertrophy and congestive heart failure. Am. J. Physiol.: Heart Circ. Physiol. 2006;291:H2344–H2353. doi: 10.1152/ajpheart.00541.2006. [DOI] [PubMed] [Google Scholar]
- 48.Zakhary DR, Moravec CS, Bond M. Reduced protein kinase A-dependent troponin-I phosphorylation in failing human hearts. Biophys. J. 1998;74:A353–A353. [Google Scholar]
- 49.Wolff MR, Buck SH, Stoker SW, Greaser ML, Mentzer RM. Myofibrillar calcium sensitivity of isometric tension is increased in human dilated cardiomyopathies: Role of altered beta-adrenergically mediated protein phosphorylation. J. Clin. Invest. 1996;98:167–176. doi: 10.1172/JCI118762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Velden J, de Jong JW, Owen VJ, Burton PBJ, Stienen GJM. Effect of protein kinase A on calcium sensitivity of force and its sarcomere length dependence in human cardiomyocytes. Cardiovasc. Res. 2000;46:487–495. doi: 10.1016/s0008-6363(00)00050-x. [DOI] [PubMed] [Google Scholar]
- 51.Burkart EM, Sumandea MP, Kobayashi T, Nili M, Martin AF, Homsher E, Solaro RJ. Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J. Biol. Chem. 2003;278:11265–11272. doi: 10.1074/jbc.M210712200. [DOI] [PubMed] [Google Scholar]
- 52.Vahebi S, Kobayashi T, Warren CM, de Tombe PP, Solaro RJ. Functional effects of Rho-kinase-dependent phosphorylation of specific sites on cardiac troponin. Circ. Res. 2005;96:740–747. doi: 10.1161/01.RES.0000162457.56568.7d. [DOI] [PubMed] [Google Scholar]
- 53.Vlahos CJ, McDowell SA, Clerk A. Kinases as therapeutic targets for heart failure. Nat. Rev. Drug Discovery. 2003;2:99–113. doi: 10.1038/nrd1009. [DOI] [PubMed] [Google Scholar]
- 54.Knott A, Purcell I, Marston S. In vitro motility analysis of thin filaments from failing and non-failing human heart: Troponin from failing human hearts induces slower filament sliding and higher Ca2+ sensitivity. J. Mol. Cell. Cardiol. 2002;34:469–482. doi: 10.1006/jmcc.2002.1528. [DOI] [PubMed] [Google Scholar]
- 55.Jin WH, Brown AT, Murphy AM. Cardiac myofilaments: from proteome to pathophysiology. Proteomics Clin. Appl. 2008;2:800–810. doi: 10.1002/prca.200780075. [DOI] [PubMed] [Google Scholar]
- 56.Sakthivel S, Finley NL, Rosevear PR, Lorenz JN, Gulick J, Kim S, VanBuren P, Martin LA, Robbins J. In vivo and in vitro analysis of cardiac troponin I phosphorylation. J. Biol. Chem. 2005;280:703–714. doi: 10.1074/jbc.M409513200. [DOI] [PubMed] [Google Scholar]
- 57.Takeishi Y, Chu GX, Kirkpatrick DM, Li ZL, Wakasaki H, Kranias EG, King GL, Walsh RA. In vivo phosphorylation of cardiac troponin I by protein kinase C beta 2 decreases cardiomyocyte calcium responsiveness and contractility in transgenic mouse hearts. J. Clin. Invest. 1998;102:72–78. doi: 10.1172/JCI2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pyle WG, Sumandea MP, Solaro RJ, De Tombe PP. Troponin I serines 43/45 and regulation of cardiac myofilament function. Am. J. Physiol.: Heart Circ. Physiol. 2002;283:H1215–H1224. doi: 10.1152/ajpheart.00128.2002. [DOI] [PubMed] [Google Scholar]
- 59.Huang L, Wolska BM, Montgomery DE, Burkart EM, Buttrick PM, Solaro RJ. Increased contractility and altered Ca2+ transients of mouse heart myocytes conditionally expressing PKC beta. Am. J. Physiol. Cell Physiol. 2001;280:C1114–C1120. doi: 10.1152/ajpcell.2001.280.5.C1114. [DOI] [PubMed] [Google Scholar]
- 60.Goldspink PH, Montgomery DE, Walker LA, Urboniene D, McKinney RD, Geenen DL, Solaro RJ, Buttrick PM. Protein kinase C epsilon overexpression alters myofilament properties and composition during the progression of heart failure. Circ. Res. 2004;95:424–432. doi: 10.1161/01.RES.0000138299.85648.92. [DOI] [PubMed] [Google Scholar]
- 61.Scruggs SB, Walker LA, Lyu T, Geenen DL, Solaro RJ, Buttrick PM, Goldspink PH. Partial replacement of cardiac troponin I with a non-phosphorylatable mutant at serines 43/45 attenuates the contractile dysfunction associated with PKC epsilon phosphorylation. J. Mol. Cell. Cardiol. 2006;40:465–473. doi: 10.1016/j.yjmcc.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 62.Nita-Lazar A, Saito-Benz H, White FM. Quantitative phosphoproteomics by mass spectrometry: Past, present, and future. Proteomics. 2008;8:4433–4443. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Population proteomics: The concept, attributes, and potential for cancer biomarker research. Mol. Cell. Proteomics. 2006;5:1811–1818. doi: 10.1074/mcp.R600006-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.