Abstract
The VLDL transport vesicle (VTV) mediates the transport of nascent VLDL particles from the ER to the Golgi and plays a key role in VLDL-secretion from the liver. The functionality of VTV is controlled by specific proteins; however, full characterization and proteomic profiling of VTV remain to be carried out. Here, we report the first proteomic profile of VTVs. VTVs were purified to their homogeneity and characterized biochemically and morphologically. Thin section transmission electron microscopy suggests that the size of VTV ranges between 100 nm to 120 nm and each vesicle contains only one VLDL particle. Immunoblotting data indicate VTV concentrate apoB100, apoB48 and apoAIV but exclude apoAI. Proteomic analysis based on 2D-gel coupled with MALDI-TOF identified a number of vesicle-related proteins, however, many important VTV proteins could only be identified using LC-MS/MS methodology. Our data strongly indicate that VTVs greatly differ in their proteome with their counterparts of intestinal origin, the PCTVs. For example, VTV contains Sec22b, SVIP, ApoC-I, reticulon 3, cideB, LPCAT3 etc. which are not present in PCTV. The VTV proteome reported here will provide a basic tool to study the mechanisms underlying VLDL biogenesis, maturation, intracellular trafficking and secretion from the liver.
Keywords: very low-density lipoprotein (VLDL), VLDL transport vesicle (VTV), apolipoprotein B (apoB), endoplasmic reticulum (ER), triacylglycerol (TAG)
1. INTRODUCTION
Elevated concentrations of circulating plasma very low-density lipoproteins (VLDL) pose a major risk for the pathogenesis of atherosclerosis leading to various cardiovascular diseases [1,2]. These atherogenic particles are produced by the liver and secreted into the blood. The biogenesis of VLDL occurs in the endoplasmic reticulum (ER) of liver cells or hepatocytes in a two-step process [3,4]. First, newly synthesized apolipoproteinB100 (apoB100) is partially lipidated to form the primordial VLDL particle and this process is facilitated by microsomal triglyceride transfer protein (MTP) [5–8]. In the second step, bulk of neutral lipids or triglycerides (TAG) are added to primordial particle to form a TAG-rich VLDL. Once formed in the lumen of ER, nascent VLDL particles move to the Golgi for their further processing [9–11]. The movement of nascent VLDLs from the ER to the Golgi is rate-determining step in their eventual secretion from the liver into the blood. This important transport event is mediated by a specialized vesicle, VLDL transport vesicle (VTV), which has emerged to be different morphologically and biochemically from other ER-derived vesicles.
The generation of VTV from hepatic ER membranes is a highly controlled process and mediated by coat-complex II (COPII) proteins [12–19]. COPII protein-complex is composed of five different cytosolic proteins viz. Sar1, Sec23-Sec24 and Sec13-Sec31 [12,14]. These proteins constitute minimal machinery for cargo-selection and vesicle formation from ER membranes [14]. The assembly of COPII complex begins with the recruitment of Sar1 to the ER membrane in its GTP-form, which is followed by serial recruitment of Sec23-Sec24 and Sec13-Sec31 as heterodimers [13]. Sar1 has two mammalian homologs, Sar1a and Sar1b [20]. Sar1a is primarily involved in the ER-to-Golgi transport of newly synthesized secretory proteins whereas Sar1b has been shown to facilitate the transport of lipoproteins from the ER to the Golgi. It has been reported that mutations in Sar1b are associated with reduced chylomicrons secretion from small intestinal epithelial cells, enterocytes leading to the pathogenesis of a rare disorder, chylomicron retention disease (CMRD) [21–25]. The role of Sar1b in the secretion of chylomicrons has been substantiated by a recent data demonstrating that over-expression of Sar1b in Caco2 cells leads to increased synthesis and secretion of chylomicrons [22]. Our published data have shown that Sar1 protein plays an important role in VLDL trafficking along the secretory pathway [26]. Using over-expression of dominant negative Sar1 (Sar1T39N) in McArdle cells, Gusarova et al showed a marked reduction in ER-exit of apoB100, a VLDL structural protein [27].
