Abstract
Developing portable and low-cost methods for quantitative detection of large protein biomarkers and small molecular toxins can play a significant role in controlling and preventing diseases or toxins outbreaks. Despite years of research, most current methods still require laboratory-based or customized devices that are not widely available to the general public for quantitative analysis. We have previously demonstrated the use of personal glucose meters (PGMs) and functional DNAs for the detection of many non-glucose targets. However, the range of targets detectable by functional DNAs is limited at the current stage. To expand the range of targets that can be detected by PGMs, we report here the use of antibodies in combination with sandwich and competitive assays for quantitative detection of protein biomarkers (PSA, with a detection limit of 0.4 ng/mL) and small molecular toxins (Ochratoxin A, with a detection limit of 6.8 ng/mL), respectively. In both assay methods, with invertase conjugates as the link, quantitative detection is achieved via the dependence between the concentrations of the targets in the sample and the glucose measured by PGMs. Given the wide availability of antibodies for numerous targets, the methods demonstrated here can expand the range of target detection by PGMs significantly.
INTRODUCTION
Affordable medical diagnostics and toxin monitoring at home or in the field are playing an increasingly important role in modern healthcare, as it can result in early detection, allow timely intervention to prevent wide-spread of diseases or toxins and facilitate personalized medicine. It will also bridge the gap between the well-offs and the poor, as well as between those living in urban area and those in rural and remote areas where access to clinical labs is limited, if not impossible.1 Toward this goal, much effort has been devoted toward developing in-home medical tests, such as the pregnancy test. However, most of these tests are qualitative based on colorimetry, even though quantitative numbers are more helpful or even required in diagnosis of many other diseases or detection of toxins. To overcome this limitation, a number of portable quantitative tests have been developed. Despite tremendous progress made in the past decades, few such devices are widely available to the public and most people have to go to hospitals or clinical labs for diagnosis.
One ideal device that can meet the above challenge is personal glucose meter (PGM).2–4 Compared to most other devices, PGMs are successful for in-home medical diagnostics not only because of their portable pocked size, low cost, simple operation and reliable quantitative results, but more importantly, also for its wide accessibility to the public worldwide. PGMs are commercially available in stores everywhere, and their recent integration in cell phones may give PGMs an even larger number of users.5 However, the current PGMs can only be used by the diabetes to monitor blood glucose. To overcome this limitation, several groups have reported modifications of PGMs in order to make them measure targets beyond glucose.6, 7 For example, the enzyme inside test strips of PGMs has been replaced with other enzymes that catalyze the redox reactions of alcohols, lactates and ornithine, instead of glucose, to quantify these substrates with the modified PGMs or other customized devices.6, 7 While these results are encouraging, it is preferable not to modify the PGMs so that any of the available PGMs at home or in the market can be used by the public. In a recent publication, we reported the use of commercial PGMs for quantitative detection of many non-glucose analytes using invertases conjugated to functional DNAs such as aptamers and DNAzymes,8 and the concept has also been applied for the development of a new method to quantify DNA using PGMs.9 In both cases, a direct relationship could be established between the concentrations of the targets in the samples and the glucose detected by PGMs. An important feature of the methods is that no modification of the PGM itself is required so that any commercial PGM can be used.
While use of functional DNA molecules10 in PGM-based detection has several advantages, such as high stability and low costs, the number of effective functional DNA molecules for useful medical targets are still limited at the current stage. In contrast, thanks to many years of research and development in both academic and industrial labs, numerous antibodies have been obtained for a much wider range of targets with excellent affinity and selectivity. In fact, medical diagnostics in clinical labs is dominated by the use of antibodies. With more than 50 years’ development since Yalow and Berson’s pioneer work on immunoassays,11 many analytical techniques have been coupled with the specific antibody-antigen interactions in these assays to transform the recognition events into physically detectable signals, including those based on nanoparticles,12 fluorescence,13–16 chemiluminescence,17 electrochemistry18–20 and mass spectrum.21 Although the use of these laboratory-based instruments and methods can improve the accuracy and sensitivity of the assays, they are not accessible to the public for diagnosis or detections at home or in the field. Colorimetric assays, such as many commercialized ELISA kits and test strips currently available in the market, are very useful for the public to detect many targets without instrumentation. However, they can only provide qualitative or semi-quantitative results based on eye observations of colors that may have variations between different people, while many diagnosis and quality control can benefit from quantitative analysis.
While it is important to develop a new method to take advantage of the much broader scope of targets that antibodies can bind for potential in-home diagnosis and detection using PGMs, the methods demonstrated previously in our group8, 9 based on the differences of DNA melting temperatures before and after the binding of the target or the DNA hybridization cannot be directly applied to PGM detection using antibodies, since no such enzymatic cleavage or conformational change upon target binding can be easily engineered into antibodies. Therefore a new challenge of signal transduction has to be overcome in order for antibodies to be used as a general platform for detecting non-glucose analytes using PGMs. Herein we present two general strategies of sandwich and competitive assays to use antibody-coated MBs for the detection of large protein biomarkers such as prostate-specific antigen (PSA) and small toxic molecules such as ochratoxin A, respectively.
