Abstract
The accumulation of somatic mutations in mitochondrial DNA (mtDNA) induced by reactive oxidative species (ROS) is regarded as a major contributor of aging and age-related degenerative diseases. ROS has also been shown to facilitate the formation of certain advanced glycation end-products in proteins and DNAs, and N2-carboxyethyl-2′-deoxyguanosine (CEdG) has been identified as a major DNA-bound AGE. Therefore, the influence of mitochondrial ROS on the glycation of mtDNA was investigated in primary embryonic fibroblasts derived from mutant mice (Sod2−/+) deficient in the mitochondrial antioxidant enzyme, manganese superoxide dismutase (MnSOD). In Sod2−/+ fibroblasts vs. wildtype fibroblasts, the CEdG content of mtDNA was increased from 1.90±1.39 pg/μg DNA to 17.14±6.60 pg/μg DNA (p<0.001). On the other hand, the CEdG content of nuclear DNA did not differ between Sod2+/+ and −/+ cells. Similarly, cytosolic proteins did not show any difference in the advanced glycation end-products or protein carbonyl contents between Sod2+/+ and −/+. Taken together, the data suggest that mitochondrial oxidative stress specifically promotes glycation of mtDNA and does not affect nuclear DNA or cytosolic proteins. Because DNA glycation can change DNA integrity and gene functions, glycation of mtDNA may play an important role in the decline of mitochondrial functions.
Keywords: Advanced glycation end-products (AGEs), N2-carboxyethyl-2′-deoxyguanosine (CEdG), DNA glycation, mitochondria, Mn superoxide dismutase (MnSOD), mitochondrial DNA, oxidative stress
Introduction
The accumulation of somatic mutations in the mitochondrial DNA (mtDNA) induced by oxidative stress is regarded as a major contributor of aging and age-related degenerative diseases [1]. Mitochondria are extremely susceptible to oxidative damage since 2–4% of the oxygen consumed by mitochondria is converted to superoxide anions by the electron transport chain [2]. Additionally, mitochondria have limited protection from oxidative stress [3]. Under normal conditions, the antioxidant enzyme manganese superoxide dismutase (MnSOD) protects mitochondria from superoxide anions that are produced as a byproduct of the respiratory chain. Two mouse lines lacking MnSOD (Sod2) have been constructed to further establish the role of mitochondrial ROS in the pathophysiology of diseases, especially in age-related disorders [4, 5]. Reduced activity of MnSOD can be correlated to a decline in mitochondrial function. Whereas complete ablation of MnSOD causes dilated cardiomyopathy, neurodegeneration, and early postnatal death [4, 6], heterozygous mutant mice (Sod2−/+) with a 50% reduction in MnSOD have a normal lifespan compared to the wildtype controls [7]. However, young Sod2−/+ mice exhibited decreased electron transport activity of Complex I, increased proton leak, reduced respiration rate, decreased respiratory control ratio for substrates metabolized by Complexes I, II, and III, lower mitochondrial membrane potential, and reduced ATP synthesis [8–10]. These findings show that a heterozygous knockout of MnSOD results in mitochondrial respiratory enzyme deficiency and that Sod2−/+ mouse is a useful model to study the molecular and pathological consequences of enhanced endogenous mitochondrial oxidative stress.
Advanced glycation end-products (AGEs) are generated by nonenzymatic reactions between sugars or sugar degradation products and free amino groups of proteins. Formation of some AGEs, such as the glycoxidation product Nε-(carboxymethyl)lysine (CML) is greatly favored by reactive oxygen species (ROS) [11, 12]. In a similar way to amino groups of proteins, the exocyclic amino group of 2′-deoxyguanosine can be glycated, and N2-carboxyethyl-2′-deoxyguanosine (CEdG) was identified as a major DNA-bound AGE [13]. CEdG is generated from a range of reactive carbonyl species (RCS), such as sugars, glyceraldehyde, or methylglyoxal [14, 15]. To date, CEdG has been detected in vitro in cultured smooth muscle cells and endothelial cells, in human urine, as well as in the human kidney and aorta [16–20]. Very recently, a method was developed to allow simultaneous quantification of AGEs bound to proteins, mtDNA, and nuclear DNA (nDNA). Therewith, significantly higher CEdG levels in mtDNA than in nDNA were detected in NIH 3T3 fibroblasts [21].
We hypothesize that one way mitochondrial ROS contributes to the accumulation of somatic mutations is by promoting the formation of mtDNA-AGEs. Therefore, the present study used primary mouse embryonic fibroblasts (MEFs) isolated from Sod2−/+ mice as a model for endogenously elevated mitochondrial oxidative stress, and the induction of intracellular AGEs was monitored by simultaneous quantification of nDNA-, mtDNA-, and protein-AGEs in Sod2+/+ and −/+ cells.
Experimental Procedures
Chemicals, reagents, and antibodies
Herring sperm DNA was purchased from Fluka (Buchs, Switzerland), sodium chloride, and Roti®-Phenol (phenol/chloroform/isoamyl alcohol [25/24/1]) were obtained from Roth (Karlsruhe, Germany). Aminoguanidine was purchased from Acros Organics (Geel, Belgium) proteinase K (1000 U/mL) and RNase A/T1 mix (RNase A: 2 mg/mL, RNase T1: 5000U/mL) from Fermentas (St Leon-Rot, Germany), and PBS from Biochrom (Berlin, Germany).