Despite similar requirement of COPII machinery for their exit from the same ER, both nascent proteins and VLDLs are transported from the ER to the Golgi in separate vesicles [26,27]. Recent studies have demonstrated that protein transport vesicle (PTV) and VTV are different in their protein compositions and utilize distinct sets of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins to form fusion-complex required for their fusion with cis-Golgi [28]. SNAREs are integral membrane proteins that facilitate the targeting of transport vesicles to their destinations and form a 4-member α-helix coiled-coil structure necessary for fusion with their target membranes [29–32]. Our data has shown that Sec22b, syntaxin 5, rBet1 and GOS28 form the SNARE-complex necessary for VTV-Golgi fusion [28]. Interestingly, the mechanisms underlying the biogenesis of intestinal ER-derived pre-chylomicron transport vesicle (PCTV) and VTV are not the same [26,33–36]. The formation of VTV from hepatic ER requires guanosine-5′-triphosphate (GTP) whereas PCTV-budding from intestinal ER is GTP-independent [26,34]. Moreover, both the VTV and PCTV utilize different SNARE proteins to fuse with and deliver their cargoes (VLDL and pre-chylomicron, respectively) to cis-Golgi [28,37–39]. These observations indicate the existence of cargo-specific vesicular transport mechanisms.
Although the role of VTV in VLDL transport and secretion is of paramount importance, these unique vesicles are poorly studied and their proteomic analysis remains to be carried out. In this study, we performed a detailed proteomic analysis to characterize VTV proteome. Identification of a complete VTV proteome would reveal proteins important in intracellular VLDL transport and their secretion, offering putative therapeutic targets for hyperlipidemia, atherosclerosis and associated complications. We report here a detailed characterization and proteomic analysis of VTV with an aim to identify proteins crucial for VLDL synthesis, intracellular transport and secretion.
2. MATERIALS AND METHODS
Materials
Sprague-Dawley rats, 150 – 200 g were procured from Harlan (Indianapolis, IN). [3H]oleic acid (9.2 Ci/mM) was purchased from Perkin Elmer Life Sciences (Boston, MA). Gel electrophoresis and immunoblotting reagents were procured from Bio-Rad (Hercules, CA). Enhanced chemiluminescence (ECL) reagents were obtained from GE Healthcare. Other reagents used were of analytical grade and purchased from local companies.
Polyclonal antibodies against mammalian Sar1 have been characterized earlier [34]. Mouse monoclonal anti-Sec22b antibodies and rabbit polyclonal anti-syntaxin 5 (Syn5) antibodies were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-GOS28 antibodies were procured from StressGen Biotechnologies (Victoria, Canada). Rabbit anti-albumin antibodies were purchased from Bethyl Laboratories Inc. (Montgomery, TX). Goat anti-calnexin antibodies, goat anti-rabbit IgGs, goat anti-mouse IgG, and goat anti-rabbit IgGs conjugated with horseradish peroxidase (HRP) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Goat anti-rabbit IgGs conjugated with agarose beads was purchased from Sigma Chemical Co. (St. Louis, MO).
Preparation of hepatic endoplasmic reticulum and cytosol
ER containing [3H]-TAG was prepared using essentially the same procedure as described earlier [26]. Briefly, freshly isolated primary rat hepatocytes were washed thoroughly with cold 0.25 M sucrose in 10 mM HEPES (pH 7.2) and incubated with [3H]oleate (100 μCi) conjugated with albumin for 30 min at 37 °C and washed twice with phosphate buffered saline (PBS) containing 2% BSA to remove the excess [3H]oleate, homogenized in Buffer A (10 mM Hepes, pH 7.2, 0.25 M sucrose, 0.5 mM EGTA and protease inhibitors) using a Parr bomb and the ER was isolated using a sucrose step gradient [26,34] that was repeated to purify the ER. The purity of the ER was determined by Western blots using marker proteins as discussed previously [26,28].
Cytosol was prepared from primary rat hepatocytes using the same methodology as described previously [26]. To remove endogenous ATP and GTP, cytosol was dialyzed twice against buffer ice-cold buffer [25 mM Hepes (pH 7.2),125 mM KCl, 2.5 mM MgCl2, 0.5 mM DTT and protease inhibitors] for 6 hours at 4 °C [26].
