Abstract
Intracellular inclusion bodies (IBs) containing ferritin and iron accumulation are hallmarks of hereditary ferritinopathy (HF). This neurodegenerative disease is caused by mutations in the coding sequence of the ferritin light chain (FTL) gene that generate FTL polypeptides with a C-terminus that is altered in amino acid sequence and length. Previous studies of ferritin formed with p.Phe167SerfsX26 mutant FTL (Mt-FTL) subunits found disordered 4-fold pores, iron mishandling, and pro-aggregative behavior, as well as a general increase in cellular oxidative stress when expressed in vivo. Herein we demonstrate that Mt-FTL is also a target of iron-catalyzed oxidative damage in vitro and in vivo. Incubation of recombinant Mt-FTL ferritin with physiological concentrations of iron and ascorbate resulted in shell structural disruption and polypeptide cleavage not seen with the wild type, as well as a 2.5-fold increase in carbonyl group formation. However, Mt-FTL shell disruption and polypeptide cleavage were completely inhibited by addition of the radical trap 5,5-dimethyl-1-pyrroline N-oxide. These results indicate an enhanced propensity of Mt-FTL toward free radical-induced, oxidative damage in vitro. We also found evidence of extensive carbonylation in IBs from a patient with HF together with isolation of a C-terminal Mt-FTL fragment, which are both indicative of oxidative ferritin damage in vivo. Our data demonstrate an enhanced propensity of mutant ferritin to undergo iron-catalyzed, oxidative damage and support this as a mechanism causing disruption of ferritin structure and iron mishandling that contributes to the pathology of HF.
Keywords: ferritin, iron, neurodegeneration, oxidative damage
INTRODUCTION
Iron is the most abundant transition metal in biological systems. The ability of iron to change its oxidation state and reduction potential through ligand binding enables it to serve multifunctional roles. However, the same physiochemical properties that allow iron to be crucial to many fundamental cellular reactions also may involve it in destructive modification of proteins (1). Specifically, Fenton- and Haber-Weiss-type reactions generate reactive oxygen species (ROS), which attack proteins and cause them to undergo iron-catalyzed, oxidative damage. The hydroxyl radical (HO•), superoxide radical anion (O2•−), and hydrogen peroxide (H2O2) can all be produced in aqueous solutions of iron salts (2, 3), depending on the absence, presence and identity of chelating ligands. Even metalloenzymes that normally perform redox reactions can “misfire” producing ROS. Virtually all amino acids are potential targets for such damage, but due to high reactivity, the HO• reacts mainly near the iron binding sites of proteins where it is generated (4). Iron-catalyzed, oxidative damage can result in carbonylated amino acid side chains, cleavage of the protein peptide bond, and even covalent crosslinking (5, 6), of which any of these could diminish protein function. Carbonylation and crossinking could, if sufficient, also hinder protein clearance and cause cellular accumulation (7, 8). In addition, the hydroxyl radical-generating system of iron and ascorbate has been used as a site-specific probe of iron binding to proteins through polypeptide cleavage near the binding site by causing formation of polypeptide fragments of predictable lengths. The finding of oxidative damage to proteins by reactive ROS in many neurodegenerative diseases such as Alzheimer disease (AD), Parkinson disease (PD) and Huntington disease (HD) has led to the hypothesis that oxidative stress plays a major role in the pathogenesis of these diseases (9–15).
Most of the iron found in living organisms is tightly complexed to proteins. Ferritin, the main intracellular iron storage protein, plays a central role in cellular iron metabolism (16). It is considered also part of the cellular antioxidant response due to its ability to store and transform potentially toxic ferrous (Fe(II)) iron into a bioavailable but non-toxic ferric (Fe(III)) form. Mammalian ferritin is a 24-mer heteropolymer (~480 kDa) that forms a spherical shell composed of two subunit types originating from ferritin light chain (FTL) and heavy chain (FTH1) polypeptides. Although close to identical in overall conformation, both subunit types have only an ~50% sequence homology and show diverse functional roles. The FTH1 subunit (~21 kDa) has the ferroxidase active site which catalyzes the oxidation of Fe(II) to Fe(III), whereas the FTL subunit (~20 kDa) plays an important role in iron nucleation to form the Fe(III) oxy core and in protein stability (16). The two ferritin polypeptides are folded into a bundle of 4 long parallel α helices (A, B, C, D) and a short C-terminal α helix (E). In the 24-mer the subunits are structurally related by 4-, 3-, and 2-fold symmetry axes and pack tightly together except at the 3-and 4-fold axes where there are small pores traversing the shell, with the C-terminal helix (helix E) outlining the 4-fold pore. Because of the conformational equivalence, the FTL and FTH1 subunits can combine at different ratios preserving the 3-dimensional structure of the 24-mer and offering the possibility of regulating independently the iron storage capacity and the number of catalytic sites (16–21).
Nucleotide duplications in exon 4 of the FTL gene (22–27) are associated with a fatal autosomal dominant neurodegenerative disease named Neuroferritinopathy (22) or hereditary ferritinopathy (HF) (23, 24, 28). HF affects the central nervous system (CNS) presenting clinically as an extra-pyramidal movement disorder accompanied by cognitive and behavioral disturbances which start between the third and sixth decade of life. Neuropathologically, the disease is characterized by the presence of intracellular ferritin inclusion bodies (IBs) and iron accumulation in glia and neurons throughout the CNS and also in other organ systems such as liver, kidney and skin (23, 24). Thus far, all known nucleotide duplications in the FTL gene give rise to the generation of FTL polypeptides that are altered in length and amino acid C-terminal sequence to different extents depending on the specific mutation (28, 29).