DMEM F12 50/50 (with L-glutamine), MEM nonessential amino acids (100 x solution), penicillin, streptomycin, and amphotericin B (PSA) antibiotic antimycotic (100 x), TrypLE express (stable trypsin replacement) and HEPES buffer (1 M) were purchased from Invitrogen (Darmstadt, Germany). Standard fetal bovine serum (collected and processed in USA) was obtained from Thermo Scientific (Logan, USA).
The following commercial kits were used: OxyBlot oxidized protein detection kit, Millipore (Schwalbach, Germany); ECL Western blot system, GE Healthcare (Munich, Germany); DC protein assay, Bio-Rad (Munich, Germany); Complete® protease inhibitor cocktail tablets, Roche Applied Science (Mannheim, Germany).
The following antibodies were applied: for CEdG-ELISA, a monoclonal CEdG antibody developed in mouse (MAb M-5.1.6, [17]) was used. Labeling was performed with a sheep anti-mouse IgG conjugated with horseradish peroxidase conjugate, A5906, from Sigma-Aldrich (Munich, Germany). For AGE Western blot the primary antibody was a rabbit polyclonal antibody to CML 100 μg (1 mg/mL), ab27684, from Abcam (Cambridge, UK), the secondary antibody was a goat anti-rabbit IgG tagged with horseradish peroxidase, A0545, from Sigma-Aldrich (Munich, Germany).
In vitro incubation of DNA under oxidative conditions
Incubation tests were performed with herring sperm DNA (1 mg/mL) in PBS using either solely DNA or reaction mixtures of the DNA with 1 mM H2O2 or 25 mU/mL xanthine oxidase (XO) and 0.4 mM xanthine. The H2O2 test solutions were varied by adding 5 mM glucose or 10 mM pyridoxamine, respectively. DNA was also incubated in the presence of the pyridoxamine. Parallel to the DNA/XO/xanthine tests, solutions were processed after addition of catalase (275 U/mL) or catalase and SOD (100 U/mL).
Controls containing only H2O2, glucose, or pyridoxamine in PBS were prepared to exclude an impact of the reagents on the subsequent ELISA step.
Aliquots of initially 10 mL of each test solution in 50 mL reaction tubes were incubated for 14 d at 37°C in a shaking water bath. Incubations were performed in triplicates. Samples of 350 μL each were drawn every other day and stored at −80°C until analysis by CEdG ELISA.
Cell culture
MEFs were derived from embryonic day 13 Sod2+/+ and −/+ fetuses by standard protocols as described [22]. Animal procedures were approved by the IACUC committees at Stanford University and VA Palo Alto Health Care System. MEFs were cultured in 50% DMEM/50% F12 medium supplemented with 10% FCS, 1% PSA, 1% non-essential amino acids, and 15 mM HEPES in humidified atmosphere containing 5% CO2 at 37°C. Cells were expanded to passage one (P1) and frozen in liquid nitrogen for long-term storage. Frozen stock of Sod2 fibroblasts were thawed as P2 and usually split 1:3 every 4–5 days when confluent. All experiments were conducted using MEFs within five passages post isolation. The cell population doubling time (Td) was monitored for passage three and four. The Td was calculated from the exponential portion of the growth curve using an online calculator (http://www.doubling-time.com).
Simultaneous isolation of nDNA, mtDNA, and cytosolic proteins from MEFs
The simultaneous isolation of nDNA, mtDNA, and cytosolic proteins was performed as described previously for NIH 3T3 fibroblasts [21] with minor modifications. For cell disruption, only freeze-thaw cycles were performed. The pelleted nuclei and mitochondria were resuspended in an extraction buffer containing 50 mM Tris (pH 8.0), 50 mM EDTA, 50 mM NaCl and 1% sodium dodecyl sulfate (SDS), and the final supernatant was collected as cytosolic protein fraction. Aminoguanidine (83 mM final concentration) was also added to the resuspended nuclei and mitochondria to prevent artificial glycation reactions during the workup. Nuclei and mitochondria were digested by consecutive additions (4x) of 30 μL proteinase K and incubation at 55°C for 3 h, overnight, 3 h, and 2 h. The DNA quality and concentrations were determined by UV measurements at 260 and 280 nm, and the protein yield was quantified using the DC protein assay.
Assay for succinate dehydrogenase (SDH) activity
The mitochondrial pellet was suspended in 1 mL of buffer M (210 mM D-mannitol, 70 mM sucrose, 5 mM HEPES, pH 7.4). The SDH assay was performed as previously described [21]. The suspended pellet, the post-mitochondrial supernatant, or buffer M alone was analyzed in a concentration of 10% v/v in the assay buffer as described. The absorption at 600 nm was determined in five-minute intervals for the first 30 min, and then approximately every 30 min for a total of 160 min. Total optical density changes after 160 min were used to determine SDH activities.
Competitive ELISA for CEdG
Prior to ELISA measurement, DNA was denatured for 5 min at 99°C to render CEdG adducts more accessible to the antibody, and, thus, increase the sensitivity. Denaturation does not lead to artificial CEdG formation (data not shown). The formation of CEdG-modifications of in vitro incubated DNA, isolated nDNA and mtDNA was then monitored by ELISA as previously described [23].