In vitro VTV formation
To generate VTVs containing [3H]TAG, we used the same methodology as discussed earlier [26]. Briefly, ER containing [3H]-TAG (500 μg protein) was incubated at 37 °C for 30 min with hepatic cytosol (1 mg protein), 1 mM GTP and an ATP-regenerating system (1 mM ATP, 5mM phosphocreatine and 5 units of creatine phosphokinase) in the absence of Golgi acceptor (total reaction volume 0.5 ml). Post-incubation, the reaction mixture was resolved on a continuous sucrose density gradient (0.1 to 1.15 M sucrose) and VTVs isolated from the light portions of the gradient. VTVs thus formed were concentrated using YM-10 centricons (Millipore).
Measurement of TAG radioactivity
Radioactivity associated with TAG and protein was determined as described previously [26,34]. TAG was extracted from vesicular fractions and the associated radioactivity was quantitated [26,34]. The single-isotope mode was used on the scintillation analyzer (TriCarb, Model 2910, Perkin Elmer).
SDS-PAGE and Immunoblotting
SDS-PAGE and immunoblotting were carried out similarly as described [26]. Briefly, protein samples were solubilized in 2x Laemmli’s buffer and separated by SDS-PAGE electrophoresis. For immunoblotting, proteins resolved by SDS-PAGE were transblotted on to nitrocellulose membranes (Bio-Rad) as described previously [26]. The membrane was then blocked with 5% (w/v) non-fat dried skimmed milk in PBS, incubated with specific primary and then secondary antibodies. Detection of protein bands was accomplished by treating the membrane with ECL reagents and exposing to Biomax film (Eastman Kodak Co., Rochester, NY) [26].
Two-dimensional (2D) gel electrophoresis
Preparation of sample
A total of 300 μg of VTV proteins were used for 2D-gel. First, 30 μg of VTV proteins were solubilized in 2D gel sample buffer [7 M urea, 2 M thiourea, 4% CHAPS, 2 mM tri-butyl phosphine (TBP), 0.5% carrier ampholyte (pH 3–10), 40 mM Tris]. Proteins were labeled Cy5 dye by mixing with 1.0 μl of diluted CyDye followed by incubation at 4 °C for 30 min in the dark. The reaction was stopped by adding 1.0 μl of 10 mM lysine to the sample and incubating in dark on ice for an additional 15 min.
The labeled sample was then mixed with 270 μg of unlabeled VTV proteins. The 2D gel sample buffer [7 M urea, 2 M thiourea, 4% CHAPS, 2 mM tri-butyl phosphine (TBP), 0.5% carrier ampholyte (pH 3–10), 40 mM Tris], 100 μl destreak solution were added to the labeling mix to make the total volume of 250 μl, mixed well and loaded onto immobilized pI gradient gel (IPG) strips (pH 3–10) (GE Healthcare) by overnight incubation at room temperature.
IEF and SDS-PAGE
Isoelectric focusing was carried out following the protocol provided by GE Healthcare and a total of 50,500 volt-hours were applied to each IPG strip. Following isoelectric focusing, the IPG strip was first treated with equilibration buffer I [0.5 M Tris-HCl pH 6.8, 6 M urea, 30% glycerol (v/v), 2% SDS, 25 mM DTT] for 10 minutes and then with buffer II [0.5 M Tris-HCl pH 6.8, 6 M urea, 30% glycerol (v/v), 2% SDS, 12 mM iodoacetamide] for 10 minutes. The IPG strip was loaded onto a gradient gel (8–18% SDS-polyacrylamide gel) for proteins separation. The SDS-gel was run at 15 °C until the dye front was running out of the gel.
Image scan and data analysis
Gel images were scanned immediately following the SDS-PAGE using Typhoon TRIO (GE Healthcare). The scanned images were then analyzed by Image Quant software (version 6.0, GE Healthcare), followed by in-gel analysis using DeCyder software version 6.5 (GE Healthcare).