We previously presented structural and functional studies on ferritin containing the mutant FTL p.Phe167SerfsX26 (Mt-FTL) polypeptide derived from the FTL(498–499)InsTC (c.497_498dupTC) mutation (30–32), in which the last 9 amino acids of the wild-type FTL (Wt-FTL) polypeptide (FERLTLKHD) are replaced by an unrelated sequence of 25 residues (SSKGSLSSTTKSLLSPATSEGPLAK). Mt-FTL subunits lack the C-terminal E-helix, which causes severe disordering of ferritin 4-fold pores (31), leading to iron mishandling and enhanced iron-mediated aggregation of the ferritin 24-mer in vitro (30–32). Transgenic mice expressing the p.Phe167SerfsX26 mutant polypeptide exhibit several pathological features of HF, including the presence of ferritin-containing IBs in neurons and glia in the CNS, and in cells of other organ systems (33). The mouse model also shows abnormal iron metabolism and increased oxidative stress leading to a general increase in protein oxidative damage, which can also be observed independently in a study of fibroblasts derived from a patient with HF (34, 35).
Here, we describe for the first time the increased 24-mer shell disruption, polypeptide cleavage, and carbonylation of Mt- versus Wt-FTL recombinant ferritins caused by addition of physiological concentrations of iron and ascorbate. This study differs from those previously done on Mt-FTL by employing very low iron concentrations and the redox cycling of iron by reductant. Oxidative damage was prevented by the addition of the free radical trap 5,5-dimethyl-1-pyrroline N-oxide (DMPO), which implies iron-catalyzed, radical formation as the causative mechanism disrupting mutant ferritin structure and function. The significance of these in vitro observations is enhanced by finding extensive protein carbonylation in ferritin IBs from a HF patient, as well as the isolation of an ~14k Da C-terminal Mt-FTL fragment from the IBs. Taken together, our data strongly support iron-catalyzed oxidation of mutant ferritin leading to irreversible structural damage and iron mishandling as a contributing factor to the pathology of HF.
MATERIALS AND METHODS
Recombinant ferritin
Wt- and Mt-FTL polypeptides were expressed in E. Coli, purified, and assembled into ferritin homopolymers (24-mers) as we described previously (30). Protein concentration was determined using the BCA reagent (Pierce) with bovine serum albumin (BSA) as standard. For iron removal, recombinant homopolymers were incubated with 1% thioglycollic acid, pH 5.5, and 2,2'-bipyridine as previously described (30, 36). Homopolymers (5 mg/ml) were dissolved in 100 mM Hepes buffer (pH 7.4) that was treated with Chelex ion exchange resin (Bio-Rad) to remove transition metals usually found in aqueous buffer as contaminants.
Fe/Asc-treatment of recombinant ferritin
Recombinant Wt- and Mt-FTL apoferritin homopolymers (0.5 µM) were separately incubated in 100 mM Hepes buffer (pH 7.4) with freshly prepared 2 mM ascorbate and ferrous ammonium sulphate (2, 4, 6 and 8 µM) previously diluted in 10 mM HCl. When used, high purity DMPO (50 mM) (Alexis) was added to the reaction mixture. Incubation of these reaction mixtures was performed in the dark at 24°C for 1 h. The iron/ascorbate (Fe/Asc)-treated homopolymers were then analyzed by native electrophoresis and western blotting, or used for carbonyl content assays as described below.
Electrophoresis and western blots
The ferritin reaction mixtures were resolved in 3–8% native gels (Invitrogen) or 4–20% gradient SDS-PAGE (Pierce). Proteins were stained with SimplyBlue (Invitrogen), or electro-blotted onto nitrocellulose membranes (Amersham Biosciences). Western blots using polyclonal antibodies 1283 (Mt-FTL C-terminus) or 1278 (Wt-FTL C-terminus) were performed as described previously (23, 30). Membranes were then incubated with peroxidase-conjugated secondary antibodies (1:5000; GE Healthcare) for 1 h. Membranes were developed using the ECL chemiluminescent detection system (GE Healthcare).
Detection of protein carbonyl formation in recombinant ferritin
After Fe/Asc treatment of ferritin in vitro, protein carbonyl groups were detected by western blot (37) using the Oxyblot protein oxidation kit (Millipore) according to the manufacturer instructions. Briefly, samples were derivatized with 2,4-dinitrophenylhydrazine (DNP-H) for 15 min at room temperature. Afterwards, the neutralization solution was added. Samples were then separated by 4–20% gradient SDS-PAGE (Pierce) and transferred onto nitrocellulose membranes (Amersham Biosciences). Membranes were blocked for 1 h in 70 mM PBS, 0.1% tween 20, and 5% nonfat dry milk, followed by 1 h incubation with rabbit anti-dinitrophenyl (anti-DNP) antibodies at 1:200. After washing, membranes were incubated with peroxidase-conjugated secondary antibody (GE Healthcare) for 1 h, washed, and developed using the ECL chemiluminescent detection system.
Protein carbonyl quantification in recombinant ferritin
Protein carbonyl concentration was measured by ELISA as described previously (38), using a commercial kit (Biocell) following the manufacturer instructions. Briefly, after Fe/Asc treatment and derivatization as described above, 3 µg of Wt- and Mt-FTL homopolymers were diluted in 300 µl of buffer containing 10 mM sodium carbonate and 150 mM NaCl (pH 9.0). Samples were adsorbed to Nunc Immuno Maxisorp Plates overnight. Wells were blocked with the diluted blocking solution (200 µl/well) for 2 h at room temperature. The diluted anti-DNP-biotin-antibody solution (200 µl per well) was added and incubated for 1 h at 37°C. After washing, 200 µl of the solution containing streptavidin-horseradish peroxidase was added and incubated for an additional hour at room temperature. After extensive washing, 200 µl per well of the chromatin reagent was added. After 10 min, the reaction was stopped with 100 µl of the stopping reagent and absorbance was recorded at 450 nm using a BioTek EL800 microplate reader (BioTek Instruments, Inc). The assay was repeated three times for each iron concentration. A six-point standard curve assayed in triplicate of reduced and oxidized BSA was included with each plate. A blank for DNP reagent in PBS without protein was subtracted from all other absorbance values.