Coomassie staining of proteins separated by SDS-PAGE
SDS-PAGE of the cytosolic proteins was performed according to Laemmli [24]. Fifteen μg of proteins per lane was loaded onto a 12% acryl amide gel and separated at 150 V for 80 min. After electrophoresis was completed, the gels were stained with Coomassie blue as previously described [21].
Western blot analysis for AGE-modified proteins
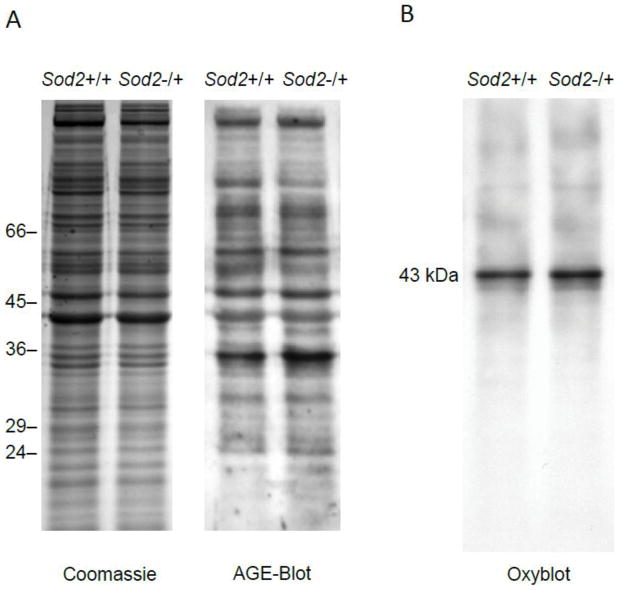
Western blotting for AGE-modified proteins was performed as previously described [21] with minor modifications. Labeling was performed using the horseradish peroxidase-conjugated rabbit IgG diluted 1:1,000 in blocking solution. Specificity of the AGE antibody for CML and Nε-(carboxyethyl)lysine (CEL) was verified by pre-incubation of the antibody with synthesized CML-bovine serum albumin and CEL-bovine serum albumin, respectively, which abolished the signals of the Western blot (data not shown). Simultaneously to each Western blot, an identically loaded acrylamide gel was stained with Coomassie blue, which served as a reference for AGE quantification (see Fig. 4A).
Fig. 4.
Analysis of the cytosolic protein fraction of Sod2+/+ and Sod2−/+ MEFs. A) Protein CML-/CEL analysis. The cytosolic protein fractions were separated on a 12% SDS-PAGE prior to blotting and reaction with a polyclonal CML/CEL-antibody (n=5 each) B) Protein carbonyl analysis. The cytosolic protein fraction of Sod2+/+ and Sod2−/+ MEFs was derivatized with DNPH and separated on a 12% SDS-PAGE prior to blotting and reaction with anti-DNPH antibody (n=5 each). Representative Western blot images are shown in A and B.
Analysis of oxidative protein modifications
OxyBlot oxidized protein detection kit was applied according to the manufacturer’s instructions. Two aliquots of each sample were analyzed; one was subjected to the derivatization reaction, the other aliquot was treated with a control solution and served as negative control. Detection of oxidized bands was performed using the ECL Western blot system with exposure times from 5 s to 5 min.
Results
In vitro glycation of DNA by oxidative species
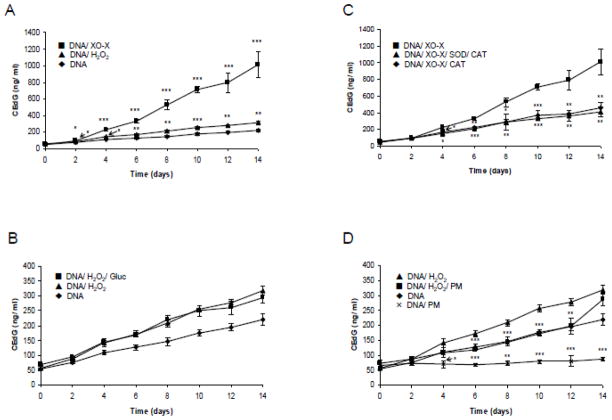
In order to test if reactive oxidative species (ROS) are able to introduce AGEs in DNA, herring sperm DNA was incubated in vitro in PBS under different oxidative conditions at 37°C for up to 14 d. Samples were taken every other day and the CEdG concentration was determined using an established CEdG ELISA method. Incubation was carried out in the presence of air oxygen, hydrogen peroxide (1 mM), and XO/xanthine system. The CEdG concentration increased with incubation time during reaction in the presence of oxygen (Fig. 1A). Starting from day 2, a significant (p < 0.01) and, from day 4, a highly significant (p < 0.001) increase of CEdG concentration compared to day 0 was measured. Addition of hydrogen peroxide and XO/xanthine further increased CEdG formation significantly compared to the incubation in the presence of air oxygen. Hydrogen peroxide and the XO/xanthine system induced a significant increase of CEdG concentration compared to the unincubated sample (0d) starting from day 2 (p < 0.01 at 2d and p < 0.001 at ≥ 4d for hydrogen peroxide; p < 0.05 at 2d and p < 0.001 at ≥ 4d). It was also investigated if co-incubation of DNA with hydrogen peroxide and physiological concentrations of glucose may further enhance CEdG formation. Hydrogen peroxide may liberate reactive carbonyl compounds from glucose which may serve as CEdG-precursors. Further, DNA-oxidation in the presence and absence of hydrogen peroxide was repeated after the addition of the AGE-inhibitor pyridoxamine. As shown in Fig. 1B and 1D, addition of 5 mM glucose did not further increase the CEdG-concentration, whereas pyridoxamine inhibited CEdG-formation in the presence and absence of hydrogen peroxide. Finally, the inhibitory effect of SOD and catalase or catalase alone on XO/xanthine -induced CEdG formation is demonstrated in Fig. 1C.
