Abstract
PTEN is a tumor suppressor that negatively regulates the PI3K-AKT signaling pathway, which is implicated in the pathogenesis of endometrial carcinoma. Sanger sequencing has been considered to be the gold standard for detection of PTEN sequence abnormalities. However, this approach fails to address the epigenetic mechanisms that contribute to functional PTEN loss. Using a study cohort of 154 endometrioid and non-endometrioid endometrial carcinomas, we performed full-length PTEN sequencing and PTEN immunohistochemistry on each tumor. PTEN sequence abnormalities were detected in a significantly lower proportion of cases (43%) than PTEN protein loss (64%, p= 0.0004). Endometrioid tumors had a significantly higher proportion of PTEN sequence abnormalities and PTEN protein loss than non-endometrioid tumors. Within the latter group, PTEN sequence abnormalities and PTEN protein loss were most frequent in undifferentiated carcinomas, followed by mixed carcinomas; they were least frequent in carcinosarcomas. Overall, at least one PTEN sequence abnormality was detected in each exon, and the greatest number of sequence abnormalities was detected in exon 8. Pure endometrioid tumors had a significantly higher frequency of sequence abnormalities in exon 7 than did the non-endometrioid tumors (p=0.0199). Importantly, no mutational hotspots were identified. While PTEN protein loss by immunohistochemistry was identified in 89% of cases with a PTEN sequence abnormality, PTEN protein loss was detected by immunohistochemistry in 44% of cases classified as PTEN wildtype by sequencing. For the first time, we demonstrate that PTEN immunohistochemistry is able to identify the majority of cases with functional PTEN loss. However, PTEN immunohistochemistry also detects additional cases with PTEN protein loss that would otherwise be undetected by gene sequencing. Therefore, for clinical purposes, immunohistochemistry appears to be a preferable technique for identifying endometrial tumors with loss of PTEN function.
Introduction
The (phosphatase and tensin homolog) PTEN gene is located at chromosome 10q23.31. The PTEN protein plays a crucial role in the control of the PI3K -AKT pathway through dephosphorylation of PIP3 at the cell membrane. Absence of functional PTEN protein leads to unopposed action of PI3K with resultant uncontrolled PIP3 production. One major effector of the PI3K -AKT pathway is mTOR, which stimulates protein synthesis, initiates entry into G1 phase of the cell cycle and interacts with proteins that regulate apoptosis (1). Loss of PTEN function has been implicated in the pathogenesis of a number of different tumors, particularly endometrial cancer. Functional PTEN loss can result from somatic mutations, abnormalities in PTEN transcriptional and post-transcriptional regulation, microRNAs, regulation of microRNAs by a PTEN pseudogene, or regulation of PTEN protein stability and degradation mechanisms (2). Somatic PTEN mutations have been detected in 34–55% of endometrial cancers (3, 4), particularly in the endometrioid histotype.
Accurate identification of functional PTEN loss is an integral part of a comprehensive evaluation of tumors with potential abnormalities in the PI3K -AKT pathway, particularly as targeted therapeutics against the pathway are entering trials and the clinic. Studies have shown that different types of PIK3CA (the catalytic subunit of PI3K) isoform derangements are required in combination with PTEN abnormalities to achieve upregulation of the PI3K -AKT pathway (5, 6). Even in tumors in which this pathway may not be the primary driver of cell proliferation, the PI3K -AKT pathway has been shown to be activated in a compensatory fashion in the setting of treatment with anti-receptor tyrosine kinase agents (2), prompting the need for therapies that target more than one signal transduction pathway. Currently, numerous clinical trials with PI3K, AKT and mTOR inhibitors are underway (http://www.ClinicalTrials.gov), several specifically involving endometrial cancer (7).
Previously reported PTEN gene sequence abnormalities are highly variable in type (frameshifts, point mutations) and can occur throughout all 9 exons (3, 4, 8). Therefore, to date, Sanger sequencing has been considered to be the gold standard for detection of PTEN mutations and subsequent loss of PTEN protein function. From a clinical perspective, the problems with this sequencing approach include high cost, labor-intensiveness, and failure to identify PTEN protein loss via epigenetic mechanisms. Alternative methods, such as immunohistochemistry, may be the preferred method of assessing functional PTEN loss in patient tumors. However, until recently, PTEN immunohistochemistry has been considered somewhat unreliable. In a tissue microarray study of four different antibodies (9), only results using antibody 6H2.1 have been shown to have a statistically significant correlation with pAKT (activated form of AKT). Stemming from historical difficulties in PTEN antibody performance, there has been no uniform scoring system for PTEN immunohistochemistry. Furthermore, direct head-to-head comparisons of PTEN immunohistochemistry and PTEN sequence analysis to date have been very limited. In one study of endometrial tumors, PTEN immunohistochemistry and PTEN sequence analysis were carried out, but each technique was applied to a different set of tumors (10). Another study of histologically normal endometrium has shown that normal endometrial glands with loss of PTEN protein expression by immunohistochemistry do have PTEN mutations (11).
The objective of our study was to perform a direct comparison of PTEN immunohistochemistry and PTEN gene sequencing on a large group of endometrial carcinomas to determine which method is clinically preferable for identifying PTEN loss. This first necessitated that we establish a PTEN immunohistochemistry scoring system that can be applied clinically. The large cohort of both endometrioid and non-endometrioid endometrial carcinomas used has been carefully characterized as to tumor grade and histotype. Given that PTEN loss can occur through a variety of mechanisms in addition to mutations, we hypothesized that immunohistochemistry would outperform the sequencing analysis as a clinically applicable method to detect PTEN loss in endometrial carcinomas.