RESULTS AND DISCUSSION
Detection of large protein biomarkers by PGM using a sandwich assay method
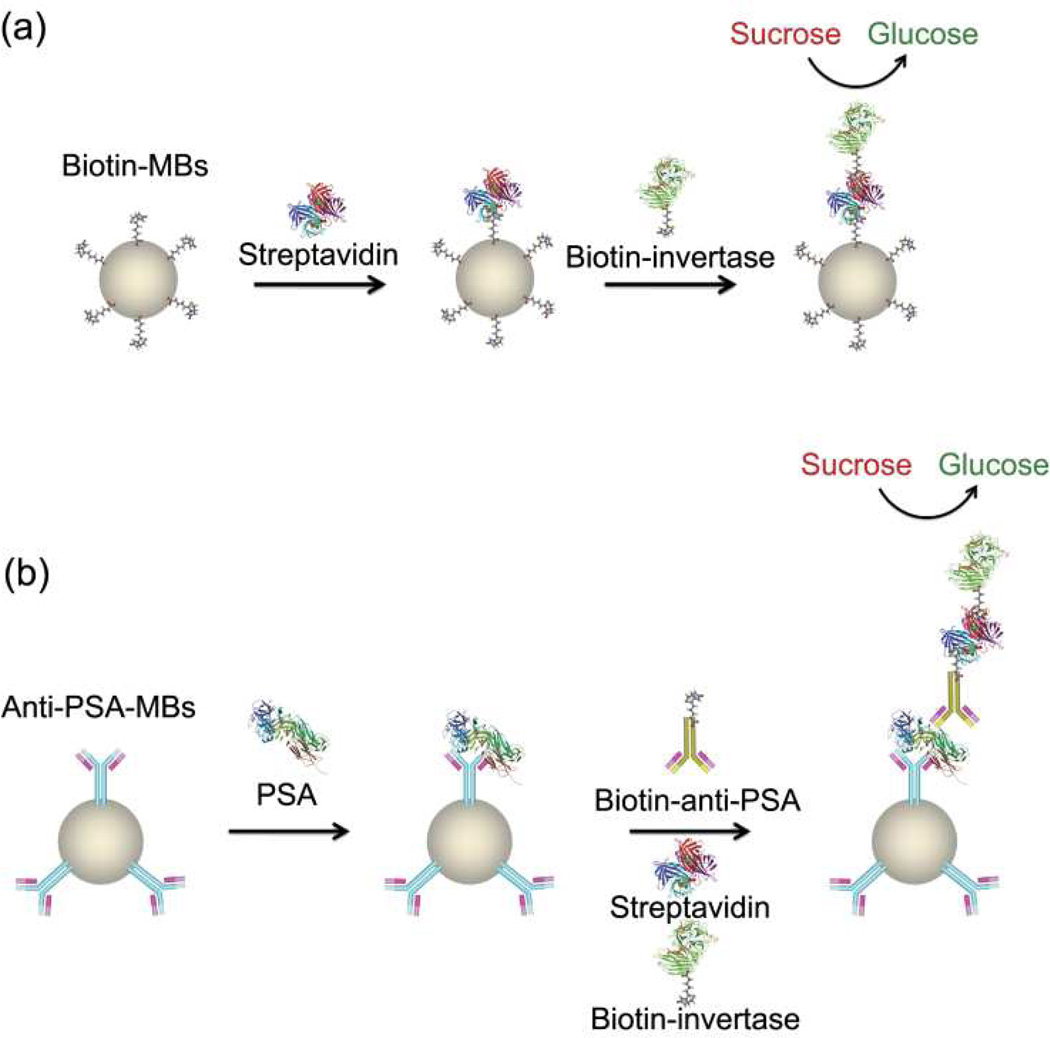
To expand the scope of targets detectable by PGMs, we first choose antibodies that can bind protein biomarkers, as protein biomarkers are ubiquitous for medical diagnostics.16, 22–24 To use antibodies for protein biomarkers in PGMs, we applied a sandwich assay method that utilized antibody-coated magnetic beads (MBs) (Figure 1). If the target (streptavidin or prostate specific antigen (PSA)) is present in the sample, it will bind to both the antibody-coated MBs and the antibody-conjugated invertase (here biotin is considered as “antibody” to bind the target strepavidin just like anti-PSA binds PSA) to form the sandwich complex (Figure 1). After magnetic separation from the solution, the resulting sandwich complex containing invertase conjugates bound on the MBs can be used to hydrolyze PGMs-inert sucrose into glucose, thus establishing direct correlation between the concentrations of the targets in the samples and the glucose monitored by PGMs, with glucose signal enhanced for the sandwich assay in the presence of the target (Figure 1). In the absence of the target, however, the formation of sandwich complex will not occur and no invertase could carry out the sucrose hydrolysis reactions after magnetic separation, therefore little glucose signal can be measured by PGMs.
Figure 1.