Fig. 1.
CEdG concentration after incubation of DNA under oxidative conditions at 37°C for 14 d: (A) herring sperm DNA (1 mg/mL) in the presence of air oxygen, hydrogen peroxide (H2O2, 1 mM), or xanthine oxidase/xanthine (XO-X, 25 mU/mL/0.4 mM); (B) DNA, DNA/H2O2 and DNA/H2O2 incubated in the presence of D-glucose (Gluc, 5 mM); (C) DNA/XO-X in absence and presence of catalase (CAT) or superoxide dismutase and catalase (SOD/CAT);(D) DNA and DNA/H2O2 in the absence and presence of pyridoxamine (PM, 10 mM). Concentrations are given as ng CEdG/mL incubation mixture. Data from each time point was measured in triplicates. Mean and standard deviation are shown. Significant differences between DNA and DNA/H2O2 or DNA/XO-X (A), between DNA/XO-X and DNA/XO-X/SOD/CAT or DNA/XO-X/CAT (C), between DNA/H2O2 and DNA/H2O2/PM (D) as well as between DNA and DNA/PM (D) are shown (* p<0.05, ** p<0.001,*** p<0.001).
Standardization of cell culture conditions
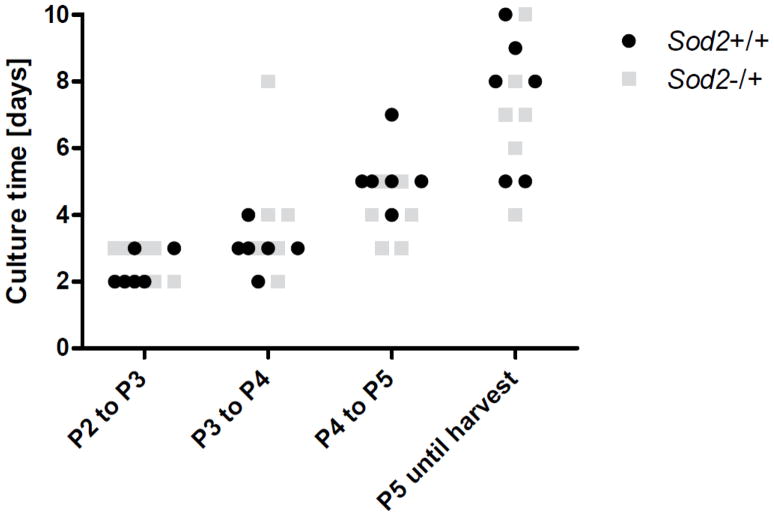
To collect MEFs for DNA and protein glycation studies, wildtype (Sod2+/+) and Sod2−/+ MEFs were cultivated and expanded continuously from P2 to P5. Growth curves were recorded for P3 and P4 and the Td was calculated. The Td for Sod2+/+ MEFs was 50.99 h for P3 and 54.86 h for P4, whereas the Td for Sod2−/+ MEFs was 39.18 h for P3 and 44.98 h for P4. However, the differences in Td were not significant between Sod2+/+ and Sod2−/+ cells. At the time of harvest, Sod2+/+ and Sod2−/+ MEFs were cultured for a total of 18.0 d and 17.7 d, respectively (Fig. 2). The Td and culture time showed that the culture conditions were similar for Sod2+/+ and Sod2−/+ fibroblasts.
Fig. 2.
Culture time of Sod2+/+ and −/+ MEFs. MEFs were thawed at P2 and split 1:3 until harvest at P5. The time required to reach confluency in culture was monitored for P2–P5. With increasing passages post isolation, the time until confluency increased. There was no significant difference between Sod2−/+ MEFs and wildtype MEFs (n=6 each).
Isolation of nDNA, mtDNA, and cytosolic proteins from Sod2 fibroblasts
Table 1 shows the yield of nDNA, mtDNA, and cytosolic proteins per cell. The protein yields amounted to 141.11 pg/cell and 145.23 pg/cell for Sod2+/+ and Sod2−/+ MEFs, respectively. From Sod2+/+ MEFs, the average yield was 4.94 pg nDNA and 0.018 pg mtDNA per cell. From Sod2−/+ fibroblasts, the average yield was 3.76 pg nDNA and 0.034 pg mtDNA per cell. There was no significant difference in the yield of nDNA, mtDNA, or cytosolic proteins between Sod2+/+ and Sod2−/+ fibroblasts. The ratio of A260/280 for nDNA and mtDNA was always greater than 1.8.
Table 1.