Materials and Methods
Case Selection
One hundred fifty-four consecutive cases of endometrial carcinoma with available fresh frozen tissue and formalin fixed, paraffin embedded tissue were retrieved from the tumor bank of the University of Texas M.D. Anderson Cancer Center Department of Pathology under the auspices of an institutional review board-approved protocol. By histotype, the study cohort was composed of 100 pure endometrioid carcinomas (6 FIGO grade 1, 78 FIGO grade 2, and 16 FIGO grade 3) and 54 non-endometrioid carcinomas consisting of 13 carcinosarcomas, 10 undifferentiated carcinomas, 4 clear cell carcinomas, 1 serous carcinoma, and 26 mixed carcinomas. Mixed carcinomas were designated as tumors with an endometrioid component and any amount of at least one other non-endometrioid histotype, including clear cell carcinoma, serous carcinoma, undifferentiated carcinoma, carcinoma with sarcomatoid features or carcinoma with neuroendocrine features. Among the 26 mixed endometrial carcinomas, 19 had a minor component of serous carcinoma. There is limited experience regarding the nature of mixed endometrial carcinomas composed of endometrioid and non-endometrioid components. The most studied combination has been that of endometrioid and serous histotypes. While the majority of studies have shown that the behavior of these tumors is similar to that of pure serous carcinoma, the cut-offs for the relative amount of serous carcinoma in a mixed tumor have been quite varied, ranging from ≥50% (12), ≥ 25% (13, 14) to <10% (15, 16). Similarly, a few studies have shown that clear cell carcinoma in combination with endometrioid carcinoma confers a poor outcome (16, 17).
Sequencing Analysis
Genomic DNA was isolated from fresh frozen tumors using standard methods. Bidirectional sequencing of PTEN exons 1–8 was performed at the Human Genome Sequencing Center at Baylor College of Medicine using intron-based, exon-specific primers. PCR reactions were performed in 8 μl containing 10 ng of genomic DNA, 0.4 μM oligonucleotide primers, and 0.7× Qiagen® PCR HotStar Taq Master Mix containing buffer and polymerase. Cycling parameters were 95°– 15 minutes, then 95° – 45 seconds, 60° – 45 seconds, and 72° – 45 seconds for 40 cycles followed by a final extension at 72° for 7 minutes After thermocycling, 5 μl of a 1:15 dilution of Exo-SAP was added to each well and reactions incubated at 37°C for 15 minutes prior to inactivation at 80° for 15 minutes. Reactions were diluted by 0.6× and 2 μl were combined with 5 μl of 1/64th Applied Biosystems® (AB) BigDye™ sequencing reaction mix and cycled as above for 25 cycles. Reactions were precipitated with ethanol, re-suspended in 0.1 mM EDTA and loaded on AB 3730XL sequencing instruments using the Rapid36 run module and 3xx base-caller. SNPs were identified using SNP Detector software. Identified mutations were verified by bidirectional re-sequencing of the original DNA sample. Sequencing of exon 9 for all tumors was performed at the University of Texas M.D. Anderson Cancer Center Molecular Diagnostics Laboratory. Two μl of DNA were used to amplify exon 9 of PTEN using M13-tagged published primers. Reaction mixtures of 50 μL contained 1 mM dNTPs, 2.5 mM MgCl2, 0.2 uM primers, 1.5 U Ampli-Taq Gold 360 Polymerase (Applied Biosystems) were amplified with the following PCR conditions: an initial 10-minute activation at 95°C followed by 40 cycles of 30 seconds at 95°C; 30 seconds at 50°C; 30 seconds at 72°C, and a final extension of 10 minutes at 72°C. PCR products were purified (Agencourt Ampure, Beckman Coulter) prior to being loaded on an ethidium bromide-stained agarose gel. Fluorescent-based automated cycle sequencing was performed by the dye-terminator method using a multi-capillary sequencer (ABI 3130 Genetic Analyzer; PE Applied Biosystems) according to manufacturer's protocol (BigDye Terminator v1.1 Ready Reaction Cycle Sequencing Kit; PE Applied Biosystems). Briefly, reaction tubes (total volume 20 μL) containing 100 ng of the purified PCR product, 3.2 pmol of either the sense or antisense M13 primer and 6 μL of the sequencing mixture were placed in a DNA thermal cycler and cycled for 25 cycles at 96°C for 10 seconds, 58°C for 5 seconds, 60°C for 4 minutes, and final hold at 4°C. Sequencing reactions were purified using the Qiagen DyeEx™ purification kit (Qiagen) as per manufacturer's protocol. The resulting data were analyzed by Seqscape software (PE Applied Biosystems).
Immunohistochemistry
Immunohistochemistry was performed on 4μm sections of formalin fixed, paraffin embedded tumors. Following deparaffinization and rehydration of the tissues sections, antigen retrieval was performed at 100 °C for 20 minutes with Tris-EDTA buffer, pH 6.0 (PTEN) or using microwave treatment in 10 mM citrate buffer, pH 6 (pS6). Endogenous peroxidase was blocked with 3% peroxide for 5 minutes. Primary PTEN antibody (Dako, clone 6H2.1) was applied at 1:100 dilution. Primary phospho-S6 Ribosomal Protein (Ser235/236) antibody (Cell Signaling) was applied at 1:50 dilution. Primary antibody detection was carried out using a polymer system (Bond Polymer Refine Detection, Leica). Staining development was achieved by incubation with DAB and DAB Enhancer (Leica for PTEN; Dako for pS6). The PTEN antibody was validated using nude mouse xenografts of a breast carcinoma cell line with a known PTEN mutation and xenografts of a breast cancer cell line with wild type PTEN.
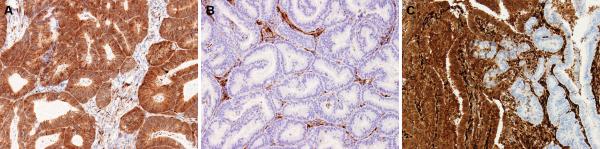
PTEN immunohistochemistry was scored by two different pathologists (BD and RB) as positive, negative and heterogeneous. In all cases, stromal cells and blood vessels had intensely positive expression for PTEN and thus served as integral internal positive controls. Tumors considered positive showed diffuse positive cytoplasmic and nuclear staining in the majority (>90%) of cells (Figure 1A). Positive staining in tumor cells was comparable to that detected in normal stromal cells. Tumors with no or only rare cells staining (<1%) were considered negative for PTEN (Figure 1B). For all negative cases, the presence of positive staining stromal cells confirmed that the immunohistochemistry reaction was working. Tumors with distinctive positive and negative staining foci were designated as having a heterogeneous staining pattern (Figure 1C). pS6 immunohistochemistry was scored as a percentage of tumor cells with positive cytoplasmic expression.
Figure 1.
(a) Positive immunohistochemistry for PTEN. Diffuse cytoplasmic staining is present in the majority (>90%) of tumor cells. (b) Negative immunohistochemistry for PTEN. No or only scattered tumor cells (<1%) have cytoplasmic staining. Note stromal cells serve as positive internal control. (c) Heterogeneous immunohistochemistry score for PTEN. Heterogeneous tumors have distinct positive and negative foci. All photomicrographs 20×.
