Abstract
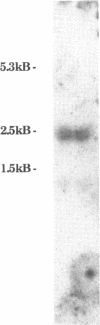
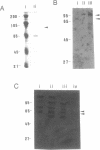
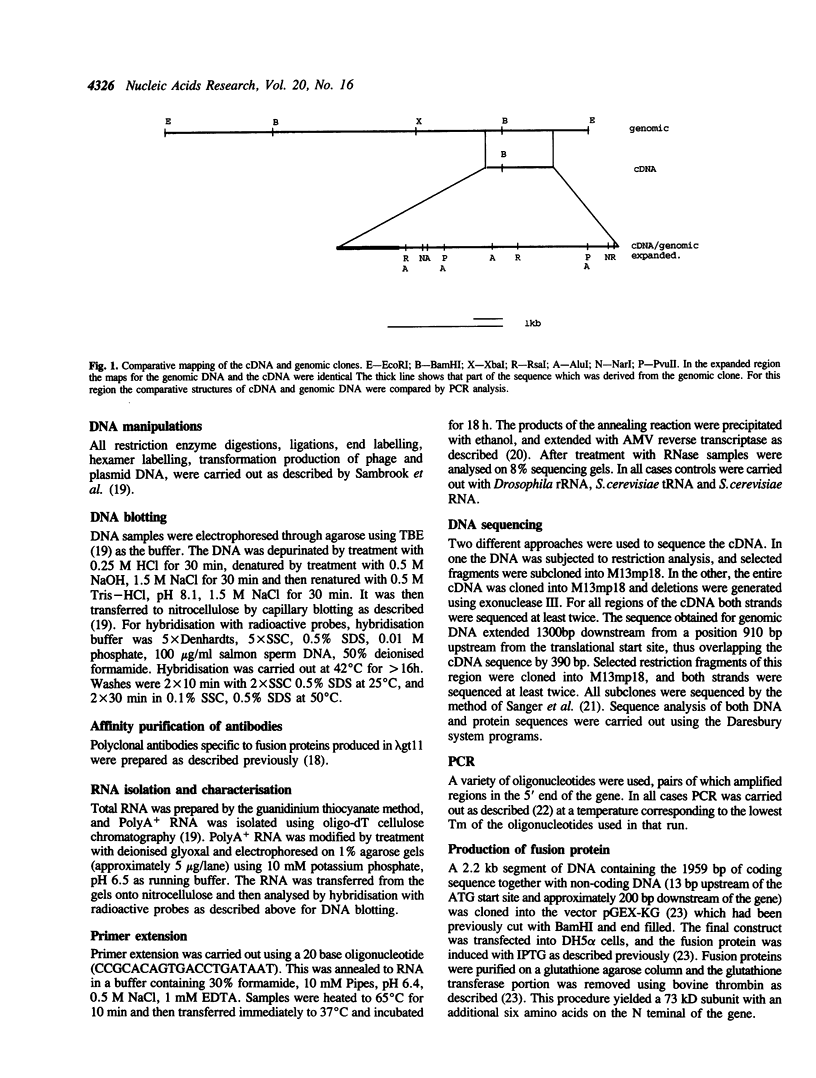
We have isolated both cDNA and genomic clones for the 73 kDa subunit of the DNA polymerase alpha primase of Drosophila melanogaster. Analysis of these clones has identified an open reading frame of 1959 bases coding for a protein of 72.5 kDa. Northern analysis has shown the mRNA for the gene to be approximately 2.5 kb, and comparison of the cDNA and the genomic clones shows that the coding region of the gene lacks introns. The 5' end of the transcript has been mapped by primer extension, and the position of the gene in the genome mapped using in situ analysis. Computer analysis has been carried out on both coding and non coding regions of the gene. The protein sequence shows some homology to the analogous subunit in the S. cerevisiae DNA polymerase alpha, however a search of the data banks failed to reveal other homologies, or provide any clues as to the function of the protein. Analysis of the non-coding regions indicates some potential control regions for the gene. The 73 kDa protein has been overproduced, but a preliminary analysis failed to reveal any enzymatic activities.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler D. A., Tseng B. Y., Wang T. S., Disteche C. M. Physical mapping of the genes for three components of the mouse DNA replication complex: polymerase alpha to the X chromosome, primase p49 subunit to chromosome 10, and primase p58 subunit to chromosome 1. Genomics. 1991 Apr;9(4):642–646. doi: 10.1016/0888-7543(91)90357-k. [DOI] [PubMed] [Google Scholar]
- Bambara R. A., Jessee C. B. Properties of DNA polymerases delta and epsilon, and their roles in eukaryotic DNA replication. Biochim Biophys Acta. 1991 Jan 17;1088(1):11–24. doi: 10.1016/0167-4781(91)90147-e. [DOI] [PubMed] [Google Scholar]
- Brooke R. G., Dumas L. B. Reconstitution of the Saccharomyces cerevisiae DNA primase-DNA polymerase protein complex in vitro. The 86-kDa subunit facilitates but is not required for complex formation. J Biol Chem. 1991 Jun 5;266(16):10093–10098. [PubMed] [Google Scholar]
- Brooke R. G., Singhal R., Hinkle D. C., Dumas L. B. Purification and characterization of the 180- and 86-kilodalton subunits of the Saccharomyces cerevisiae DNA primase-DNA polymerase protein complex. The 180-kilodalton subunit has both DNA polymerase and 3'----5'-exonuclease activities. J Biol Chem. 1991 Feb 15;266(5):3005–3015. [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterill S. M., Reyland M. E., Loeb L. A., Lehman I. R. A cryptic proofreading 3'----5' exonuclease associated with the polymerase subunit of the DNA polymerase-primase from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5635–5639. doi: 10.1073/pnas.84.16.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterill S., Chui G., Lehman I. R. DNA polymerase-primase from embryos of Drosophila melanogaster. DNA primase subunits. J Biol Chem. 1987 Nov 25;262(33):16105–16108. [PubMed] [Google Scholar]
- Cotterill S., Chui G., Lehman I. R. DNA polymerase-primase from embryos of Drosophila melanogaster. The DNA polymerase subunit. J Biol Chem. 1987 Nov 25;262(33):16100–16104. [PubMed] [Google Scholar]
- Foiani M., Santocanale C., Plevani P., Lucchini G. A single essential gene, PRI2, encodes the large subunit of DNA primase in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jul;9(7):3081–3087. doi: 10.1128/mcb.9.7.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi S., Longhese M. P., Piseri A., Santocanale C., Lucchini G., Plevani P. Mutations in conserved yeast DNA primase domains impair DNA replication in vivo. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3877–3881. doi: 10.1073/pnas.88.9.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991 Feb 1;192(2):262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Snyder M., Chang L. M., Davis R. W., Campbell J. L. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985 Nov;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Kaguni L. S., Lehman I. R. Eukaryotic DNA polymerase-primase: structure, mechanism and function. Biochim Biophys Acta. 1988 Jul 13;950(2):87–101. doi: 10.1016/0167-4781(88)90001-2. [DOI] [PubMed] [Google Scholar]