Yield of cytosolic proteins, total nDNA, and mtDNA isolated simultaneously from wildtype and Sod2−/+ fibroblasts.
| Protein/Cell [pg] | nDNA/Cell [pg] | mtDNA/Cell [pg] | |
|---|---|---|---|
| Sod2+/+ | 141.11 ± 7.30 | 4.94 ± 1.91 | 0.018 ± 0.008 |
| Sod2−/+ | 145.23 ± 68.26 | 3.76 ± 1.13 | 0.034 ± 0.009 |
Protein yield was measured using the DC protein assay. DNA concentration was determined by absorption at 260 nm. Cells were counted using a Neubauer counting chamber after trypan blue staining. At least three isolation procedures were performed for one data point. Each data point represents the mean value ± standard deviation.
Characterization of the mitochondrial fraction
To ensure that the mitochondrial pellet indeed contained the mitochondria, the isolated fraction was analyzed for mitochondrial SDH activity. The post-mitochondrial supernatant was analyzed in parallel as a negative control. Addition of the substrate 2,6-dichlorophenolindophenol (DCPIP) to the mitochondrial fraction resulted in a color reduction of 72% within 160 min, which verifies SDH activity (data not shown). The post-mitochondrial supernatant, on the other hand, did not show SDH activity.
Determination of the CEdG content of nDNA and mtDNA
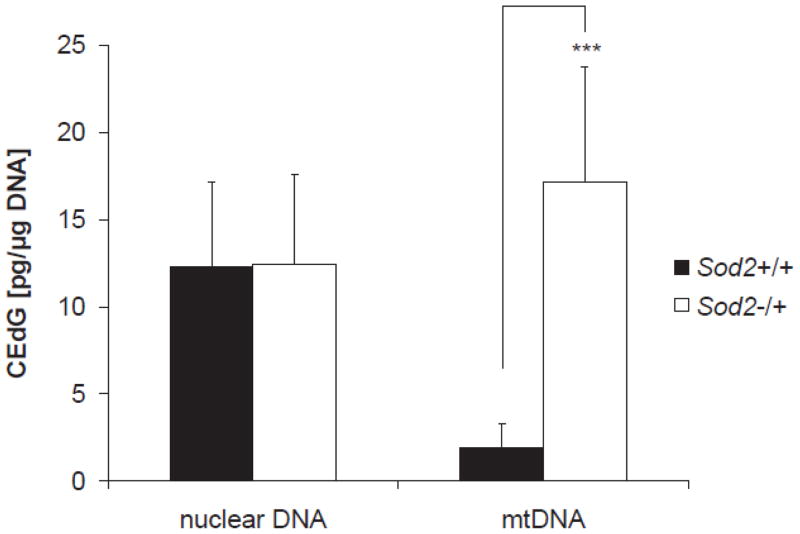
In order to measure DNA-AGEs, CEdG modifications in nDNA and mtDNA isolated from Sod2+/+ and Sod2−/+ MEFs were quantified using an established CEdG ELISA method. Nuclear DNA isolated from Sod2+/+ and Sod2−/+ fibroblasts showed CEdG levels of 12.27±4.90 pg/μg nDNA and 12.47±5.13 pg/μg nDNA, respectively. No significant difference in CEdG content was observed in nDNA. Mitochondrial DNA isolated from Sod2+/+ fibroblasts showed low levels of CEdG modifications of 1.90±1.39 pg/μg mtDNA. A significant increase in CEdG content to 17.14±6.60 pg/μg mtDNA was observed in Sod2−/+ MEFs (n=5, p<0.001, Fig. 3). The results indicated that proportionally more mtDNA is glycated in Sod2−/+ cells when compared to Sod2+/+ controls.
Fig. 3.
CEdG content of nDNA and mtDNA. Nuclear and mtDNA were isolated simultaneously from wildtype and Sod2−/+ MEFs. CEdG concentration was determined using an established CEdG ELISA method. Data are presented as mean ± SD. Wildtype cells–nDNA: 12.27±4.90 pg/μg DNA, n=6; mtDNA: 1.90±1.39 pg/μg DNA, n=5; Sod2−/+ cells nDNA: 12.47±5.13 pg/μg DNA, n=5; mtDNA: 17.14±6.60 pg/μg DNA, n=5 (*** p<0.001)
Analysis of protein AGEs and protein carbonyls
The cytosolic protein fractions isolated from Sod2+/+ and Sod2−/+ MEFs were analyzed for their AGE content with Western blot using an antibody that detects both CML and CEL. In the cytosolic protein fraction of both MEFs, AGEs were detected over the whole range of proteins. There was no significant difference in AGE pattern or intensity between wildtype and Sod2−/+ MEFs.
Additionally, oxidative modifications of cytosolic proteins were analyzed by Western blot after derivatization with 2,4-dinitrophenylhydrazine (DNPH). Protein samples without DNPH derivatization were used as negative control. As shown in Fig. 4B, there are two dominant bands at 42–43 kDa in both lanes. After quantification of the band intensities and normalization to the corresponding lanes on the Coomassie gel, no significant differences in protein carbonyl content were observed between Sod2+/+ and Sod2−/+ MEFs.