(a) Proof-of-concept sandwich assay for the detection of streptavidin by biotin coated MBs and invertase conjugates using PGMs. (b) Sandwich assay of PSA by anti-PSA-coated MBs and invertase conjugates using PGMs.
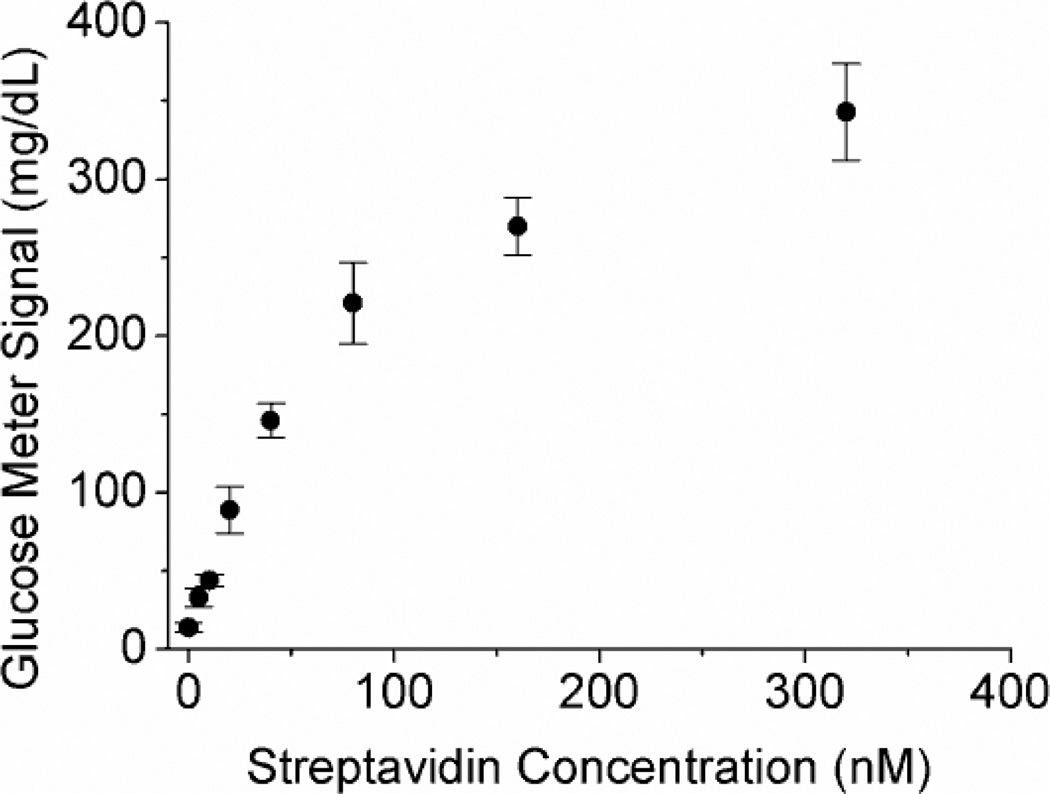
To demonstrate the general concept of sandwich assays using PGMs, we first use biotin as the binder “antibody” to detect the model protein target, streptavidin (Figure 1a). Biotin was immobilized onto amine-modified MBs using an N-Hydroxysuccinimide-PEG4-Biotin linker via the amine-NHS (N-Hydroxysuccinimide) coupling in the reaction buffer (0.2 M NaCl, 0.1 M sodium phosphate buffer, pH 7.3). In the presence of streptavidin, biotin-invertase conjugates were captured on the MBs due to the formation of the sandwich complex. The MBs were then placed into a sucrose solution in Buffer A (0.2 M NaCl, 0.1 M sodium phosphate buffer, pH 7.3, 0.05% Tween-20) to yield glucose for quantification in PGMs. As shown in Figure 2 the presence of streptavidin in the samples resulted in elevated glucose production, and the concentration of streptavidin in the sample was dependent on the glucose concentration measured by PGMs, showing an approximate linear relationship within 0~80 nM until the signal gradually reached saturation from 80 to 320 nM. The detection limit was determined to be 4.0 nM according to the definition of 3σb/slope (σb, standard deviation of the blank samples). The assay was very selective to streptavidin over bovine serum albumin (BSA) in the buffer because of the specific biotin-streptavidin interaction.
Figure 2.
The relationship between PGM signals and streptavidin concentrations in the samples for the sandwich assay of streptavidin using PGMs.