Mass Spectrometry and protein identification
Spot picking, trypsin digestion and MALDI-TOF
The spots of interest were picked up by Ettan Spot Picker (GE Healthcare) based on the in-gel analysis and spot picking design by DeCyder software. The gel spots were washed a few times then digested in-gel with modified porcine trypsin protease (Promega). The digested tryptic peptides were desalted using a Zip-tip C18 (Millipore). Peptides were eluted from the Zip-tip with 0.5 μl of matrix solution (α-cyano-4-hydroxycinnamic acid (5 mg/ml in 50% acetonitrile, 0.1% trifluoroacetic acid, 25 mM ammonium bicarbonate) and spotted on a MALDI plate (model ABI 01-192-6-AB).
MALDI-TOF MS was performed on AB SCIEX TOF/TOF™ 5800 System (AB SCIEX). MALDI-TOF mass spectra were acquired in reflectron positive ion mode, averaging 4000 laser shots per spectrum.
NanoLC-MS/MS
VTV proteins (150 μg) were precipitated by methanol then resuspended in 50 mM ammonium bicarbonate. DTT was added to the final concentration of 10 mM and incubated at 60 °C for 30 min, followed by cooling down to room temperature, iodoacetamide was added to final concentration of 10 mM and incubated in dark for 30 min at room temperature. The proteins were then digested by Trypsin (Promega) overnight at 37 °C. NanoLC was carried out using a Dionex Ultimate 3000 (Milford, MA). Mobile phase solvents A and B were 0.1% TFA (v/v) in water and 0.1% TFA (v/v) in 80% acetonitrile, respectively. Trypic peptides were loaded onto a pre-column cartridge (Dionex) and then separated using a nanoLC column (Dionex) applying a 150-min acetonitrile gradient (ranging from 5% to 60%). Fractions were collected at 20-second intervals followed by mass spectrometry analysis on AB SCIEX TOF/TOFT 5800 System (AB SCIEX). We carried out LC-MS/MS analysis two times and the data was reproducible. We carried out LC-MS/MS analysis two times and the data was reproducible.
Database search
Both of the resulting peptide mass and the associated fragmentation spectra were submitted to GPS Explorer workstation equipped with MASCOT search engine (Matrix science, http://www.matrixscience.com) to search the database of National Center for Biotechnology Information non-redundant (NCBInr). Searches were performed without constraining protein molecular weight or isoelectric point, with variable carbamidomethylation of cysteine and oxidation of methionine residues, and with one missed cleavage also allowed in the search parameters. Candidates with either protein score C.I.% (Confidence Interval) or Ion C.I.% greater than 95 were considered significant.
Electron microscopy
To study the morphology of VTVs, we first used the negative-staining technique of electron microscopy as discussed previously [26,28,34]. In brief, a formvar-carbon coated nickel grid was placed on a drop of concentrated VTV fraction for 2–3 minutes, rinsed with PBS and water. The grid was then stained with 0.5% aqueous uranyl acetate, air-dried and examined using a FEI Morgagni 268(D) transmission electron microscope (FEI, Hillsboro, Oregon) at 10,000x magnification.
To visualize VTVs by thin section electron microscopy, VTVs were concentrated and then pelleted by centrifugation (3000 ×g for 10 minutes using a table-top microcentrifuge at 4°C). The pelleted VTVs were fixed first in 2.5% glutaraldehyde and then in 1% osmium tetroxide buffered with 0.2 M imidazole, pH 7.2. Fixed VTVs were embedded in Spur medium and sections were stained with uranyl acetate and lead citrate. The thin sections were probed utilizing a FEI Morgagni 268(D) transmission electron microscope (FEI, Hillsboro, Oregon) at 6,000x magnification.
Statistical analysis
Comparisons between means were carried out using a statistical package supplied by GraphPad Software (Instat, GraphPad Software, San Diego, CA) using a two-tailed t test.