Statistical Analysis
Statistical analyses were performed using SPSS 17.0 (Chicago, IL). Statistical comparisons were carried out using the Fisher's exact test. A p-value of <0.05 was considered statistically significant.
Results
An overview of PTEN sequencing and PTEN immunohistochemistry results is summarized in Table 1. More detailed sequencing (Tables 2 and 3) and immunohistochemistry (Tables 4, 5 and 6) data are provided in subsequent tables. As shown in Table 1, more endometrial carcinomas had loss of PTEN protein detected by immunohistochemistry (64%) than had a PTEN sequence abnormality (43%; p=0.0004). Endometrioid tumors had a significantly higher proportion of PTEN sequence abnormalities (51%) and PTEN protein loss by immunohistochemistry (75%) than non- endometrioid tumors (28% for sequencing, p=0.0064; 43% for immunohistochemistry, p=0.0001). Grade of endometrioid tumor did not significantly correlate with the likelihood of a PTEN sequence abnormality or PTEN protein loss being detected. Undifferentiated endometrial carcinomas have been previously shown to have a prognosis significantly worse than that of grade 3 endometrioid endometrial carcinoma (18). However, based on PTEN sequencing and PTEN immunohistochemistry, the undifferentiated tumors are more comparable to the endometrioid tumors (Table 1). The undifferentiated tumors had a significantly higher proportion of cases with PTEN sequence abnormality (60%) compared to the remainder of the cases in the non-endometrioid group (20%, p= 0.0199). Also, undifferentiated tumors had a significantly higher proportion of cases with PTEN protein loss (80%) compared to the remainder of the cases in the non-endometrioid group (20%, p=0.0125). Carcinosarcomas are known to have an aggressive clinical course, similar to that of uterine serous carcinoma. The carcinosarcomas had a notably lower proportion of cases with PTEN sequence abnormality (8%) than the remainder of the cases in the non-endometrioid group (34%, p=0.0830) and a significantly lower proportion of cases with PTEN protein loss (15%) compared to the remainder of the cases in the non-endometrioid group (51%, p=0.0277) (Table 1). Of the 13 carcinosarcoma cases, a PTEN mutation was detected in only one case. This case was scored as heterogeneous by immunohistochemistry. Mixed endometrioid-non-endometrioid tumors had a lower proportion of PTEN sequence abnormalities (31%) than pure endometrioid tumors (51%, p= 0.0794) and also a significantly lower proportion of PTEN protein loss (42%) than pure endometrioid tumors (75%, p=0.0037).
Table 1.
An overview of the relationship of PTEN sequencing results and PTEN status by immunohistochemistry according to histological tumor type and grade.
| Tumor Histotype and Grade (n) | PTEN Sequence Abnormality Detected n(%) | PTEN Protein Loss by Immunohistochemistry n(%)1 |
|---|---|---|
| All cases (154) | 66 (43%) | 98 (64%) |
| Endometrioid (100) | 51 (51%) | 75 (75%) |
| Grade 1 (6) | 2 (33%) | 4 (67%) |
| Grade 2 (78) | 38 (49%) | 62 (79%) |
| Grade 3 (16) | 11 (69%) | 9 (56%) |
| Non-endometrioid (54) | 15 (28%) | 23 (43%) |
| Mixed (26) | 8 (31%) | 11 (42%) |
| Undifferentiated (10) | 6 (60%) | 8 (80%) |
| Carcinosarcoma (13) | 1 (8%) | 2 (15%) |
| Clear Cell (4) | 0 (0%) | 2 (50%) |
| Serous (1) | 0 (0%) | 0 (0%) |
Immunohistochemical loss of PTEN protein included tumors scored as negative and heterogeneous.
Table 2.
Detailed PTEN sequencing results according to tumor histological type.
| ENDOMETRIOID | NON-ENDOMETRIOID | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Type of Sequence Abnormality | 1Number of sequence abnormalities | Sequence abnormality as a % of all cases (n=100) | Sequence abnormality as a % of the total number of sequence abnormalities detected (n=70) | 2Number of sequence abnormalities | Sequence abnormality as a % of all non-endometrioid cases (n=54) | Sequence abnormality as a % of the total number of sequence abnormalities detected (n=19) |
| Insertions and deletions | 32 | 32.0% | 45.7% | 10 | 18.5% | 52.6% |
|
| ||||||
| 4 amino acid deletion | 1 | 1.0% | 1.4% | 0 | 0.0% | 0.0% |
| 1 amino acid deletion | 1 | 1.0% | 1.4% | 0 | 0.0% | 0.0% |
| Exon boundary, insertion | 0 | 0.0% | 0.0% | 1 | 1.9% | 5.3% |
| Exon boundary, deletions | 1 | 1.0% | 1.4% | 0 | 0.0% | 0.0% |
| Frameshift, insertion | 9 | 9.0% | 12.9% | 5 | 9.3% | 26.3% |
| 3Frameshift, deletion** | 20 | 20.0% | 28.6% | 4 | 7.4% | 21.1% |
|
| ||||||
| Point Mutations | 38 | 38.0% | 54.3% | 9 | 16.7% | 47.4% |
|
| ||||||
| 3Missense mutation* | 25 | 25.0% | 35.7% | 4 | 7.4% | 21.1% |
| Nonsense mutation | 10 | 10.0% | 14.3% | 4 | 7.4% | 21.1% |
| Splice region point mutation | 2 | 2.0% | 2.9% | 1 | 1.9% | 5.3% |
| Splice site point mutation | 1 | 1.0% | 1.4% | 0 | 0.0% | 0.0% |
|
| ||||||
| All sequence abnormalities | 70 | 70.0% | 100.0% | 19 | 35.2% | 100.0% |
51 endometrioid tumors in the study had a total of 70 sequence abnormalities for an average of 1.4 sequence abnormalities per case.
15 non-endometrioid cases in the study had a total of 19 sequence abnormalities for an average of 1.3 sequence abnormalities per case.
Endometrioid tumors had a significantly higher frequency of
missense mutations (p=0.0088) than the non-endometrioid tumors and showed a trend toward a higher frequency of
frameshift deletions (p=0.0606).
Table 3.