The proof-of-concept experiment shown above using PGMs to detect streptavidin encouraged us to apply the successful methodology to develop PGM-based sandwich assays to detect protein biomarkers of interest in medical diagnostics, i.e., prostate specific antigen (PSA),12, 25–28 which is a FDA-approved biomarker for prostate cancer.29
As shown in Figure 1b, the monoclonal anti-PSA antibody-conjugated MBs were prepared first and then added to the samples containing different concentrations of PSA, and another biotinylated polyclonal anti-PSA antibody was subsequently added to form a sandwich complex. Here, the monoclonal antibody immobilized on MBs could bind PSA with high affinity and specificity but only on one binding site per PSA, thus the polyclonal antibody, which binds PSA at different sites compared to the monoclonal one, was required for the successful formation of the sandwich complex. In addition, the different binding sites of the two antibodies minimized the potential competitive binding that could cause the release of bound PSA from MBs when the second antibody was added in excess. After the formation of the sandwich complex in the presence of PSA, addition of streptavidin and biotin-invertase conjugates further made biotin-invertase conjugates captured onto the MBs. After magnetic separation, the bound biotin-invertase conjugates on MBs could catalyze the production of glucose from sucrose, and the amount of glucose detected by the PGM can be used to calculate the concentration of PSA in the samples (Figure 1b).
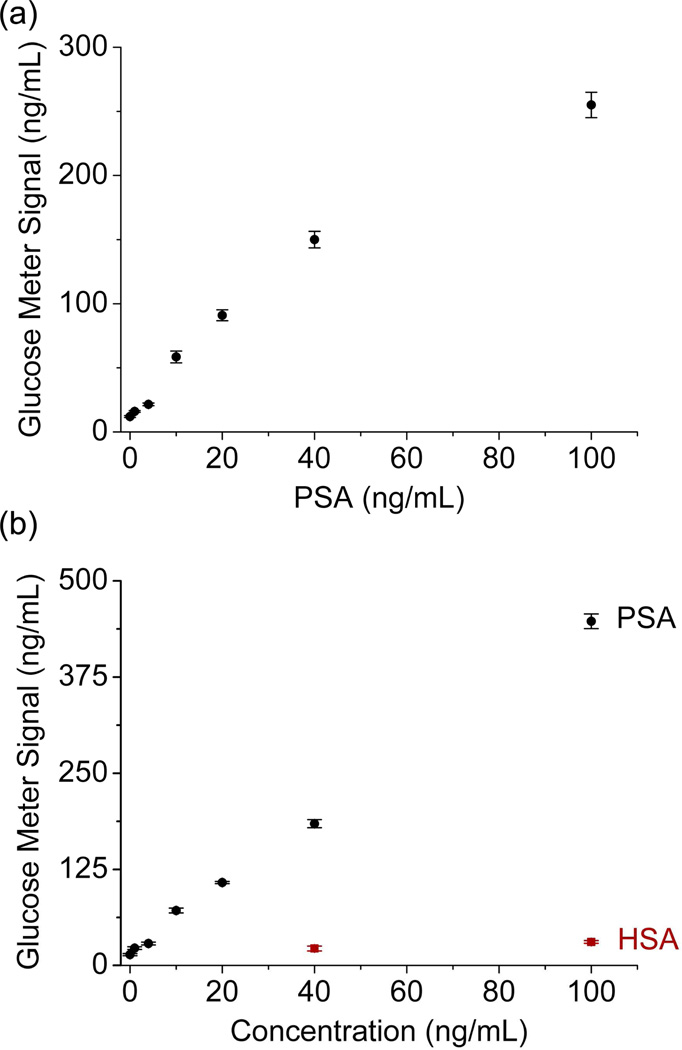
The PSA detection using this method was carried out in both Buffer B (Phosphate buffered saline (PBS, pH 7.0, 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.8 mM KH2PO4), 0.025% Tween-20, 1 g/L BSA) and 25% human serum diluted by Buffer B. As shown in Figure 3, in both cases, increasing amounts of PSA in the sample resulted in a higher glucose read out in the PGMs, with a detection range at least within 0~100 ng/mL PSA. Detection limits as low as 0.4 and 1.5 ng/mL were obtained from the data for the PSA detections in Buffer B (Figure 3a) and 25% human serum (Figure 3b), respectively, by the definition of 3σb/slope (σb, standard deviation of the blank samples). The detection limits achieved by the method indicate the promise for the detection of PSA in potential diagnosis applications of prostate cancer, since the cutoff level of normal blood PSA concentration is 4 ng/mL.12, 25–28 Moreover, because high concentrations of bovine serum albumin (BSA) were used in the assay buffers and human serum (HSA) as high as 100 mg/L gave neglectable response in PGMs under the same condition, the assay was selective to PSA over BSA or HSA as control proteins.
Figure 3.
The relationship between PGM signals and PSA concentrations in the samples for the sandwich assays of PSA using PGMs: (a) in Buffer B; (b) in 25% human serum.