Isolation of ferritin IBs from brain
Inclusion bodies were isolated from post-mortem tissue from an individual with HF as described previously (23). Briefly, fresh frozen samples from the putamen were dissected, minced into 1 to 3 mm pieces and placed in Dulbecco’s PBS (D-PBS; Sigma) and protease inhibitors (Complete; Roche Molecular Biochemical) on ice. Tissue was then washed by re-suspension in D-PBS and clarified by centrifugation. The soluble material was stored as soluble fraction. The insoluble material was re-suspended and digested with collagenase and DNase I. After digestion, the insoluble undigested material was subjected to 3 cycles of detergent wash. The insoluble material was then solubilized in formic acid, centrifuged, and the supernatant dried under nitrogen gas to form the insoluble fraction.
Carbonyl detection in brain
Carbonyl detection was performed on paraffin-embedded sections of a patient with HF (23) as described previously (39). Briefly, brain sections were sequentially deparaffinized in xylene and rehydrated in descending ethanol, followed by blocking of endogenous peroxidase. For DNP labeling, sections were incubated with 0.1% DNP-H (Sigma) in 2 N HCl for 1 h at room temperature. Sections were rinsed in buffer containing 50 mM Tris-HCl and 150 mM NaCl (pH 7.6) (TBS), followed by blocking of non-specific binding sites for 1 h at room temperature in 10% normal goat serum (NGS) in TBS. After rinsing, sections were incubated with monoclonal antibody (LO-DNP-2; Zymed) against dinitrophenyl dinitrophenol (1:100) in 1% NGS/TBS for 16 h at 4°C. Sections were then rinsed with 1% NGS/TBS followed by detection using a secondary biotinylated goat anti-rat antibody and streptavidin–horseradish peroxidase (BD Biosciences). Development of the sections was directly monitored under an ×10 objective. Slides were counter-stained with hematoxylin and images were captured by a digital camera coupled to a Leica DM4000B microscope (Leica Microsystems). The specificity of the antibodies was verified by omitting the DNP-H incubation step.
RESULTS
Enhanced Fe/Asc-mediated shell disruption and polypeptide cleavage of recombinant mutant ferritin
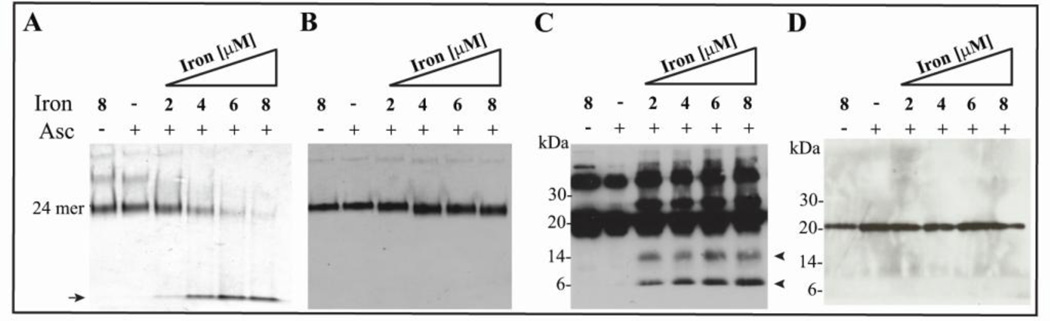
Because ferritin that contain Mt-FTL subunits has a reduced ability to incorporate and nucleate iron relative to the corresponding wild type (30–32), we determined whether addition of iron and ascorbate together could catalyze oxidative modification of Mt-FTL. The Fe/Asc treatment led to a significant decrease in the amount of 24-mers Mt-FTL homopolymers as observed by native electrophoresis in an iron concentration-dependent manner (Fig. 1A). No significant change in the amount of 24-mers of Mt-FTL homopolymers was observed when the protein was incubated with iron or ascorbate separately. Furthermore, Wt-FTL homopolymers did not show any significant change in the amount of 24-mers when incubated under the same conditions as the mutant (Fig. 1B). In addition to the band corresponding to 24-mers, native gels also showed a lower band, apparently originating from the disrupted 24-mer shells, that correlated with increasing iron concentration with Mt-FTL (Fig. 1A, arrow) but not with Wt-FTL homopolymers (Fig. 1B). When the experiment was repeated under identical conditions and proteins were examined on SDS-PAGE, we observed along with the full-length Mt-FTL polypeptide (~21 kDa), the presence of C-terminal fragments of ~14 and ~6 kDa (Fig. 1C, arrowheads). An additional band was also seen at ~27 kDa (Fig. 1C) suggesting covalent crosslinking between full-length Mt-FTL polypeptides (~21 kDa) and fragments of ~6 kDa. In agreement with the results shown in Fig. 1B, no fragments were observed when Wt-FTL homopolymers were analyzed under identical conditions on SDS-PAGE (Fig. 1D). The ~ 40 kDa band seen with SDS-PAGE in Fig 1C is often found in purified Mt-FTL homopolymer preparations and can be attributed to a dimeric form of the polypeptide (23).
Figure 1. Shell disruption and polypeptide cleavage of recombinant Mt-FTL homopolymers in the presence of iron and ascorbate.
Wt- and Mt-FTL apoferritin homopolymers (0.5 µM) were incubated with increasing concentrations of ferrous ammonium sulphate (2, 4, 6 and 8 µM) in Hepes buffer (100 mM, pH 7) in the presence of ascorbate (2 mM) for 1 h in the dark at 24°C. Homopolymers (0.5 µM) were also incubated with iron (8 µM) but without ascorbate (lane 1), and with ascorbate (2 mM) without iron (lane 2). Native-PAGE of Mt-FTL (A) and Wt-FTL (B) homopolymers stained with Coomasie blue. Western blot analysis of Mt-FTL homopolymers (C) and Wt-FTL homopolymers (D) after SDS-PAGE using an antibody specific for the C-terminus of Mt-and Wt-FTL (23).