3. RESULTS
Purity of subcellular fractions
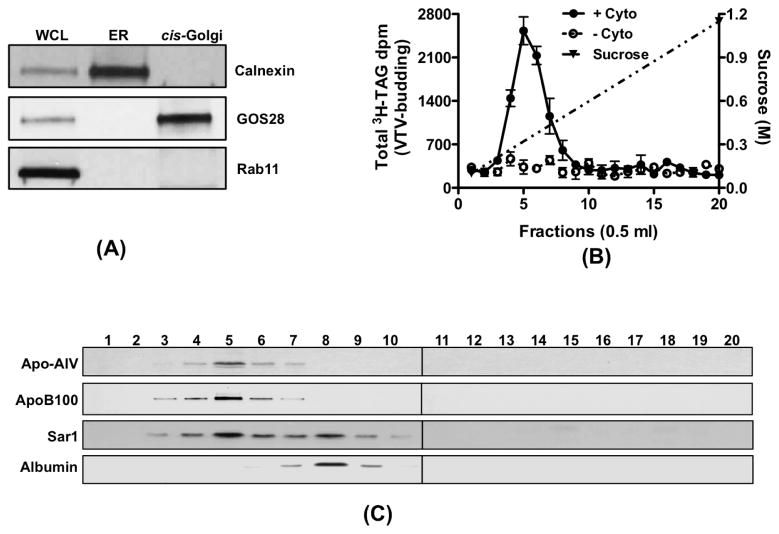
Since our in vitro assays entirely depend on various sub-cellular organelles, it was imperative to establish the purity of our sub-cellular organelles isolated from primary rat hepatocytes prior to their use in various assays. Hepatic ER membranes contained calnexin, an ER resident protein [40], but did not have recognizable GOS28, a cis-Golgi protein and rab11, an endosomal/lysosomal protein as determined by Western blots (Fig. 1A). As shown in figure 1A, the cis-Golgi fraction was free from ER resident protein, calnexin and endosomal/lysosomal marker protein, rab11. The cis-Golgi membranes were, however, enriched with GOS28 (Fig. 1A). We next checked whether our cis-Golgi contains TGN38, a trans-Golgi protein. TGN38 was concentrated in trans-Golgi fraction but very little amount of TGN38 was present in cis-Golgi (data not shown), which is consistent with our previous observations [26]. These results suggest that our sub-cellular organelle preparations were not contaminated and were suitable for our in vitro assays.
Figure 1.
VTV-budding and their distribution in continuous sucrose density gradient. (A) Protein samples containing liver whole-cell lysate (WCL), ER and cis-Golgi (each sample contains 30 μg of protein) were separated by SDS-PAGE (12% gel), transblotted on to a nitrocellulose membrane and probed with specific antibodies against calnexin, GOS28, and Rab11. For details, see methods. Proteins were detected using ECL. (B) Hepatic ER containing [3H]TAG was incubated at 37 °C with or without rat liver cytosol in the presence of an ATP-regenerating system. The reaction mix was resolved on a continuous sucrose gradient (0.1–1.15 M) and fractions of 0.5 ml were collected. The [3H]TAG dpm from each fraction (500 μl) was counted. Values are the means ± S.E.M. (n = 4) (C) Distribution of apo-AIV, apoB100, Sar1 and albumin across the continuous sucrose gradient. Hepatic ER, cytosol and an ATP-regenerating system were incubated for 30 min at 37 °C. The incubation mix was resolved on a continuous sucrose gradient an equal volume (100 μl) of each fraction was separated by SDS-PAGE (5–15% gels for apoB). The proteins were transferred on to nitrocellulose membrane and probed with antibodies against indicated proteins. Proteins were detected using ECL. The data are representative of four experiments.
Isolation and characterization of VTV
VTV were prepared from purified hepatic ER membranes employing the same in vitro ER-budding assay that we have established previously [26]. Using [3H]-TAG containing hepatic ER as donor; the incubation at 37° C for 30 minutes in the presence of hepatic cytosol, GTP and an ATP regenerating system resulted in the appearance of [3H]-TAG-rich VTV in the expected light density fractions. The distribution of [3H]-TAG dpm in the continuous sucrose density gradient is shown in figure 1B. As expected, the maximal [3H]-TAG dpm appeared in the light density fractions of the gradient suggesting that these fractions contain putative TAG-rich VTVs. In an attempt to support our results, we explored the distribution of other VLDL markers, apoB100 and apoA-IV in the continuous sucrose gradient. Figure 1C shows that both apoB100 and apoA-IV are present in light density fractions reassuring that these fractions contain putative VTVs.