Detection of small molecular targets by PGM using a competitive assay method
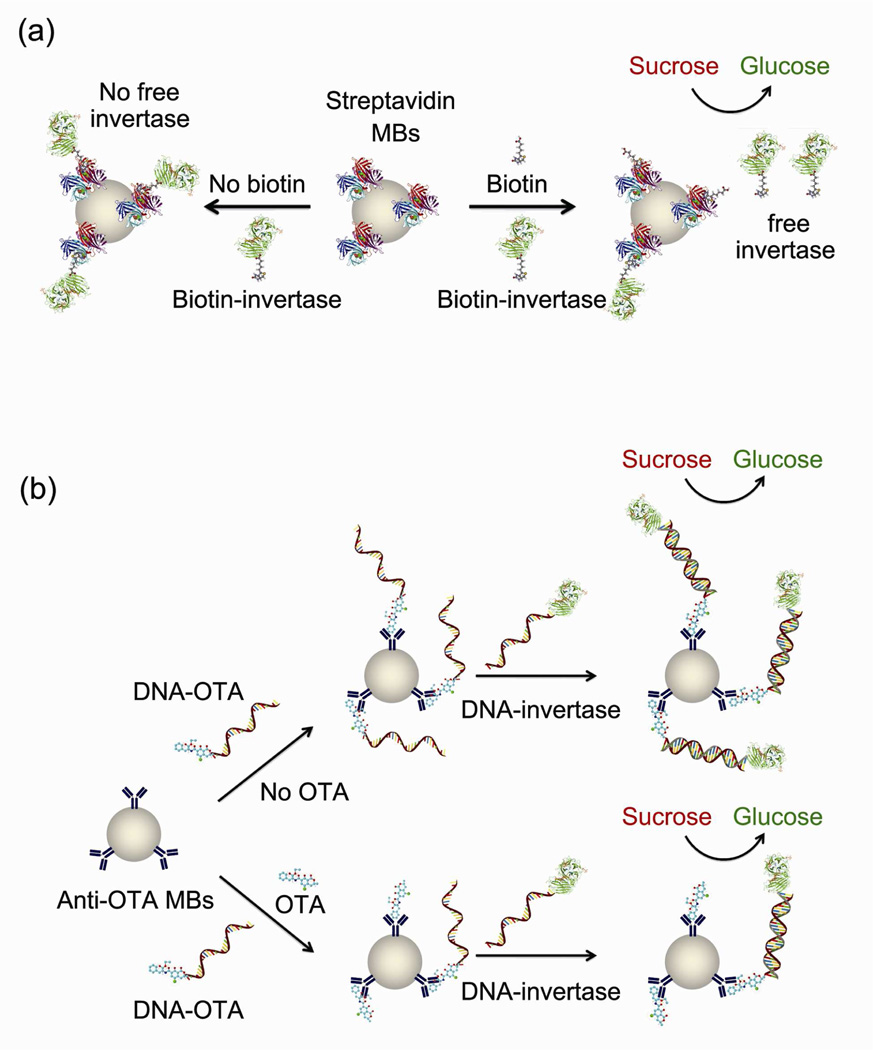
The above sandwich assay method can be very efficient when the target has multiple binding sites for at least two separated antibodies. For example, each streptavidin in Figure 1a has at least two (ideally four) binding sites for biotin, so the sandwich complex can be formed via the biotin-streptavidin-biotin mode. Other macromolecular analytes such as proteins biomarkers, nucleic acids and polysaccharides can also be assayed using the similar mechanism. However, when the target of interest has only one binding site, especially in the case of small molecular targets, sandwich assays are generally not applicable. To overcome this limitation for the detection of these mono-epitope targets, we used a competitive assay method (Figure 4). In a typical competitive assay, the target competes with its enzyme-labeled analogue to bind to an antibody. The more targets in the sample, the less enzyme-labeled analogues are bound to the antibody and yield corresponding changes in the signal readout (Figure 4). Note that the target and its enzyme-labeled analogues are allow to interact with the antibody simultaneously, thus the method is defined as competitive here rather than replacement.
Figure 4.
(a) Proof-of-concept competitive assay of biotin by streptavidin-coated MBs and biotin-invertase conjugates using PGMs. (b) Competitive assay of OTA by anti-OTA-coated MBs, DNA-OTA and DNA-invertase conjugates using PGMs.