Radical trap prevents shell disruption and polypeptide cleavage of recombinant mutant ferrtin
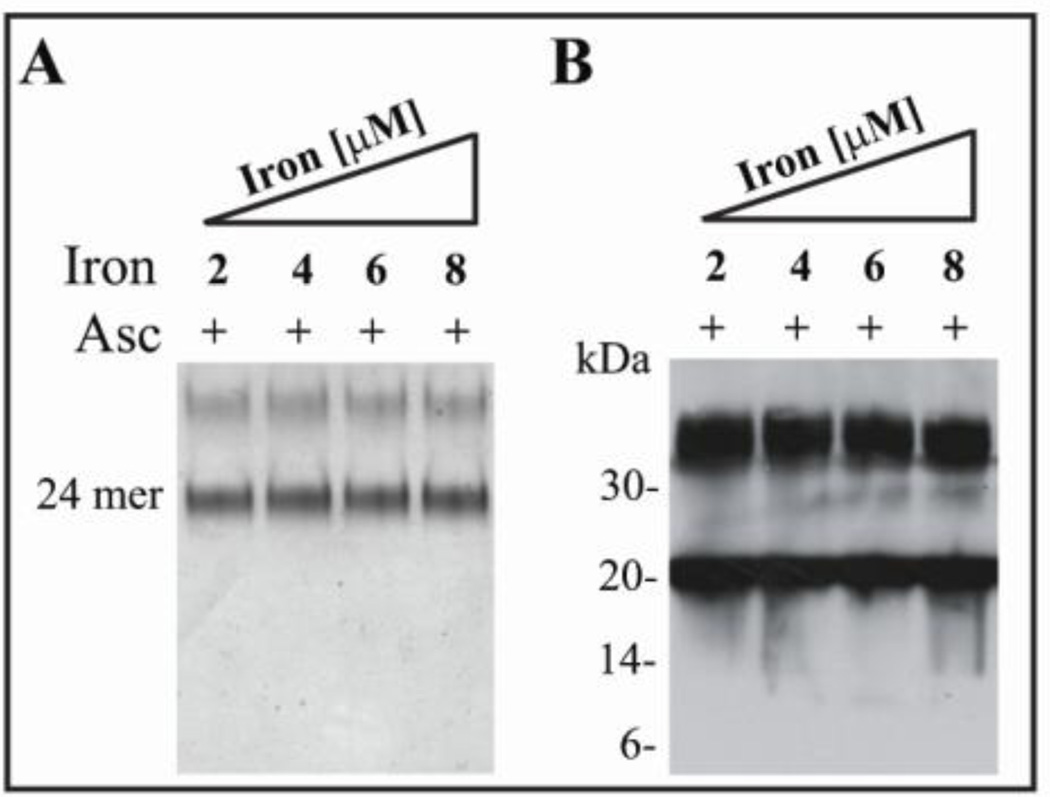
The nitroxide radical trap DMPO can trap both hydroxyl and protein free radicals in vitro and in vivo (40), thus preventing or at least localizing overall protein oxidative damage. We have previously shown that systemic administration of DMPO to FTL-transgenic mice leads to the generation of DMPO adducts associated with ferritin IBs (34). Addition of DMPO to the Fe/Asc-containing reaction mixture completely prevented Mt-FTL 24-mer shell disruption and polypeptide cleavage at all iron concentrations used previously, as revealed by non-denaturing electrophoresis (Fig. 2A) and SDS PAGE (Fig. 2B). The denaturing gel showed bands corresponding to monomers (~21 kDa) and dimers (~42 kDa) of Mt-FTL subunits (Fig. 2B), but no bands of lower molecular weight fragments as were observed when DMPO was absent (Fig. 1C).
Figure 2. Prevention of shell disruption and polypeptide cleavage of recombinant Mt- FTL homopolymers by DMPO.
Mt-FTL apoferritin homopolymers (0.5 µM) were incubated with increasing concentrations of iron (2, 4, 6 and 8 µM) in the presence of ascorbate (2 mM) and DMPO (50 mM). Samples were electrophoresed with native PAGE and stained with Coomasie blue (A) or in SDS-PAGE and blotted using an antibody specific for the C-terminus of Mt-FTL (B).
Enhanced Fe/Asc-mediated carbonyl formation in recombinant mutant ferritin
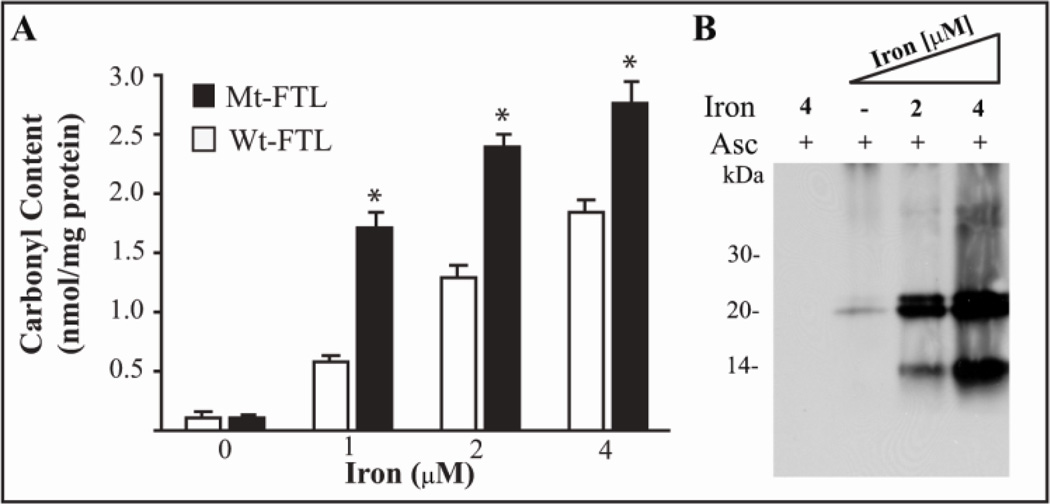
Carbonylation of recombinant Wt- and Mt-FTL homopolymers was examined to determine any difference in propensity for iron-catalyzed oxidation by exposure to iron with ascorbate. Quantitative analysis of protein carbonylation by ELISA showed a statically significant, iron-dependent increase in carbonyl group formation in recombinant Mt-FTL homopolymers compared to Wt-FTL homopolymers (Fig. 3A). Analysis of the samples by western blot after SDS-PAGE showed strong immunoreactivity against carbonylated residues in the Mt-FTL full-length polypeptide (~21 kDa) and a C-terminal fragment of ~14 kDa (Fig. 3B) compared to a weak immunoreactive signal in full-length Wt-FTL (not shown), localizing more specifically the enhanced propensity in the mutant.