Summary of PTEN exon sequence abnormalities (insertions, deletions, and point mutations) and endometrial carcinoma type.
| ENDOMETRIOID | NON-ENDOMETRIOID | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| All Sequence Abnormalities Detected n(%)1 | Insertions and deletions | Point Mutations | All Sequence Abnormalities Detected n(%)1 | Insertions and deletions | Point Mutations | |
| Exon 1 | 9 (12.9%) | 5 | 4 | 5 (26.3%) | 2 | 3 |
| Exon 2 | 2 (2.9%) | 0 | 2 | 0 (0%) | 0 | 0 |
| Exon 3 | 2 (2.9%) | 0 | 2 | 0 (0%) | 0 | 0 |
| Exon 4 | 1 (1.4%) | 0 | 1 | 1 (5.3%) | 1 | 0 |
| Exon 5 | 12 (17.1%) | 4 | 8 | 4 (21.1%) | 1 | 3 |
| Exon 6 | 10 (14.3%) | 5 | 5 | 0 (0%) | 0 | 0 |
| 1 Exon 7 | 14 (20.0%) | 10 | 4 | 1 (5.3%) | 1 | 0 |
| Exon 8 | 16 (22.9%) | 7 | 9 | 5 (26.3%) | 3 | 2 |
| Exon 9 | 0 (0%) | 0 | 0 | 1 (5.3%) | 1 | 0 |
| Exon 3 boundary | 1 (1.4%) | 1 | 0 | 0 (0%) | 0 | 0 |
| Exon 8 boundary | 0 (0%) | 0 | 0 | 1 (5.3%) | 1 | 0 |
| Splice region 6/7 | 2 (2.9%) | 0 | 2 | 1 (5.3%) | 0 | 1 |
| Splice region 7/8 | 1 (1.4%) | 0 | 1 | 0 (0%) | 0 | 0 |
|
| ||||||
| All | 70 | 32 | 38 | 19 | 10 | 9 |
Exon 7 sequence abnormalities were significantly more common in endometrioid carcinomas compared to the non-endometrioid group (p=0.0199).
Table 4.
Relationship between PTEN immunohistochemistry and tumor histology.
| Immunohistochemistry Score | Endometrioid n (%) | Non-Endometrioid n (%) |
|---|---|---|
| Positive | 25 (25%) | 31 (57%) |
| Loss | 75 (75%) | 23 (43%) |
| Heterogeneous | 18 (18%) | 9 (17%) |
| Negative | 57 (57%) | 14 (26%) |
|
| ||
| All Cases | 100 | 54 |
Table 5.
Mean percentage of tumor cells positive for pS6 in relation to PTEN immunohistochemistry (IHC) and sequencing results
| PTEN IHC Positive | PTEN IHC Heterogeneous | PTEN IHC Negative | All cases | |
|---|---|---|---|---|
|
| ||||
| PTEN Sequence Abnormality Detected | 15% | 18.5% | 45.8% | 36.0% |
|
| ||||
| PTEN Sequence Abnormality Not Detected | 31.5% | 46.2% | 55.0% | 46.7% |
|
| ||||
| All cases | 30.4% | 34.3% | 51.3% | 42.9% |
Table 6.
Relationship between PTEN immunohistochemistry and PTEN sequencing.
| Immunohistochemistry Score | Sequence Abnormality Detected n (%) | Sequence Abnormality Not Detected n (%) |
|---|---|---|
| Positive | 7 (11%) | 49 (56%) |
| Loss | 59 (89%) | 39 (44%) |
| Heterogeneous | 15 (22%) | 12 (14%) |
| Negative | 44 (67%) | 27 (30%) |
|
| ||
| All Cases | 66 | 88 |
PTEN sequencing revealed a wide variety of different sequence abnormalities detected (Table 2). Endometrioid tumors had a comparatively higher proportion of missense mutations (p=0.0088) and frameshift deletions (p=0.0606) than the non-endometrioid tumors. The non- endometrioid tumors did not have any significant association with any particular PTEN sequence abnormality. Endometrioid tumors and non-endometrioid tumors had a comparable average number of PTEN sequence abnormalities per case (1.4 and 1.3, respectively). From a total of 89 PTEN sequence abnormalities, only 6 (6.7%) were homozygous, while the remainder was hemizygous.
Table 3 summarizes the PTEN sequence abnormalities by exon for the endometrioid and nonendometrioid carcinomas. Overall, at least one PTEN sequence abnormality was detected in each exon, and the greatest number of sequence abnormalities was detected in exon 8. Endometrioid tumors had a significantly higher frequency of sequence abnormalities in exon 7 than did the non-endometrioid tumors (p=0.0199). The presence of sequence aberrations across all exons highlights the impracticality of focused sequencing efforts on just one or a few exons to evaluate tumors clinically.
Table 4 summarizes the results of 3-tiered PTEN immunohistochemistry scoring in the endometrioid and non-endometrioid carcinomas. More PTEN positive cases were observed in the non-endometrioid group. The percentages of endometrioid and non-endometrioid tumors with the heterogeneous pattern were comparable (18% and 17%, respectively). A significantly greater percentage of cases with immunohistochemical PTEN loss (negative and heterogeneous scored cases combined) was detected in endometrioid carcinomas (75%) compared to the non-endometrioid carcinomas (43%; p= 0.0001).