- Kaguni L. S., Rossignol J. M., Conaway R. C., Banks G. R., Lehman I. R. Association of DNA primase with the beta/gamma subunits of DNA polymerase alpha from Drosophila melanogaster embryos. J Biol Chem. 1983 Aug 10;258(15):9037–9039. [PubMed] [Google Scholar]
- Lucchini G., Brandazza A., Badaracco G., Bianchi M., Plevani P. Identification of the yeast DNA polymerase I gene with antibody probes. Curr Genet. 1985;10(4):245–252. doi: 10.1007/BF00365620. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Francesconi S., Foiani M., Badaracco G., Plevani P. Yeast DNA polymerase--DNA primase complex; cloning of PRI 1, a single essential gene related to DNA primase activity. EMBO J. 1987 Mar;6(3):737–742. doi: 10.1002/j.1460-2075.1987.tb04815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nasheuer H. P., Moore A., Wahl A. F., Wang T. S. Cell cycle-dependent phosphorylation of human DNA polymerase alpha. J Biol Chem. 1991 Apr 25;266(12):7893–7903. [PubMed] [Google Scholar]
- Pearson B. E., Nasheuer H. P., Wang T. S. Human DNA polymerase alpha gene: sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991 Apr;11(4):2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussak C. E., Almazan M. T., Tseng B. Y. Mouse primase p49 subunit molecular cloning indicates conserved and divergent regions. J Biol Chem. 1989 Mar 25;264(9):4957–4963. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Elledge S., Sweetser D., Young R. A., Davis R. W. Lambda gt 11: gene isolation with antibody probes and other applications. Methods Enzymol. 1987;154:107–128. doi: 10.1016/0076-6879(87)54073-3. [DOI] [PubMed] [Google Scholar]
- Venturelli D., Travali S., Calabretta B. Inhibition of T-cell proliferation by a MYB antisense oligomer is accompanied by selective down-regulation of DNA polymerase alpha expression. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5963–5967. doi: 10.1073/pnas.87.15.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. H., Johnson A. L., Lowndes N. F., Johnston L. H. The yeast DNA ligase gene CDC9 is controlled by six orientation specific upstream activating sequences that respond to cellular proliferation but which alone cannot mediate cell cycle regulation. Nucleic Acids Res. 1991 Jan 25;19(2):359–364. doi: 10.1093/nar/19.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. W., Wahl A. F., Yuan P. M., Arai N., Pearson B. E., Arai K., Korn D., Hunkapiller M. W., Wang T. S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988 Jan;7(1):37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff E., Natalie D., Nolan J. M., Lee M., Hsieh T. Structure of the Drosophila DNA topoisomerase II gene. Nucleotide sequence and homology among topoisomerases II. J Mol Biol. 1989 Jan 5;205(1):1–13. doi: 10.1016/0022-2836(89)90361-6. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Hayashi Y., Hirose F., Matsuoka S., Moriuchi T., Shiroishi T., Moriwaki K., Matsukage A. Molecular cloning and structural analysis of mouse gene and pseudogenes for proliferating cell nuclear antigen. Nucleic Acids Res. 1991 May 11;19(9):2403–2410. doi: 10.1093/nar/19.9.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Nishida Y., Moriuchi T., Hirose F., Hui C. C., Suzuki Y., Matsukage A. Drosophila proliferating cell nuclear antigen (cyclin) gene: structure, expression during development, and specific binding of homeodomain proteins to its 5'-flanking region. Mol Cell Biol. 1990 Mar;10(3):872–879. doi: 10.1128/mcb.10.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]