Figure 3. Protein carbonyl group formation in recombinant ferritin by treatment with iron and ascorbate.
Wt- and Mt-FTL apoferritin homopolymers (0.5 µM) were incubated with increasing concentrations of iron (0, 1, 2, and 4 µM) in the presence of ascorbate (2 mM) and then derivatized with DNP-H. Carbonyl content was quantified by ELISA. Asterisks indicate statistically significant differences (p<0.05) between Mt- and Wt-FTL carbonyls content (A). Carbonyl groups were detected on Mt-FTL samples by western blot after SDS-PAGE using an antibody specific for DNP As controls, recombinant ferritins were incubated with iron (4 uM) and ascorbate (2 mM) but not treated with DNPH (lane 1) (B).
Carbonylation and polypeptide cleavage in ferritin IBs from HF brain
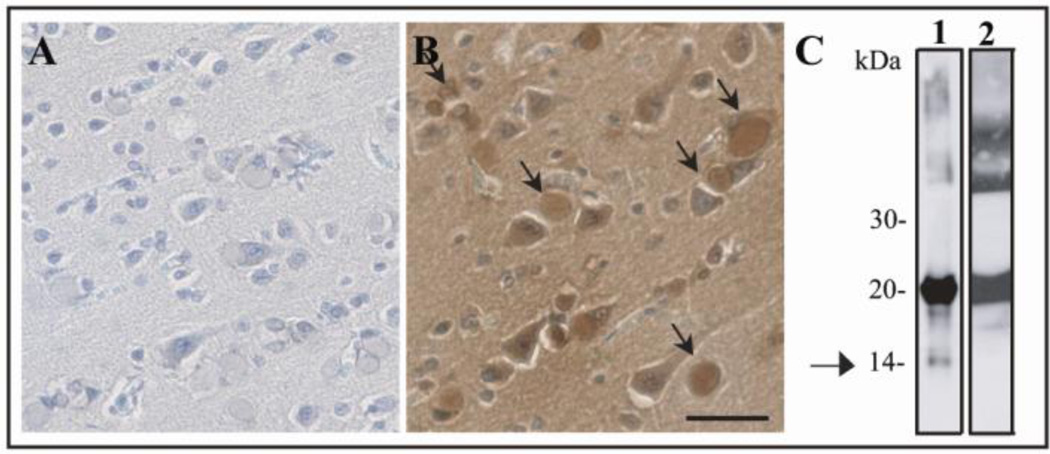
The presence of oxidative damage in Mt-FTL in vivo was assessed by immunohistochemistry. No immunostaining was observed when the tissue was not labeled with DNP (Fig. 4A). Carbonyl group immunopositivity was observed in ferritin IBs after labeling with DNP (Fig. 4B, arrows). To further characterize ferritin aggregates, IBs were isolated as previously described (23, 33), purified into soluble and insoluble fractions, run on SDS-PAGE, and blotted against an antibody specific for the C-terminus of Mt-FTL (23). While both soluble and insoluble fractions contained full-length Mt-FTL polypeptide (~21 kDa), only the insoluble fraction showed the presence of a Mt-FTL C-terminal fragment of ~14 kDa (Fig. 4C, arrow). An ~14 kDa C-terminal polypeptide was also observed with Fe/Asc treatment of recombinant Mt-FTL homopolymers as described above.
Figure 4. Protein carbonylation and Mt-FTL polypeptide cleavage in IBs from a HF patient carrying the c.497_498dupTC mutation.
Immunohistochemistry for carbonyl group detection was performed in post-mortem brain tissue from a patient with HF, in which sections were not treated (A) or treated (B) with DNP-H. Carbonyl immunoreactivity was found co-localized with ferritin IBs (arrows). Bar: 25 µm. Western blot analysis (C) after SDS-PAGE of insoluble (lane 1) and soluble (lane 2) fractions obtained from IBs isolated from the putamen of the same patient blotted using an antibody specific for the C-terminus of Mt-FTL (23). The arrow indicates the presence of an ~14 kDa Mt-FTL C-terminal fragment visible only in the insoluble fraction.
DISCUSSON
Hereditary ferritinopathy is caused by expression of a mutant form of the FTL polypeptide and is characterized pathologically by accumulation of iron and formation of intracellular IBs containing ferritin (29). Proper cellular iron handling and storage, which includes the controlled oxidation of Fe(II), is the role of ferritin itself and any misfunction may cause iron mishandling, which can lead to pathological consequences. Ferritin that contains the Mt-FTL subunit is unable to appropriately sequester and store iron in vitro and apparently also in vivo as evident by increased levels of iron in brain from Mt-FTL transgenic mice (34) and fibroblasts from a patient with HF (35). Previously we had proposed that in HF, this altered iron metabolism caused a compensatory over-production of ferritin polypeptides, which contributed to the formation of ferritin aggregates sequestered into IBs (28). Biochemical analysis of IBs and the use of antibodies raised to detect specifically the C-terminus of mutant and wild type FTL, demonstrated that IBs contain all three ferritin polypeptides (Mt-FTL, Wt-FTL and FTH1) (23), both in patients with HF and in a transgenic model (23, 33) implying involvement further than just that of the mutant polypeptide. The enhanced aggregation and precipitation of ferritin 24-mers that contained Mt-FTL over those with Wt-FTL subunits was demonstrated previously in vitro under routine iron-loading conditions using Fe (II) of ~1 mM concentration without additional added reductant, and such studies were explained by aggregation of ferritin 24-mers through iron bridging of mutant, unraveled C-termini (30). Considering both the overproduction of ferritin and increased iron levels in HF, this mechanism is consistent with a process contributing to the formation of IBs. However, other mechanisms proceeding in parallel may also contribute to IB formation and ferritin loss of function.