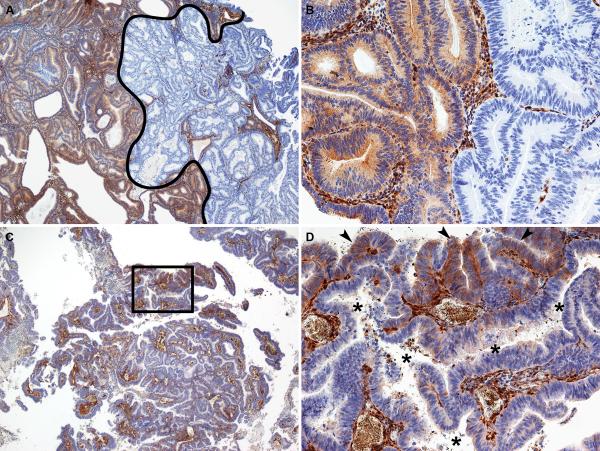
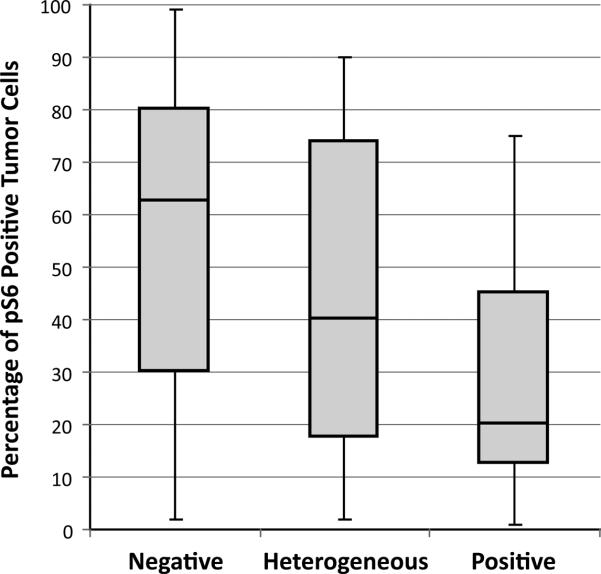
We observed two general patterns of heterogeneous PTEN expression; a geographic pattern containing large and well demarcated positive and negative areas (Figure 2, A and B), and a focal pattern, where the majority of the tumor is PTEN negative, with focal, less well-demarcated areas of positive staining (Figure 2, C and D). The nature of the PTEN immunohistochemistry heterogeneous cases is uncertain. A trivial explanation for this group is that it represents uneven immunohistochemical staining in tumor sections that may not have undergone optimal histological processing. There are several arguments against this explanation, however. One, in these heterogeneous cases, we uniformly observed very strong, positive PTEN immunohistochemical expression in adjacent stromal cells and blood vessels. Second, collaborators at Memorial Sloan-Kettering Cancer Center have observed an identical staining pattern when using the same PTEN antibody (19). Finally, activation of the PI3K-AKT pathway, such as occurs with PTEN protein loss, results in elevated levels of pS6 (20). Accordingly, the PTEN negative tumors had significantly higher pS6 scores compared to the PTEN positive tumors (p=0.037) (Figure 3). Endometrial carcinomas with a heterogeneous PTEN immunohistochemistry staining pattern have an intermediate level of pS6 compared to PTEN positive and negative tumors (Figure 3). pS6 immunohistochemistry scores in relation to PTEN immunohistochemistry and PTEN sequence abnormalities are presented in Table 5. The mean pS6 immunohistochemistry score was comparable between the groups which had no PTEN sequence abnormalities (mean = 46.7%) compared to the group in which PTEN sequence abnormalities were detected (mean = 36.0%). Note that in both the PTEN mutant and PTEN wild-type groups, mean pS6 is highest in the tumors with negative PTEN immunohistochemistry.
Figure 2.
Representative photomicrographs of different patterns of PTEN heterogeneous immunohistochemistry. (A and B) Geographic pattern of PTEN heterogeneous immunohistochemical expression. In A, the portion of the tumor to the right of the black line represents a large area that is negative for PTEN immunohistochemistry, while the area to the left is positive for PTEN immunohistochemistry. A higher magnification of the positive-negative interface is shown in B. For the geographic pattern, the interface between the positive and negative areas is typically sharp and abrupt. (C and D) Focal pattern of PTEN heterogeneous immunohistochemical expression. In this case, the vast majority of the tumor is PTEN negative. D represents a higher magnification view of the area outlined by the rectangle in C. In D, it is clear that small foci of the tumor are immunohistochemically positive for PTEN (arrowheads), while a large, irregular malignant gland immediately adjacent is PTEN negative (asterisks in gland lumen). Note that in this pattern, the interface between PTEN positive and negative areas is more subtle. Note also that the intervening tumor stroma has uniformly strong PTEN staining throughout the tumor. A and C, 4×; B and D, 20×.
Figure 3.
Percentage of immunopositive tumor cells for phosphorylated S6 in PTEN negative, heterogeneous, and positive groups. Rectangular bars represent the 25th, 50th (median), and 75th percentiles, while the whiskers represent the range. There was a significant (p=0.037) difference in the percentage of pS6 positive tumor cells between the PTEN negative and positive tumors.
Table 6 provides a direct comparison of immunohistochemistry and sequencing. In 89% of the cases with a detected PTEN sequence abnormality, PTEN protein loss was identified by immunohistochemistry (22% heterogeneous and 67% negative). However, the absence of a PTEN sequence abnormality was corroborated by positive expression of PTEN by immunohistochemistry in only 56% of cases. In other words, in 44% of the tumors in which no PTEN sequence abnormality was detected, immunohistochemistry identified partial (14% heterogeneous scored cases) or complete (30% negative scored cases) PTEN protein loss. Thus, when compared head-to-head, immunohistochemistry identifies more endometrial carcinomas with PTEN loss, and this correlates with pathway activation, at least as indicated by pS6 staining.
Discussion
Endometrial cancer is the most prevalent malignancy of the gynecologic tract in the developed world. While patients with early stage disease and tumors of endometrioid histotype tend to have a good prognosis, patients with non-endometrioid tumors or recurrent endometrioid tumors do not respond well to conventional chemotherapy and have poor outcomes. The PI3K-AKT pathway is frequently activated in endometrial carcinoma, often in part due to functional PTEN loss. Therefore, accurate identification of this aberration is an important component of patient triage and selection in order to achieve optimal therapeutic benefit from targeted agents. Given the limited life expectancy of endometrial cancer patients being considered for targeted therapies, the laboratory tools used to gauge potential eligibility of targeted therapy must therefore be not only accurate, but inexpensive and accessible, as well as simple, reproducible, and able to be performed in a narrow time window.
Other key components of the PI3K-AKT pathway, such as AKT (21) or PI3K (22, 23), are altered primarily by mutation, resulting in pathway activation. In contrast, loss of PTEN may be due to gene mutations, gene methylation (24, 25), and a variety of other causes including abnormalities in PTEN post-transcriptional regulation, actions of micro RNAs, as well as due to aberrant mechanisms of PTEN protein stability and degradation (2). While immunohistochemistry would seem to be the best method to assess functional PTEN loss in endometrial tumors, it has historically been considered as having somewhat poor reproducibility (26–33). This has, in large part, been due to inconsistent performance of one PTEN antibody compared to another, as well as the variation of scoring systems between studies. However, we have recently shown that the 3-tiered PTEN immunohistochemical scoring system summarized in this study is highly reproducible (19) and that PTEN immunohistochemistry scores are concordant with reverse phase protein lysate array results in 93% of cases (23). In the current study, 64% of all endometrial carcinomas had PTEN protein loss demonstrated by immunohistochemistry. In comparison, AKT mutations have been identified in only 2%–4% of endometrial carcinomas (21, 34), and PIK3CA mutations in 24–52% of endometrioid endometrial tumors (22, 35, 36) and 15%–33% non-endometrioid endometrial tumors (22, 37). Therefore, adding PTEN immunohistochemistry to the clinical evaluation of these tumors would allow for the identification of more endometrial cancer patients with activation of the PI3K-AKT pathway; importantly, these are patients who might be potentially responsive to treatment with PI3K pathway inhibitors. The importance of such accurate patient identification is underscored by the fact that it has recently been demonstrated that patients with advanced cancers with PIK3CA mutations have better response to pharmacological inhibitors of PI3K/AKT/mTOR(38).