Discussion
In order to investigate whether ROS induces DNA glycation, DNA was incubated under various oxidative conditions in vitro. Even air oxygen was sufficient to induce CEdG formation. It is well established that metal traces present in PBS induce oxidation in the presence of oxygen [25]. The addition of hydrogen peroxide as well as of the XO/xanthine system led to a further significant increase in CEdG-concentration. XO produces superoxide and hydrogen peroxide [26]. In a secondary reaction, superoxide can be spontaneously or enzymatically converted into hydrogen peroxide by dismutation. Co-incubation of DNA/XO/xanthine with SOD and catalase significantly suppressed CEdG formation, confirming that the measured CEdG formation was indeed caused by ROS. Co-incubation with catalase alone inhibited CEdG formation to a similar extent as the combination of both enzymes indicating that hydrogen peroxide is the reactive agent leading to DNA glycation. Superoxide, in contrast, must first be converted to hydrogen peroxide which then induces CEdG formation. An increase of CEdG concentration with increasing incubation time could be observed under all tested conditions. Most likely, RCS are formed during the reaction of ROS with DNA by oxidative degradation of deoxyribose. The RCS then reacts with the guanine base to yield CEdG. The addition of glucose did not further enhance CEdG concentration indicating that deoxyribose from the DNA is a considerably more efficient carbonyl precursor than glucose. Pyridoxamine effectively suppressed CEdG formation. Pyridoxamine is a well established AGE-inhibitor, which acts most likely by trapping reactive carbonyl compounds by its amino group [27]. In vivo, the administration of pyridoxal phosphate inhibited CEdG accumulation in glomeruli of diabetic rats [28]. Pyridoxal phosphate and pyridoxamine are both B6 vitamers, which can be converted into each other in vivo. Our findings showed that ROS, in particular hydrogen peroxide and superoxide as a precursor of hydrogen peroxide, are able to induce DNA glycation in vitro. As a next step, we investigated if endogenous oxidative stress enhances intracellular glycation. Mitochondria are the main source of cellular ROS, because 2–4% of the oxygen consumed by mitochondria is converted to superoxide anions by the electron transport chain [2]. Superoxide anions may then react to generate more reactive ROS, such as hydrogen peroxide and hydroxyl radicals [29].
Partial knockout of MnSOD activity has been established as a suitable model to study the role of endogenous mitochondrial oxidative stress in vitro and in vivo. Heterozygous MnSOD knockout mice (Sod2−/+) show increased levels of ROS and, at the same time, have no compensatory response from other antioxidant enzymes [30]. The detrimental significance of mitochondrial electron transport chain-derived superoxide is well illustrated by the lethal phenotype of mice lacking the mitochondrial matrix superoxide dismutase (i.e. MnSOD) [4]. Therefore, endogenous glycation of nDNA, mtDNA, and cytosolic proteins was analyzed in MEFs with reduced MnSOD activities. Nuclear DNA from both Sod2+/+ and −/+ MEFs showed similar CEdG levels (Fig. 3), indicating that reduced levels of mitochondrial MnSOD did not cause increased oxidative stress in the nucleus. This conclusion is supported by a previous study reporting similar levels of 8-OH-deoxyguanosine in wildtype and Sod2−/+ mice [10].
Parallel to nDNA analyses, mtDNA was examined for glycation adducts. The CEdG content of mtDNA extracted from Sod2−/+ cells was eight times higher than that in Sod2+/+ cells, suggesting that the increased mitochondrial oxidative stress in Sod2−/+ cells promoted mtDNA glycation. Consistent with this finding, a previous study reported a 30% increase in the 8-OH deoxyguanosine levels of mtDNA from the liver of Sod2−/+ mice compared to control animals [10]. The present results demonstrated that DNA glycation is an important second mechanism – in addition to DNA oxidation – that leads to ROS induced damage of mtDNA. It is assumed that elevated superoxide concentration accelerates DNA damage by leaching iron from iron-sulfur cluster containing proteins and subsequent hydroxyl radical generation [31, 32]. The notion is supported by the observation that the activities of Complex I and aconitase, both containing iron-sulfur clusters, are significantly reduced in the liver mitochondria of Sod2−/+ mice [33, 34]. Unlike nDNA, mtDNA lacks extensive protection by histones and only possesses a limited capacity for DNA repair [35]. It has been well documented that mtDNA accumulates alterations (deletions, rearrangements, or point mutations) with age in various model systems as well as in humans [36]. Since deletions and point mutations can be a result of glycation reactions [37], the observed accumulation of CEdG triggered by mitochondrial oxidative stress could be involved in the decline in mitochondrial function.
Cytosolic protein AGEs were monitored in Sod2+/+ and Sod2−/+ cells by CML/CEL-Western blot analysis, and cytosolic protein carbonyls by Western blot after derivatization with DNPH. There was no significant increase in AGE modifications as a result of the partial reduction in MnSOD. In contrast, it has been reported that mitochondrial protein carbonyls are increased in Sod2−/+ mice compared to wildtype controls [10]. These findings are consistent with an analysis of skin collagen of Sod2+/+ and −/+ mice for biomarkers of aging, in which CML and pentosidine were found to change with age to a similar extent in both strains of mice [7]. It has been reported that MnSOD deficiency in Sod2−/+ mice does not influence the activity of the cytosolic superoxide dismutase, Cu,ZnSOD. Consequently, the increase in free radical formation is limited to mitochondria.