The current study examined differences in behavior between mutant and wild type recombinant ferritins, but under the substantially different reaction conditions of low micromolar iron concentrations and excess available reductant. The iron concentrations chosen were low physiological (41) and the ascorbate concentration was slightly below the average of that in the brain (42). Our work clearly uncovered an enhanced propensity of Mt-FTL over Wt-FTL homopolymers toward iron-catalyzed damage in the form of 24-mer shell disruption, carbonyl formation, polypeptide cleavage, and some covalent crosslinking. These forms of damage are self-consistent in that protein charge change and decreased motional constraints during carbonylation and polypeptide cleavage are expected to contribute to protein structural disruption. The observed shell disruption of Mt-FTL and not Wt-FTL homopolymers is significant in that this clearly can lead to dysfunctional iron storage, but it is also surprising in that the ferritin 24-mer is in general a very stable structure and tolerant to perturbations such as denaturant exposure, high temperature, and even minor proteolysis (30). Furthermore the FTL subunit is designed to aid in overall ferritin 24-mer stability and to protect ferritin from the aforementioned iron loading-induced aggregation (30), and it is the FTH1 subunit that is generally found to be susceptible to backbone cleavage during turnover (43). These factors amplify the negative cellular consequences of Mt-FTL subunit overproduction and incorporation into the ferritin 24-mers. Importantly, addition of the radical trap DMPO to the Fe/Asc reaction mixture prevented completely the Mt-FTL homopolymers from undergoing shell disruption and polypeptide cleavage, which directly implicates iron-catalyzed, radical generation as the causative agent for ferritin damage. Specifically, DMPO is a well-known nitroxide spin trap that reacts with a variety of radicals like HO• in solution preventing them from reacting and modifying protein (44). Furthermore, radicals that have been transferred to the protein form DMPO–protein radical adducts preventing polypeptide cleavage and crosslinking. Prevention of shell collapse and polypeptide cleavage by DMPO is consistent with its action as a radical trap both of HO• in solution and of protein-based radicals. The latter is in agreement with our previous in vivo data showing the presence of trapped radicals after systemic administration of DMPO to Mt-FTL transgenic mice (34). Here, ferritin-DMPO adducts were localized in inclusions as judged by immunohistochemistry and were detected on western blots after immunoprecipitation of adducts from tissue samples (34).
The current study employed iron concentrations sub-stoichiometric to the number of ferritin subunits. Under these conditions, iron should associate with a limited number of tight-binding protein sites, which could in principle be on the ferritin shell outer surface or the interior. Radical generation and damage should favor specific sites of iron association that are accessible both to iron and the more bulky ascorbate. Each ferritin subunit contains two helices (B and D) that face toward the interior of the shell with a portion of the latter forming the interior of the 3-fold pore, and metal binding is known to occur on sites along both of these helices in FTL by X-ray diffraction (45). The iron nucleation site is located on the B helix (approximately residue 60) of each FTL subunit and is composed of 4 glutamic acid residues containing carboxylic acids, which are strong iron binders (46). Formation of HO• and polypeptide cleavage at the nucleation site residues would result in an ~14 kDa C-terminal fragment as we observed by SDS PAGE. The observed ~6 kDa C-terminal fragment could result from HO• diffusion to and cleavage in the D helix at a site in close proximity to the nucleation site (approximately residue 140), or it could result by direct iron binding to carboxylic acids, HO• generation, and cleavage near residue 140, because of the three carboxylic acid residues in a row there. Alternatively, there are pore and shell carboxylic acids between residues 130 and 140 on the D helix that could serve as iron binding sites for HO• generation. In any case, the length of both observed ferritin fragments can be accounted for by iron binding to the Mt-FTL homopolymer at known metal binding sites. Polypeptide cleavage at these sites with Mt-FTL but not Wt-FTL homopolymers is also consistent with more restricted access by ascorbate to the shell interior with Wt-FTL homopolymers, but less restriction with the mutant. Here the disordered Mt-FTL 4-fold pores would allow ascorbate into the shell interior to redox cycle the iron and freely generate HO• in contrast to the pores of Wt-FTL, which are relatively small, i.e., ~2 Å for the 4-fold and ~3.4 Å for the 3-fold pores (47). Since the potential cleavage site(s) is located N-terminus from the altered C-terminus of Mt-FTL polypeptides, we hypothesize that polypeptide cleavage may occur in all mutant forms of FTL associated with HF and may play a role in the pathogenesis of HF (29).
The finding of enhanced propensity for Mt-FTL homopolymers to undergo iron-catalyzed, oxidative damage suggested that similar results might be found in IBs from HF brain. Increased levels of protein carbonyls have been found with many neurodegenerative diseases such as AD and PD, and often correlate well with the progression of the disease (9–11, 44, 48–50). We found that the IBs from human HF brain exhibited extensive carbonylation in agreement with our in vitro Mt-FTL results. Our data support our previous finding of extensive protein oxidation in the brain of Mt-FTL transgenic mice (34). These data are significant for the pathogenesis of HF because highly carbonylated proteins are more resistant to proteolytic degradation by the ubiquitin-proteasome system (51, 52) and likely to accumulate. In addition, an ~14 kDa C-terminal fragment was isolated from the insoluble fraction purified from IBs from the HF patient paralleling the C-terminal cleavage product produced from Mt-FTL homopolymers with Fe/Asc treatment. Our in vitro and in vivo results when taken together suggest that IBs contain in part iron catalyzed, oxidatively damaged mutant ferritin from the structurally disrupted shells and cleavage fragments characterized here, in addition to aggregated shells caused by iron bridging of the C-termini, as described earlier (23, 30).