There is presently no standardized system for PTEN immunohistochemistry interpretation. Some investigators have used the histologic score (H-score) (9), a complex semi-quantitative scoring method. We developed a scoring system that reflected the intrinsic nature of the staining patterns we were observing empirically with this antibody. Upon a thorough review of the literature, we found that one group (29, 30, 33) used a similar positive-negative-heterogeneous scoring system, although with a different PTEN antibody. Whereas the positive and negative PTEN immunohistochemistry categories are self-explanatory, the biological nature of the PTEN heterogeneous tumors is currently not known. There is no mouse model or cell line model that recapitulates this heterogeneous expression pattern. Whether this pattern is representative of subclones within the tumor or of differential post transcriptional regulation is not known. However, PTEN protein loss even in a portion of the tumor may mean that a patient could potentially derive some benefit from PI3K-AKT pathway inhibitors. Interestingly, in our study, the PTEN heterogeneous group had higher expression of pS6 than did the PTEN positive group (Figure 3). Future correlation with molecular analyses and clinical outcomes may help to clarify the significance of these results. It is also important to note that, while this simple scoring system seems appropriate for endometrial carcinoma, it may not be applicable to other tumors from other organ systems. Molecular regulation mechanisms of PTEN protein expression in other tissue types may be different from that in the endometrium, potentially resulting in different immunohistochemical staining patterns. Finally, our study highlights the importance of the use of whole tissue sections, rather than tissue microarrays, in evaluation of PTEN immunohistochemical expression. Cores in tissue microarrays may contain insufficient internal positive control stroma, and tissue microarray cores may be too small to capture both the positive and negative portions of the tumor, particularly in the case of geographic pattern of heterogeneous PTEN immunohistochemical expression (Figure 2, A and B).
While many studies in the past have examined either PTEN sequence abnormalities (3, 4, 8, 39) or PTEN expression by immunohistochemistry (26, 27, 29–33, 40), our study is the first to date to provide detailed information on the correlation of PTEN immunohistochemistry with PTEN sequence abnormalities and tumor histotype. In terms of correlation of the PTEN immunohistochemistry and PTEN sequence analysis results, both testing modalities detected significantly more frequent PTEN loss in endometrioid tumors relative to the non-endometrioid tumors (Table 1). However, for both the endometrioid and non-endometrioid groups, immunohistochemistry detected a substantially greater proportion of cases with PTEN loss than did the sequencing analysis. Overall, of the tumors in which PTEN sequence abnormality was not detected, 44% had partial (heterogeneous score) or complete (negative score) PTEN loss by immunohistochemistry (Table 6). The prevalence of sequence abnormalities in our study (43%) is comparable to that of 34–55% in the published literature (3, 4). By histotype, we found that PTEN mutations occurred more frequently in endometrioid-type endometrial carcinomas (51%) compared to non-endometrioid carcinomas (28%). Similarly, Tashiro et al.(41) found PTEN mutations in 50% of the endometrioid subtype and no PTEN mutations in the serous subtype. Risinger et al. (42) identified PTEN mutations in 37% of endometrioid carcinomas and in only 5% of serous/clear cell endometrial carcinomas. Sun et al. (43) detected PTEN mutations in 26% of the endometrioid subtype and no PTEN mutations in the serous subtype. Finally, Rudd et al. (22) identified PTEN mutations is as many as 79% endometrioid carcinomas and 11% of non-endometrioid (serous and clear cell) carcinomas.
The PTEN status of non-endometrioid endometrial tumors such as undifferentiated carcinoma, carcinosarcoma, and mixed endometrial carcinomas has not been previously reported. It is interesting to note that mixed carcinomas (all of which had an endometrioid component) as a group contained a lower proportion of PTEN sequence abnormalities and PTEN protein loss relative to the pure-endometrioid group, but a higher proportion of the same relative to the remainder of cases in the non-endometrioid group. Undifferentiated carcinomas showed an even more dramatic rate of PTEN sequence abnormalities and PTEN protein loss compared to the other members of the non-endometrioid group. Therefore, while non-endometrioid tumors are generally thought of as having infrequent PTEN abnormalities, our results suggest that mixed endometrial carcinomas and undifferentiated endometrial carcinomas in particular should not be excluded from clinical testing for PTEN loss.
Of further interest is the fact that the vast majority (94%) of detected PTEN sequence abnormalities in our study were hemizygous and yet resulted in either partial or complete PTEN protein loss by immunohistochemistry. Similar observations have been made in other studies of endometrial cancer, as well as in hematologic malignancies (44) and breast cancer (45). A possible explanation for this is that sequence abnormalities alone do not account for all cases of PTEN protein loss in tumors. Rather, epigenetic mechanisms, such as promoter methylation, micro RNAs or increased protein degradation in the hemizygous state may also regulate PTEN expression. PTEN promoter methylation has been observed in approximately 19% of endometrial cancers (46). In addition, recent studies in non-small cell lung cancer (47) and hepatocellular carcinoma (48) have found that miRNA −21 post-transcriptionally down-regulates the expression of PTEN.
In summary, for the first time, we demonstrate a detailed correlation between PTEN sequencing abnormalities and PTEN immunohistochemistry. PTEN immunohistochemistry is able to identify the majority of cases with genetic PTEN loss and also detects additional cases with functional PTEN loss otherwise undetected by mutational analysis. In particular, mixed carcinomas and undifferentiated carcinomas have relatively frequent loss of PTEN. In addition, compared to PTEN full-length sequencing, PTEN immunohistochemistry is quicker, less costly and labor intensive and requires routinely obtained formalin-fixed, paraffin-embedded tissues. Finally, we propose a simple scoring system for PTEN immunohistochemistry, which may act as a primer for correlating PTEN levels of expression in endometrial tumors with patient response to PI3K/AKT/mTOR inhibitors.