RCS are generated endogenously by autoxidation of carbohydrates or lipid peroxidation. This process might be accelerated by increased mitochondrial superoxide concentration that is found in Sod2−/+ mice. Therefore, cytosolic protein extracts were further analyzed for oxidative modifications. The cytosolic protein extracts of Sod2+/+ and Sod2−/+ MEFs did not show a significant difference in the carbonyl content. The results are comparable to a previous study reporting similar levels of carbonyl groups in the cytosolic proteins extracted from the liver of Sod2−/+ mice and controls [10].
Conclusion
The present findings indicate that enhanced endogenous mitochondrial oxidative stress in Sod2−/+ cells results in an accumulation of glycation products specifically in mtDNA. It is well known that increased DNA glycation can lead to reductions in gene function or alterations in DNA sequence [23]. Since intact mtDNA is important for normal mitochondrial functions, our results suggest that oxidative-stress-induced mtDNA glycation could be involved in a decline in mitochondrial function.
Highlights.
Reactive oxygen species are important precursors for DNA-glycation in vitro
Primary embryonic fibroblasts were obtained from mutant mice deficient in MnSOD
Mitochondrial DNA-glycation is increased in Sod2−/+ compared to Sod2+/+ fibroblasts
Nuclear DNA-glycation is not affected by MnSOD deficiency
MnSOD deficiency does not influence glycation and oxidation of cytosolic proteins
Acknowledgments
We thank Xinli Wang for excellent animal care. This project was supported by the Alexander von Humboldt Foundation (CONNECT), the German Academic Exchange Service (DAAD), the Bavaria California Technology Center (BaCaTeC), funding from NIH (AG24400), and by the resources and facilities at the VA Palo Alto Health Care System.
Abbreviations
- AGEs
advanced glycation end products
- CEdG
N2-carboxyethyl-2′-deoxyguanosine
- CEL
Nε-(carboxyethyl)lysine
- CML
Nε-(carboxymethyl)lysine
- DCPIP
2,6-dichlorphenolindophenol
- DNPH
2,4-dinitrophenylhydrazine
- MEFs
mouse embryonic fibroblasts
- MnSOD
manganese superoxide dismutase
- mtDNA
mitochondrial DNA
- nDNA
nuclear DNA
- P
passage
- PSA
penicillin, streptomycin, and amphotericin B
- RCS
reactive carbonyl species
- ROS
reactive oxygen species
- SDH
succinate dehydrogenase
- SDS
sodium dodecyl sulfate
- Td
cell population doubling time
- XO
xanthine oxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miquel J, Economos AC, Fleming J, Johnson JE., Jr Mitochondrial role in cell aging. Exp Gerontol. 1980;15:575–591. doi: 10.1016/0531-5565(80)90010-8. [DOI] [PubMed] [Google Scholar]
- 2.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 5.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang TT, Carlson EJ, Kozy HM, Mantha S, Goodman SI, Ursell PC, Epstein CJ. Genetic modification of prenatal lethality and dilated cardiomyopathy in Mn superoxide dismutase mutant mice. Free Radic Biol Med. 2001;31:1101–1110. doi: 10.1016/s0891-5849(01)00694-3. [DOI] [PubMed] [Google Scholar]
- 7.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 8.Mansouri A, Muller FL, Liu Y, Ng R, Faulkner J, Hamilton M, Richardson A, Huang TT, Epstein CJ, Van Remmen H. Alterations in mitochondrial function, hydrogen peroxide release and oxidative damage in mouse hind-limb skeletal muscle during aging. Mech Ageing Dev. 2006;127:298–306. doi: 10.1016/j.mad.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Kokoszka JE, Coskun P, Esposito LA, Wallace DC. Increased mitochondrial oxidative stress in the Sod2 (+/−) mouse results in the age-related decline of mitochondrial function culminating in increased apoptosis. Proc Natl Acad Sci U S A. 2001;98:2278–2283. doi: 10.1073/pnas.051627098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams MD, Van Remmen H, Conrad CC, Huang TT, Epstein CJ, Richardson A. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J Biol Chem. 1998;273:28510–28515. doi: 10.1074/jbc.273.43.28510. [DOI] [PubMed] [Google Scholar]
- 11.Dunn JA, Patrick JS, Thorpe SR, Baynes JW. Oxidation of glycated proteins: age-dependent accumulation of N epsilon-(carboxymethyl)lysine in lens proteins. Biochemistry. 1989;28:9464–9468. doi: 10.1021/bi00450a033. [DOI] [PubMed] [Google Scholar]
- 12.Glomb MA, Monnier VM. Mechanism of protein modification by glyoxal and glycolaldehyde, reactive intermediates of the Maillard reaction. J Biol Chem. 1995;270:10017–10026. doi: 10.1074/jbc.270.17.10017. [DOI] [PubMed] [Google Scholar]
- 13.Ochs S, Severin T. Reaction of 2′-deoxyguanosine with glyceraldehyde. Liebigs Ann Chem. 1994:851–853. [Google Scholar]
- 14.Seidel W, Pischetsrieder M. Immunochemical detection of N2-[1-(1-carboxy)ethyl]guanosine, an advanced glycation end product formed by the reaction of DNA and reducing sugars or L-ascorbic acid in vitro. Biochim Biophys Acta. 1998;1425:478–484. doi: 10.1016/s0304-4165(98)00101-9. [DOI] [PubMed] [Google Scholar]