In conclusion, we have (1) demonstrated an enhanced propensity for Mt-FTL homopolymers to undergo iron-catalyzed, oxidative damage from exposure to low concentrations of iron (with ascorbate), (2) found markers for oxidative damage in HF brain, and (3) identified a Mt-FTL ~14 kDa C-terminal fragment both in vitro with Fe/Asc-treated recombinant ferritin, and in vivo from IBs from a HF patient. Apparently, Mt-FTL is highly vulnerable to iron-catalyzed oxidation at physiological concentrations of iron, leading to shell disruption and carbonylation, and thus to both the inability to appropriately store iron and the production of modified protein, which is in general difficult to clear. These molecular level insights into the pathogenesis of HF may provide important clues into the role of ferritin and iron in more common neurodegenerative diseases such as AD, PD and HD and may increase our knowledge of the cellular metabolism of ferritin in the brain in health and disease.
Highlights.
Mutations in ferritin light chain gene make the molecule susceptible to oxidation
The oxidation of mutant ferritin causes loss of ferritin structure
Iron-catalyzed oxidation of mutant ferritin causes polypeptide cleavage
Oxidative modifications may contribute inclusion body formation and neuropathology
Oxidative damage of mutant ferritin is a novel pathogenic mechanism in ferritinopathy
ACKNOWLEDGMENTS
The work was supported by National Institutes of Health Grants NS050227 and NS063056 (to R. V.). The authors are thankful to Luis Barbeito, and Joseph Beckman for insightful comments and helpful discussions, and Bernardino Ghetti and Marie Bernadette Delisle for tissue samples.
ABBREVIATIONS
The abbreviations used are:
- ROS
reactive oxygen species
- AD
Alzheimer disease
- PD
Parkinson disease
- HD
Huntington disease
- FTL
ferritin light chain
- FTH1
ferritin heavy chain
- Wt-FTL
wild–type ferritin light chain
- Mt-FTL
mutant ferritin light chain
- CNS
central nervous system
- IBs
inclusion bodies
- DMPO
5,5-dimethyl-1-pyrroline N-oxide
- BSA
bovine serum albumin
- DNP-H
2,4-dinitrophenylhydrazine
- DNP
dinotrophenyl
- TBS
Tris-buffered saline
- NGS
normal goat serum
- Fe/Asc
iron and ascorbate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Stadtman ER, Berlett BS. J Biol Chem. 1991;266:17201–17211. [PubMed] [Google Scholar]
- 2.Fenton HJH. J Chem Soc Trans. 1896;69:546–562. [Google Scholar]
- 3.Haber F, Weiss J. Naturwiss. 1932;51:948–950. [Google Scholar]
- 4.Stadtman ER. Free Radical Biol. Med. 1990;9:315–325. doi: 10.1016/0891-5849(90)90006-5. [DOI] [PubMed] [Google Scholar]
- 5.Stadtman ER, Berlett BS. Drug Metab Rev. 1998;30:225–243. doi: 10.3109/03602539808996310. [DOI] [PubMed] [Google Scholar]
- 6.Berlett BS, Stadtman ER. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 7.Farout L, Friguet B. Antioxid Redox Signal. 2006;8(1–2):205–216. doi: 10.1089/ars.2006.8.205. [DOI] [PubMed] [Google Scholar]
- 8.Grune T, Jung T, Merker K, Davies KJ. J Biochem Cell Biol. 2004;12:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Sultana R, Perluigi M, Butterfield DA. Antioxid Redox Signal. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 10.Butterfield DA, Perluigi M, Sultana R. Eur J Pharmacol. 2006;545:39–50. doi: 10.1016/j.ejphar.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Danielson SR, Andersen JK. Free Radic Biol Med. 2008;44:1787–1794. doi: 10.1016/j.freeradbiomed.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keeney PM, Xie J, Capaldi RA, Bennett JP., Jr J Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Free Radic Biol Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, Calabrese V, Cini C, De Marco C, Butterfield DA. Mol Cell Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Stack EC, Matson WR, Ferrante RJ. Ann N Y Acad Sci. 2008;1147:79–92. doi: 10.1196/annals.1427.008. [DOI] [PubMed] [Google Scholar]
- 16.Harrison PM, Arosio P. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 17.Theil EC, Matzapetakis M, Liu X. J Biol Inorg Chem. 2006;11:803–810. doi: 10.1007/s00775-006-0125-6. [DOI] [PubMed] [Google Scholar]
- 18.Theil EC. J Nutr. 2003;133:S1549–S1553. [Google Scholar]
- 19.Liu X, Theil EC. Acc Chem Res. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 20.Hempstead PD, Yewdall SJ, Fernie AR, Lawson DM, Artymiuk PJ, Rice DW, Ford GC, Harrison PM. J Mol Biol. 1997;268:424–448. doi: 10.1006/jmbi.1997.0970. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Li C, Ellenburg M, Soistman E, Ruble J, Wright B, Ho JX, Carter DC. Acta Crystallogr. Sect D Biol Crystallogr. 2006;62:800–806. doi: 10.1107/S0907444906018294. [DOI] [PubMed] [Google Scholar]
- 22.Curtis AR, Fey C, Morris CM, Bindoff LA, Ince PG, Chinnery PF, Coulthard A, Jackson MJ, Jackson AP, McHale DP, Hay D, Barker WA, Markham AF, Bates D, Curtis A, Burn J. Nat Genet. 2001;28:350–354. doi: 10.1038/ng571. [DOI] [PubMed] [Google Scholar]