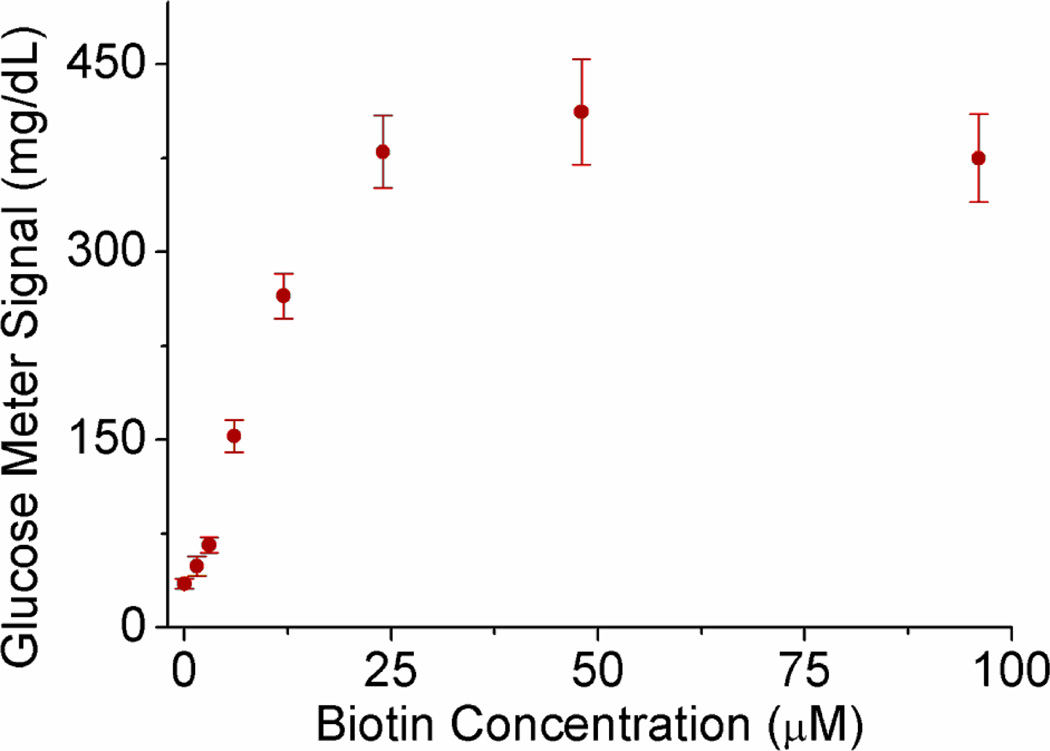
To demonstrate the general design of the competitive assay, we first use biotin and streptavidin as the pair of target-“antibody” for a proof of concept. Upon the addition of biotin as the target in the competitive assay, the amount of biotin-invertase conjugates bound to the streptavidin-coated MBs was reduced because the streptavidins on the surface of MBs were partially bound by the free biotin in the sample (Figure 4a). The final concentration of unbound biotin-invertase conjugates remaining in the solution after removal of MBs should be dependent on the concentration of biotin in the sample. Therefore, the glucose produced by the unbound biotin-invertase conjugates was detected by PGMs and the readout could be used for the quantification of biotin in the sample. The detection can also be realized by measuring the glucose produced by the biotin-invertase conjugates bound on the MBs, but we did not choose it because a signal-off glucose signal was observed instead of signal-on in this case. As expected, in the presence of increasing amount of biotin up to 96 µM, the final solution could catalyze the formation of more glucose from sucrose under the same condition, as detected by the PGM (Figure 5). The detection limit based on 3σb/slope (σb, standard deviation of the blank samples) from the data in the titration curve was as low as 1.5 µM. Other small organic molecules, such as uridine nucleotide and buffer components, did not give any signal or interfere with the biotin detection because of the highly specific interaction between biotin and streptavidin.
Figure 5.
The relationship between PGM signals and biotin concentrations in the samples for the competitive assays of biotin using PGMs
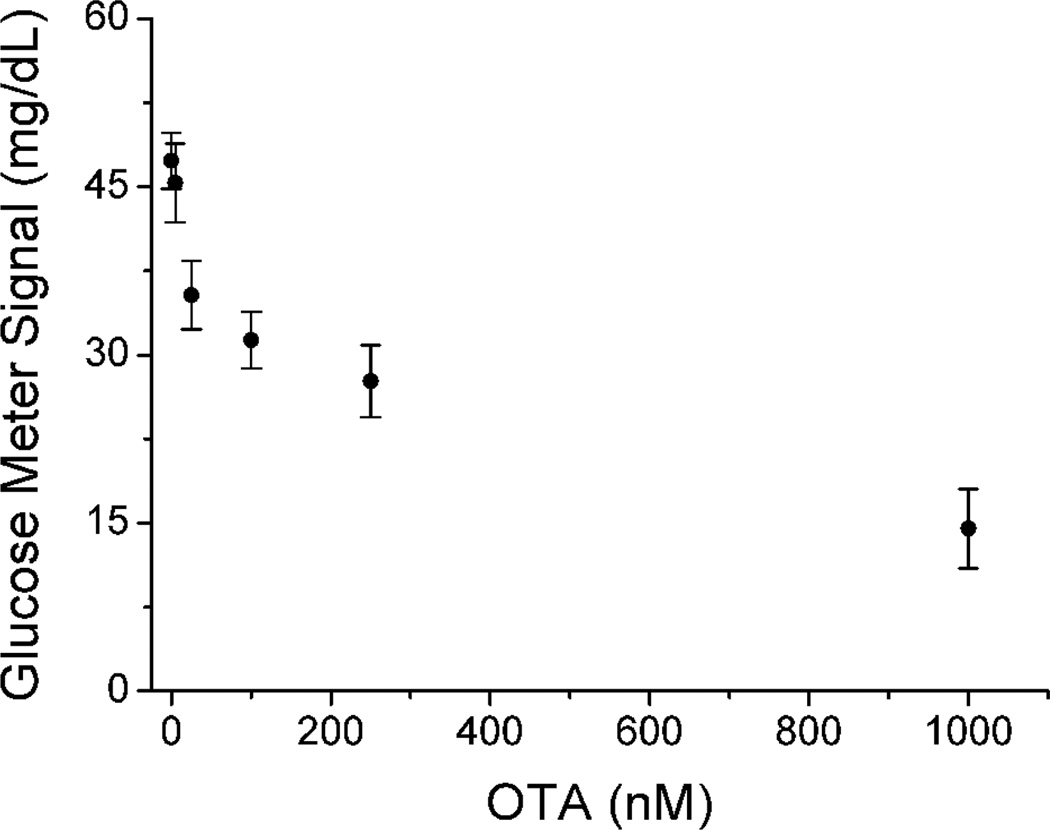
Having demonstrated the competitive assay method for detecting biotin as a model, we then investigated the application of the method to detect a small molecular toxin, Ochratoxin A (OTA),30–32 which is one of the most abundant food-contaminating mycotoxins worldwide.33, 34 Because OTA lacks multiple binding sites for antibodies, it cannot be detected via the sandwich assay method shown in Figure 1. Alternatively, the competitive assay (Figure 4b) can be used. We found that the activity of invertase was partially inhibited in the presence of high concentrations of OTA during conjugation, so the direct contact of concentrated OTA and invertase should be minimized to avoid false positive or negative results. Therefore, instead of directly conjugating OTA and invertase to yield the OTA-invertase as the enzyme-labeled OTA analogue, we propose to conjugate OTA with DNA first to form DNA-OTA conjugate (Figure S1 Supporting Information) and then the bound DNA-OTA conjugate on the surface of the MBs can further capture another complementary DNA-invertase conjugate for production of PGM detectable glucose (Figure 4b). In the presence of OTA, the target competed with its