To show that VTV fractions contain an ER-derived vesicle marker protein and exclude PTV proteins, we immunoblotted the same membrane for Sar1 and albumin. As shown in figure 1C, the presence of Sar1, an ER-derived vesicle marker, in lighter fractions where apoB100 and apoA-IV are distributed strongly suggests that VTVs are derived from the ER. The presence of Sar1 in the mid-portion of the gradient indicates the distribution of PTVs in these fractions, which is consistent with previous observations [26]. To determine that VTV fractions exclude ER-resident proteins, we immunoblotted for calnexin, an ER-resident protein, our results indicated that calnexin was not present in the sucrose density fractions (data not shown). We have shown previously that albumin, a secretory protein exits the hepatic ER in PTV [26]. Consistent with our previous finding, albumin is distributed in mid-portion of the gradient, the expected place for PTVs (Fig. 1C).
To confirm that we have isolated bona fide VTVs for proteomic analysis, we immunoblotted for Sec22b, a functional v-SNARE for VTV. As shown in figure 2A, Sec22b is concentrated in VTV fractions as compared to equal amount of hepatic ER protein sample. Another v-SNARE protein localized to ER-derived vesicles, Ykt6, was not found in VTV fractions whereas hepatic ER contains Ykt6 (Fig. 2A). To show the purity of our VTV sample, we probed for other SNARE proteins vti1a and VAMP7. Our results suggest that vti1a and VAMP7 are not present in VTV (Fig. 2A), which is consistent with our previous findings [28].
Figure 2.
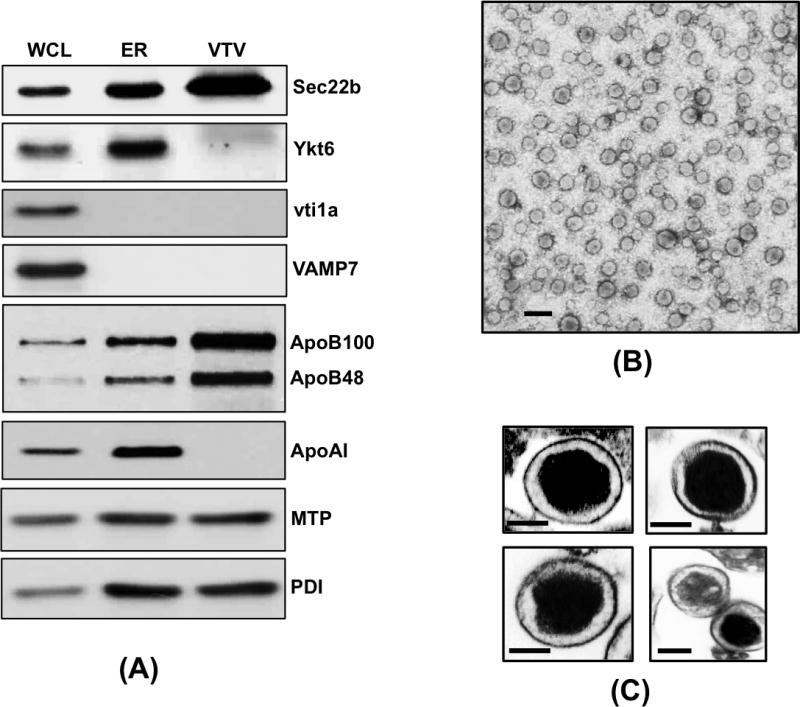
Biochemical and morphological characterization of VTVs. (A) Samples of hepatic whole-cell lysate (WCL), ER and VTVs (each sample contains 35 μg of protein) were separated by 12% SDS-PAGE (for ApoB100 and ApoB48 5–15% gel was used), transblotted on to a nitrocellulose membrane and probed with specific antibodies against the indicated proteins. Protein detection was done by ECL reagents. The data are representative of four independent experiments. (B) VTVs were generated from hepatic ER as in figure 1B. The VTVs (fractions 4–6) were examined by EM using the negative-staining technique (see the Methods for details). Bar size 200 nm. (C) Thin section transmission microscopy of VTVs. VTV were stained and processed for thin section EM (6000x). The size of VTVs ranges between 100–120 nm in diameter and VTVs contain osmophilic lipid (see Methods for details). Bar size 50 nm.