- 23.Vidal R, Ghetti B, Takao M, Brefel-Courbon C, Uro-Coste E, Glazier BS, Siani V, Benson MD, Calvas P, Miravalle L, Rascol O, Delisle MB. J Neuropathol Exp Neurol. 2004;63:363–380. doi: 10.1093/jnen/63.4.363. [DOI] [PubMed] [Google Scholar]
- 24.Mancuso M, Davidzon G, Kurlan RM, Tawil R, Bonilla E, Di Mauro S, Powers JM. J Neuropathol Exp Neurol. 2005;64:280–294. doi: 10.1093/jnen/64.4.280. [DOI] [PubMed] [Google Scholar]
- 25.Ohta E, Nagasaka T, Shindo K, Toma S, Nagasaka K, Ohta K, Shiozawa Z. Neurology. 2008;70:1493–1494. doi: 10.1212/01.wnl.0000310428.74624.95. [DOI] [PubMed] [Google Scholar]
- 26.Kubota A, Hida A, Ichikawa Y, Momose Y, Goto J, Igeta Y, Hashida H, Yoshida K, Ikeda S, Kanazawa I, Tsuji S. Mov Disord. 2009;24:441–445. doi: 10.1002/mds.22435. [DOI] [PubMed] [Google Scholar]
- 27.Devos D, Tchofo PJ, Vuillaume I, Destee A, Batey S, Burn J, Chinnery PF. Brain. 2009;132:e109. doi: 10.1093/brain/awn274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidal R, Delisle MB, Ghetti B. J Neuropathol Exp Neurol. 2004;63:787–800. doi: 10.1093/jnen/63.8.787. [DOI] [PubMed] [Google Scholar]
- 29.Vidal R, Delisle MB, Rascol O, Ghetti B. In: Neurodegeneration: The Molecular Pathology of Dementia and Movement Disorders. Dickson DW, Weller RO, editors. 2011. pp. 461–466. [Google Scholar]
- 30.Baraibar MA, Barbeito AG, Muhoberac BB, Vidal R. J Biol Chem. 2008;283:31679–31689. doi: 10.1074/jbc.M805532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baraibar MA, Muhoberac BB, Garringer HJ, Hurley TD, Vidal R. J Biol Chem. 2010;285:1950–1956. doi: 10.1074/jbc.M109.042986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhoberac BB, Baraibar MA, Vidal R. Biochim Biophys Acta. 2011;1812:544–548. doi: 10.1016/j.bbadis.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidal R, Miravalle L, Gao X, Barbeito AG, Baraibar MA, Hekmatyar SK, Widel M, Bansal N, Delisle MB, Ghetti B. J Neurosci. 2008;28:60–67. doi: 10.1523/JNEUROSCI.3962-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbeito AG, Garringer HJ, Baraibar MA, Gao X, Arredondo M, Nuñez MT, Smith MA, Ghetti B, Vidal R. J Neurochem. 2009;109:1067–1078. doi: 10.1111/j.1471-4159.2009.06028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbeito AG, Levade T, Delisle MB, Ghetti B, Vidal R. Mol Neurodegener. 2010;10:5–50. doi: 10.1186/1750-1326-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levi S, Luzzago A, Cesareni G, Cozzi A, Franceschinelli F, Albertini A, Arosio P. J Biol Chem. 1988;263:18086–18092. [PubMed] [Google Scholar]
- 37.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 38.Buss H, Chan TP, Sluis KB, Domigan NM, Winterbourn CC. Free Radic Biol Med. 1997;23:361–366. doi: 10.1016/s0891-5849(97)00104-4. [DOI] [PubMed] [Google Scholar]
- 39.Smith MA, Sayre LM, Anderson VE, Harris PL, Beal MF, Kowall N, Perry G. J Histochem Cytochem. 1998;46:731–735. doi: 10.1177/002215549804600605. [DOI] [PubMed] [Google Scholar]
- 40.Liu R, Li B, Flanagan SW, Oberley LW, Gozal D, Qiu M. J Neurochem. 2002;80:488–500. doi: 10.1046/j.0022-3042.2001.00720.x. [DOI] [PubMed] [Google Scholar]
- 41.Hallgren B, Sourander P. Neurochemistry. 1958;3:41–51. doi: 10.1111/j.1471-4159.1958.tb12607.x. [DOI] [PubMed] [Google Scholar]
- 42.Rice M. TINS. 2000;23:209–216. [Google Scholar]
- 43.Miyazaki E, Kato J, Kobune M, Okumura K, Sasaki K, Shintani N, Arosio P, Niitsu Y. Gut. 2001;50:413–419. doi: 10.1136/gut.50.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews SC, Treffry A, Harrison PM. Biochem J. 1987;245:447–453. doi: 10.1042/bj2450447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Ellenburg M, Soistmann E, Ruble J, Wright B, Ho J, Carter D. Acta Crystallog. Sect. D. 2006;62:800–806. doi: 10.1107/S0907444906018294. [DOI] [PubMed] [Google Scholar]
- 46.Meissler G, Tarr D. Inorganic Chemistry. 4th edition. Upper Saddle River, NJ: Pearson Prentice Hall; 2011. pp. 194–198. [Google Scholar]
- 47.Crichton R. Iron Metabolism: From molecular Mechanisms to Clinical Consequences. 3rd edition. West Sussex, United Kingdom: John Wiley and Sons; 2009. pp. 183–222. [Google Scholar]
- 48.Zecca L, Youdim MB, Riederer P, Connor JR, Crichton RR. Nat Rev Neurosci. 2004;5:863–873. doi: 10.1038/nrn1537. [DOI] [PubMed] [Google Scholar]
- 49.O'Connell MJ, Baum H, Peters TJ. Biochem J. 1986;240:297–300. doi: 10.1042/bj2400297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen LA, Gutierrez L, Weiss A, Leichtmann-Bardoogo Y, Zhang DL, Crooks DR, Sougrat R, Morgenstern A, Galy B, Hentze MW, Lazaro FJ, Rouault TA, Meyron-Holtz EG. Blood. 2010;116:1574–1584. doi: 10.1182/blood-2009-11-253815. [DOI] [PubMed] [Google Scholar]
- 51.Catalgol B, Grune T. Curr Pharm Des. 2009;15:3043–3051. doi: 10.2174/138161209789058129. [DOI] [PubMed] [Google Scholar]
- 52.Rudeck M, Volk T, Sitte N, Grune T. IUBMB Life. 2000;49:451–456. doi: 10.1080/152165400410317. [DOI] [PubMed] [Google Scholar]