analogue of DNA-OTA conjugate in binding to the anti-OTA-conjugated MBs. The more OTA present in the sample, the less DNA-OTA conjugates captured onto the MBs. Subsequently, less DNA-invertase conjugates could bind with the DNA-OTA on the MBs via complementary DNA hybridization to catalyze the production of glucose from sucrose (Figure 4b). Therefore, the glucose concentration detected by the PGM was inversely related to the amount of DNA-OTA conjugates on the surface of MBs, which could be used for the calculation of the OTA concentration in the sample. This method was found to be able to detect OTA at least in the range of 0~1000 nM, and the detection limit was about 17 nM (6.8 ng/mL) according to the definition of 3σb/slope (σb, standard deviation of the blank samples) (Figure 6). The sensitivity of our method is comparable to the safe level of 3~10 ng/g in food (Official Journal of the European Communities, Commission Regulation (EC) No 472/2002) assuming that OTA from each 1 g of food can be finally extracted into 1 mL solution sample. Other small molecules such as uridine nucleotide and biotin did not interfere with the OTA detection, suggesting the selectivity in the antibody-antigen interaction.
Figure 6.
The relationship between PGM signals and OTA concentrations in the samples for the competitive assays of OTA using PGMs.
CONCLUSIONS
In conclusion, we have expanded the range of PGM-detectable targets significantly by using antibodies in the sandwich and competitive assays in measuring large protein biomarkers such as PSA and small molecular toxins such as OTA, respectively. The key to transform the binding event between the targets and their antibodies into a PGM-detectable signal is the use of invertase conjugates. Since the concentration of the invertase conjugates bound to the surface of the MBs are dependent on the concentration of the targets in the sample, the concentration of glucose produced from sucrose by invertase conjugates can then be used to calculate the concentration of targets in the sample after measurement by PGM. Since antibodies for a wide range of protein biomarkers for common diseases and toxins causing environment safety issues are readily available, the methodology shown here can be applied as a general platform for the portable and quantitative detection of many other disease biomarkers and toxins using PGMs. Because of the low cost, simple operation and wide availability of PGMs to the public, the methodology here can have significant impact on in-home personalized diagnosis and detections for the public.
Supplementary Material
ACKNOWLEDGMENT
We thank the U.S. National Institutes of Health (ES16865), Department of Energy (DE-FG02-08ER64568), and National Science Foundation (CTS-0120978) for financial support.
Footnotes
Supporting Information Available. Experimental details including materials, reagents and protocols, this material is free of charge on the World Wide Web at http://pubs.acs.org.