Acknowledgements
NIH 2P50 CA098258-06 SPORE in Uterine Cancer (RRB) and Stand Up to Cancer/American Association for Cancer Research Dream Team Translational Cancer Research Grant, Grant No. SU2C-AACR- DT0209 (RRB, BTH, and GBM), and M.D. Anderson Cancer Center Support Grant (CCSG) CA016672 from National Institutes of Health. This original research was presented in part at the 99th United States and Canadian Academy of Pathology Annual Meeting, March 20–26, 2010, Washington, DC. The authors also thank Michelle Hildebrandt, Ph.D., Department of Epidemiology, M.D. Anderson Cancer Center, for her helpful discussion regarding this manuscript and to Su-Su Xie, M.D., for technical assistance. Drs. Broaddus and Hennessy contributed equally to this work as senior authors.
Footnotes
Conflict of Interest: The authors report no conflict of interest.
References
- 1.Le Tourneau C, Faivre S, Serova M, Raymond E. mTORC1 inhibitors: is temsirolimus in renal cancer telling us how they really work? Br J Cancer. 2008;99:1197–1203. doi: 10.1038/sj.bjc.6604636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S, Yu D. PI(3)king apart PTEN's role in cancer. Clin Cancer Res. 2010;16:4325–4330. doi: 10.1158/1078-0432.CCR-09-2990. [DOI] [PubMed] [Google Scholar]
- 3.Risinger JI, Hayes AK, Berchuck A, Barrett JC. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997;57:4736–4738. [PubMed] [Google Scholar]
- 4.Kong D, Suzuki A, Zou TT, et al. PTEN1 is frequently mutated in primary endometrial carcinomas. Nat Genet. 1997;17:143–144. doi: 10.1038/ng1097-143. [DOI] [PubMed] [Google Scholar]
- 5.Wee S, Wiederschain D, Maira SM, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torbett NE, Luna-Moran A, Knight ZA, et al. A chemical screen in diverse breast cancer cell lines reveals genetic enhancers and suppressors of sensitivity to PI3K isoform-selective inhibition. Biochem J. 2008;415:97–110. doi: 10.1042/BJ20080639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slomovitz BM, Lu KH, Johnston T, et al. A phase 2 study of the oral mammalian target of rapamycin inhibitor, everolimus, in patients with recurrent endometrial carcinoma. Cancer. 2010;116:5415–5419. doi: 10.1002/cncr.25515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bussaglia E, del Rio E, Matias-Guiu X, Prat J. PTEN mutations in endometrial carcinomas: a molecular and clinicopathologic analysis of 38 cases. Hum Pathol. 2000;31:312–317. doi: 10.1016/s0046-8177(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 9.Pallares J, Bussaglia E, Martinez-Guitarte JL, et al. Immunohistochemical analysis of PTEN in endometrial carcinoma: a tissue microarray study with a comparison of four commercial antibodies in correlation with molecular abnormalities. Mod Pathol. 2005;18:719–727. doi: 10.1038/modpathol.3800347. [DOI] [PubMed] [Google Scholar]
- 10.Mutter GL, Lin MC, Fitzgerald JT, et al. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J Natl Cancer Inst. 2000;92:924–930. doi: 10.1093/jnci/92.11.924. [DOI] [PubMed] [Google Scholar]
- 11.Mutter GL, Ince TA, Baak JP, et al. Molecular identification of latent precancers in histologically normal endometrium. Cancer Res. 2001;61:4311–4314. [PubMed] [Google Scholar]
- 12.Williams KE, Waters ED, Woolas RP, Hammond IG, McCartney AJ. Mixed serous-endometrioid carcinoma of the uterus: pathologic and cytopathologic analysis of a high-risk endometrial carcinoma. Int J Gynecol Cancer. 1994;4:7–18. doi: 10.1046/j.1525-1438.1994.04010007.x. [DOI] [PubMed] [Google Scholar]
- 13.Sherman ME, Bitterman P, Rosenshein NB, Delgado G, Kurman RJ. Uterine serous carcinoma. A morphologically diverse neoplasm with unifying clinicopathologic features. Am J Surg Pathol. 1992;16:600–610. doi: 10.1097/00000478-199206000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Carcangiu ML, Chambers JT. Uterine papillary serous carcinoma: a study on 108 cases with emphasis on the prognostic significance of associated endometrioid carcinoma, absence of invasion, and concomitant ovarian carcinoma. Gynecol Oncol. 1992;47:298–305. doi: 10.1016/0090-8258(92)90130-b. [DOI] [PubMed] [Google Scholar]
- 15.Lim P, Al Kushi A, Gilks B, Wong F, Aquino-Parsons C. Early stage uterine papillary serous carcinoma of the endometrium: effect of adjuvant whole abdominal radiotherapy and pathologic parameters on outcome. Cancer. 2001;91:752–757. [PubMed] [Google Scholar]
- 16.Quddus MR, Sung CJ, Zhang C, Lawrence WD. Minor serous and clear cell components adversely affect prognosis in “mixed-type” endometrial carcinomas: a clinicopathologic study of 36 stage-I cases. Reprod Sci. 2010;17:673–678. doi: 10.1177/1933719110368433. [DOI] [PubMed] [Google Scholar]
- 17.Carcangiu ML, Chambers JT. Early pathologic stage clear cell carcinoma and uterine papillary serous carcinoma of the endometrium: comparison of clinicopathologic features and survival. Int J Gynecol Pathol. 1995;14:30–38. doi: 10.1097/00004347-199501000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Altrabulsi B, Malpica A, Deavers MT, et al. Undifferentiated carcinoma of the endometrium. Am J Surg Pathol. 2005;29:1316–1321. doi: 10.1097/01.pas.0000171003.72352.9a. [DOI] [PubMed] [Google Scholar]