- 15.Frischmann M, Bidmon C, Angerer J, Pischetsrieder M. Identification of DNA adducts of methylglyoxal. Chem Res Toxicol. 2005;18:1586–1592. doi: 10.1021/tx0501278. [DOI] [PubMed] [Google Scholar]
- 16.Schneider M, Georgescu A, Bidmon C, Tutsch M, Fleischmann EH, Popov D, Pischetsrieder M. Detection of DNA-bound advanced glycation end-products by immunoaffinity chromatography coupled to HPLC-diode array detection. Mol Nutr Food Res. 2006;50:424–429. doi: 10.1002/mnfr.200500217. [DOI] [PubMed] [Google Scholar]
- 17.Schneider M, Thoss G, Hubner-Parajsz C, Kientsch-Engel R, Stahl P, Pischetsrieder M. Determination of glycated nucleobases in human urine by a new monoclonal antibody specific for N2-carboxyethyl-2′-deoxyguanosine. Chem Res Toxicol. 2004;17:1385–1390. doi: 10.1021/tx049929d. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Nakamura S, Miyazaki S, Morita T, Suzuki M, Pischetsrieder M, Niwa T. N2-carboxyethyl-2′-deoxyguanosine, a DNA glycation marker, in kidneys and aortas of diabetic and uremic patients. Kidney Int. 2006;69:388–392. doi: 10.1038/sj.ki.5000064. [DOI] [PubMed] [Google Scholar]
- 19.Synold T, Xi B, Wuenschell GE, Tamae D, Figarola JL, Rahbar S, Termini J. Advanced glycation end products of DNA: quantification of N2-(1-Carboxyethyl)-2′-deoxyguanosine in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Chem Res Toxicol. 2008;21:2148–2155. doi: 10.1021/tx800224y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Cao H, Wang Y. Quantification of N2-carboxymethyl-2′-deoxyguanosine in calf thymus DNA and cultured human kidney epithelial cells by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope dilution method. Chem Res Toxicol. 2010;23:74–81. doi: 10.1021/tx900286c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breyer V, Becker C-M, Pischetsrieder M. Intracellular glycation of nuclear DNA, mitochondrial DNA, and cytosolic proteins during senescence-like growth arrest. DNA Cell Biol. 2011;30:681–689. doi: 10.1089/dna.2011.1236. [DOI] [PubMed] [Google Scholar]
- 22.Huang TT, Yasunami M, Carlson EJ, Gillespie AM, Reaume AG, Hoffman EK, Chan PH, Scott RW, Epstein CJ. Superoxide-mediated cytotoxicity in superoxide dismutase-deficient fetal fibroblasts. Arch Biochem Biophys. 1997;344:424–432. doi: 10.1006/abbi.1997.0237. [DOI] [PubMed] [Google Scholar]
- 23.Breyer V, Frischmann M, Bidmon C, Schemm A, Schiebel K, Pischetsrieder M. Analysis and biological relevance of advanced glycation end-products of DNA in eukaryotic cells. FEBS J. 2008;275:914–925. doi: 10.1111/j.1742-4658.2008.06255.x. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed MU, Thorpe SR, Baynes JW. Identification of N epsilon-carboxymethyllysine as a degradation product of fructoselysine in glycated protein. J Biol Chem. 1986;261:4889–4894. [PubMed] [Google Scholar]
- 26.Kelley EE, Khoo NK, Hundley NJ, Malik UZ, Freeman BA, Tarpey MM. Hydrogen peroxide is the major oxidant product of xanthine oxidase. Free Radic Biol Med. 2010;48:493–498. doi: 10.1016/j.freeradbiomed.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voziyan PA, Metz TO, Baynes JW, Hudson BG. A post-Amadori inhibitor pyridoxamine also inhibits chemical modification of proteins by scavenging carbonyl intermediates of carbohydrate and lipid degradation. J Biol Chem. 2002;277:3397–3403. doi: 10.1074/jbc.M109935200. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura S, Li H, Adijiang A, Pischetsrieder M, Niwa T. Pyridoxal phosphate prevents progression of diabetic nephropathy. Nephrol Dial Transplant. 2007;22:2165–2174. doi: 10.1093/ndt/gfm166. [DOI] [PubMed] [Google Scholar]
- 29.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 30.Van Remmen H, Salvador C, Yang H, Huang TT, Epstein CJ, Richardson A. Characterization of the antioxidant status of the heterozygous manganese superoxide dismutase knockout mouse. Arch Biochem Biophys. 1999;363:91–97. doi: 10.1006/abbi.1998.1060. [DOI] [PubMed] [Google Scholar]
- 31.Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benov L. How superoxide radical damages the cell. Protoplasma. 2001;217:33–36. doi: 10.1007/BF01289410. [DOI] [PubMed] [Google Scholar]
- 33.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 34.Huang TT, Carlson EJ, Raineri I, Gillespie AM, Kozy H, Epstein CJ. The use of transgenic and mutant mice to study oxygen free radical metabolism. Ann N Y Acad Sci. 1999;893:95–112. doi: 10.1111/j.1749-6632.1999.tb07820.x. [DOI] [PubMed] [Google Scholar]
- 35.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seidel W, Pischetsrieder M. DNA-glycation leads to depurination by the loss of N2-carboxyethylguanine in vitro. Cell Mol Biol (Noisy-le-grand) 1998;44:1165–1170. [PubMed] [Google Scholar]