REFERENCES
- 1.Daar AS, Thorsteinsdottir H, Martin DK, Smith AC, Nast S, Singer PA. Nat. Genet. 2002;32:229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 2.Clark LC, Lyons C. Ann. N.Y. Acad. Sci. 1962;102:29–45. doi: 10.1111/j.1749-6632.1962.tb13623.x. [DOI] [PubMed] [Google Scholar]
- 3.Heller A, Feldman B. Chem. Rev. 2008;108:2482–2505. doi: 10.1021/cr068069y. [DOI] [PubMed] [Google Scholar]
- 4.Montagnana M, Caputo M, Giavarina D, Lippi G. Clin. Chim. Acta. 2009;402:7–13. doi: 10.1016/j.cca.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Carroll AE, Marrero DG, Downs SM. Diabetes Technol. Ther. 2007;9:158–164. doi: 10.1089/dia.2006.0002. [DOI] [PubMed] [Google Scholar]
- 6.Nie Z, Deiss F, Liu X, Akbulut O, Whitesides GM. Lab Chip. 2010;10:3163–3169. doi: 10.1039/c0lc00237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuma H, Sato N, Usui K. Anal. Chim. Acta. 2002;456:219–226. [Google Scholar]
- 8.Xiang Y, Lu Y. Nat. Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang Y, Lu Y. Anal. Chem. 2012 [Google Scholar]
- 10.Liu JW, Cao ZH, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yalow RS, Berson SA. Nature. 1959;184:1648–1649. doi: 10.1038/1841648b0. [DOI] [PubMed] [Google Scholar]
- 12.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, Mirkin CA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kingsmore SF, Patel DD. Curr. Opin. Biotechnol. 2003;14:74–81. doi: 10.1016/s0958-1669(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 14.Burgess STG, Kenyon F, O'Looney N, Ross AJ, Kwan MC, Beattie JS, Petrik J, Ghazal P, Campbell CJ. Anal. Biochem. 2008;382:9–15. doi: 10.1016/j.ab.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Ember SWJ, Schulze H, Ross AJ, Luby J, Khondoker M, Giraud G, Terry JG, Ciani I, Tlili C, Crain J, Walton AJ, Mount AR, Ghazal P, Bachmann TT, Campbell CJ. Anal. Bioanal. Chem. 2011;401:2549–2559. doi: 10.1007/s00216-011-5332-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhang HQ, Zhao Q, Li XF, Le XC. Analyst. 2007;132:724–737. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- 17.Zhao LX, Sun L, Chu XG. Trac-Trend. Anal. Chem. 2009;28:404–415. [Google Scholar]
- 18.Yang L-MC, Diaz JE, McIntire TM, Weiss GA, Penner RM. Anal. Chem. 2008;80:933–943. doi: 10.1021/ac071470f. [DOI] [PubMed] [Google Scholar]
- 19.Yang L-MC, Diaz JE, McIntire TM, Weiss GA, Penner RM. Anal. Chem. 2008;80:5695–5705. doi: 10.1021/ac8008109. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Wang T, Jiang J, Shen G, Yu R. Clin. Chem. 2009;55:964–971. doi: 10.1373/clinchem.2008.116582. [DOI] [PubMed] [Google Scholar]
- 21.Li F, Zhao Q, Wang CA, Lu XF, Li XF, Le XC. Anal. Chem. 2010;82:3399–3403. doi: 10.1021/ac100325f. [DOI] [PubMed] [Google Scholar]
- 22.Edwards R. Immunodiagnostics. Oxford: Oxford University Press; 1999. [Google Scholar]
- 23.Pavski V, Le XC. Curr. Opin. Biotechnol. 2003;14:65–73. doi: 10.1016/s0958-1669(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 24.Chen PR, He C. Curr. Opin. Chem. Biol. 2008;12:214–221. doi: 10.1016/j.cbpa.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Munge B, Patel V, Jensen G, Bhirde A, Gong JD, Kim SN, Gillespie J, Gutkind JS, Papadimitrakopoulos F, Rusling JF. J. Am. Chem. Soc. 2006;128:11199–11205. doi: 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao YP, Wei TF, Lefebvre PA, An H, He LX, Kunkel GT, Muller UR. Anal. Chem. 2006;78:2055–2059. doi: 10.1021/ac051798d. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Dai Q, Austin L, Coutts J, Knowles G, Zou JH, Chen H, Huo Q. J. Am. Chem. Soc. 2008;130:2780–2782. doi: 10.1021/ja711298b. [DOI] [PubMed] [Google Scholar]
- 28.Nie H, Liu S, Yu R, Jiang J. Angew. Chem. Int. Ed. 2009;48:9862–9866. doi: 10.1002/anie.200903503. [DOI] [PubMed] [Google Scholar]
- 29.Oesterling JE. J. Urol. 1991;145:907–923. doi: 10.1016/s0022-5347(17)38491-4. [DOI] [PubMed] [Google Scholar]
- 30.Ngundi MM, Shriver-Lake LC, Moore MH, Lassman ME, Ligler FS, Taitt CR. Anal. Chem. 2005;77:148–154. doi: 10.1021/ac048957y. [DOI] [PubMed] [Google Scholar]
- 31.Liu BH, Tsao ZJ, Wang JJ, Yu FY. Anal. Chem. 2008;80:7029–7035. doi: 10.1021/ac800951p. [DOI] [PubMed] [Google Scholar]
- 32.Yu FY, Vdovenko MM, Wang JJ, Sakharov IY. J. Agric. Food. Chem. 2011;59:809–813. doi: 10.1021/jf103261u. [DOI] [PubMed] [Google Scholar]
- 33.Pohland AE, Nesheim S, Friedman L. Pure Appl. Chem. 1992;64:1029–1046. [Google Scholar]
- 34.Pfohl-Leszkowicz A, Manderville RA. Mol. Nutr. Food Res. 2007;51:61–99. doi: 10.1002/mnfr.200600137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.