- 19.Garg K, Broaddus RR, Soslow RA, et al. Pathologic Scoring of PTEN Immunohistochemistry in Endometrial Carcinoma is Highly Reproducible. Int J Gynecol Pathol. 2011 doi: 10.1097/PGP.0b013e3182230d00. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCampbell AS, Harris HA, Crabtree JS, et al. Loss of inhibitory insulin receptor substrate-1 phosphorylation is an early event in mammalian target of rapamycin-dependent endometrial hyperplasia and carcinoma. Cancer Prev Res (Phila) 2010;3:290–300. doi: 10.1158/1940-6207.CAPR-09-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoji K, Oda K, Nakagawa S, et al. The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br J Cancer. 2009;101:145–148. doi: 10.1038/sj.bjc.6605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudd ML, Price JC, Fogoros S, et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011;17:1331–1340. doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung LW, Hennessy BT, Li J, et al. High Frequency of PIK3R1 and PIK3R2 Mutations in Endometrial Cancer Elucidates a Novel Mechanism for Regulation of PTEN Protein Stability. Cancer Discov. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvesen HB, MacDonald N, Ryan A, et al. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int J Cancer. 2001;91:22–26. doi: 10.1002/1097-0215(20010101)91:1<22::aid-ijc1002>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 25.Goel A, Arnold CN, Niedzwiecki D, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004;64:3014–3021. doi: 10.1158/0008-5472.can-2401-2. [DOI] [PubMed] [Google Scholar]
- 26.An HJ, Lee YH, Cho NH, et al. Alteration of PTEN expression in endometrial carcinoma is associated with down-regulation of cyclin-dependent kinase inhibitor, p27. Histopathology. 2002;41:437–445. doi: 10.1046/j.1365-2559.2002.01455.x. [DOI] [PubMed] [Google Scholar]
- 27.Sarmadi S, Izadi-Mood N, Sotoudeh K, Tavangar SM. Altered PTEN expression; a diagnostic marker for differentiating normal, hyperplastic and neoplastic endometrium. Diagn Pathol. 2009;4:41. doi: 10.1186/1746-1596-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Q, Ye F, Xia X, et al. Correlation between PTEN expression and PI3K/Akt signal pathway in endometrial carcinoma. J Huazhong Univ Sci Technolog Med Sci. 2009;29:59–63. doi: 10.1007/s11596-009-0112-6. [DOI] [PubMed] [Google Scholar]
- 29.Uegaki K, Kanamori Y, Kigawa J, et al. PTEN-positive and phosphorylated-Akt-negative expression is a predictor of survival for patients with advanced endometrial carcinoma. Oncol Rep. 2005;14:389–392. [PubMed] [Google Scholar]
- 30.Kanamori Y, Kigawa J, Itamochi H, et al. PTEN expression is associated with prognosis for patients with advanced endometrial carcinoma undergoing postoperative chemotherapy. Int J Cancer. 2002;100:686–689. doi: 10.1002/ijc.10542. [DOI] [PubMed] [Google Scholar]
- 31.Kapucuoglu N, Aktepe F, Kaya H, et al. Immunohistochemical expression of PTEN in normal, hyperplastic and malignant endometrium and its correlation with hormone receptors, bcl-2, bax, and apoptotic index. Pathol Res Pract. 2007;203:153–162. doi: 10.1016/j.prp.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Salvesen HB, Stefansson I, Kretzschmar EI, et al. Significance of PTEN alterations in endometrial carcinoma: a population-based study of mutations, promoter methylation and PTEN protein expression. Int J Oncol. 2004;25:1615–1623. [PubMed] [Google Scholar]
- 33.Terakawa N, Kanamori Y, Yoshida S. Loss of PTEN expression followed by Akt phosphorylation is a poor prognostic factor for patients with endometrial cancer. Endocr Relat Cancer. 2003;10:203–208. doi: 10.1677/erc.0.0100203. [DOI] [PubMed] [Google Scholar]
- 34.Cohen Y, Shalmon B, Korach J, et al. AKT1 pleckstrin homology domain E17K activating mutation in endometrial carcinoma. Gynecol Oncol. 2010;116:88–91. doi: 10.1016/j.ygyno.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Velasco A, Bussaglia E, Pallares J, et al. PIK3CA gene mutations in endometrial carcinoma: correlation with PTEN and K-RAS alterations. Hum Pathol. 2006;37:1465–1472. doi: 10.1016/j.humpath.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Hayes MP, Wang H, Espinal-Witter R, et al. PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin Cancer Res. 2006;12:5932–5935. doi: 10.1158/1078-0432.CCR-06-1375. [DOI] [PubMed] [Google Scholar]
- 37.Hayes MP, Douglas W, Ellenson LH. Molecular alterations of EGFR and PIK3CA in uterine serous carcinoma. Gynecol Oncol. 2009;113:370–373. doi: 10.1016/j.ygyno.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janku F, Tsimberidou AM, Garrido-Laguna I, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10:558–565. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minaguchi T, Yoshikawa H, Oda K, et al. PTEN mutation located only outside exons 5, 6, and 7 is an independent predictor of favorable survival in endometrial carcinomas. Clin Cancer Res. 2001;7:2636–2642. [PubMed] [Google Scholar]
- 40.Alkushi A, Kobel M, Kalloger SE, Gilks CB. High-grade endometrial carcinoma: serous and grade 3 endometrioid carcinomas have different immunophenotypes and outcomes. Int J Gynecol Pathol. 2010;29:343–350. doi: 10.1097/PGP.0b013e3181cd6552. [DOI] [PubMed] [Google Scholar]
- 41.Tashiro H, Blazes MS, Wu R, et al. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 42.Risinger JI, Hayes K, Maxwell GL, et al. PTEN mutation in endometrial cancers is associated with favorable clinical and pathologic characteristics. Clin Cancer Res. 1998;4:3005–3010. [PubMed] [Google Scholar]
- 43.Sun H, Enomoto T, Fujita M, et al. Mutational analysis of the PTEN gene in endometrial carcinoma and hyperplasia. Am J Clin Pathol. 2001;115:32–38. doi: 10.1309/7JX6-B9U9-3P0R-EQNY. [DOI] [PubMed] [Google Scholar]
- 44.Dahia PL, Aguiar RC, Alberta J, et al. PTEN is inversely correlated with the cell survival factor Akt/PKB and is inactivated via multiple mechanismsin haematological malignancies. Hum Mol Genet. 1999;8:185–193. doi: 10.1093/hmg/8.2.185. [DOI] [PubMed] [Google Scholar]
- 45.Perren A, Weng LP, Boag AH, et al. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol. 1999;155:1253–1260. doi: 10.1016/S0002-9440(10)65227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salvesen HB, MacDonald N, Ryan A, et al. Methylation of hMLH1 in a population-based series of endometrial carcinomas. Clin Cancer Res. 2000;6:3607–3613. [PubMed] [Google Scholar]
- 47.Zhang JG, Wang JJ, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 48.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]