Next we carried out electron microscopy (EM) to visualize putative VTVs morphologically. The VTVs fractions were concentrated, stained with uranyl acetate and examined using negative staining EM technique. Figure 2B shows an electron micrograph of pure vesicles. We determined the diameter of these vesicles and found that most of the vesicles range from 100 to 120 nm in their size (Fig. 2B & 2C). Examination of VTVs by transmission electron microscopy (TEM) of thin sections showed that VTVs are closed compartments containing one VLDL particle per vesicle (Fig. 2C). Taken together, these biochemical and morphological data re-affirm that our vesicular fractions contain bona fide VLDL transport vesicles that can be used for proteomic analysis.
Proteomic analysis of VTV
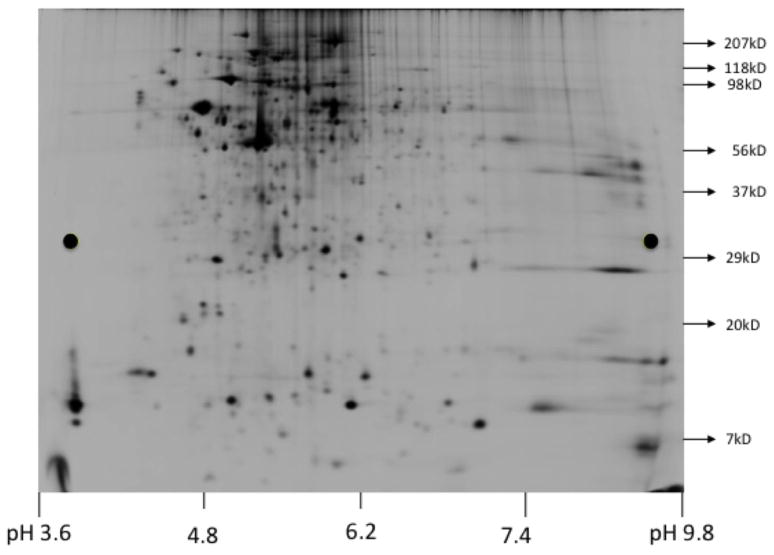
To characterize VTVs proteome, our first approach was to resolve purified VTVs by two-dimensional gel electrophoresis (2D gel) and analyze each protein spot using MALDI-TOF MS. This approach provided us an initial identification of VTV proteins. 2D gel was performed (see Methods) to resolve Cy5 labeled VTV proteins and the gel was scanned immediately using Typhoon TRIO (GE Healthcare) as shown in figure 3. Protein spots were excised using an automated spot picker (Ettan, GE Healthcare) and MALDI-TOF MS was performed on each protein spot. Using MALDI-TOF data, we searched online protein database using MASCOT (Matrix Science, http://www.matrixscience.com) search engine against database of NCBInr to identify the proteins. As presented in Table I, we identified 28 proteins, however, a number of proteins as depicted in figure 3 could not be identified confidently utilizing this methodology.
Figure 3.
Two-dimensional SDS-PAGE of VTVs. The VTV proteins (300 μg) labeled with Cy5 dye were solubilized in 2D-gel buffer (see Methods). Isoelectric focusing was carried out using a total of 50,500 volt-hours. Post-isoelectric focusing, proteins were first equilibrated (see Methods) and then separated by 8–18% SDS-PAGE. Gel was scanned immediately using Typhoon TRIO (GE Healthcare).
Table I.
VTV-proteins identified with high confidence employing 2D-gel and MALDI-TOF mass spectrometry
| VTV Proteins | NCBI database Accession # | Mr (kDa) | pI value | Peptides matched |
|---|---|---|---|---|
| Intracellular Trafficking Proteins | ||||
| Rab1a | gi|45433570 | 23.58 | 5.93 | 8 |
| Reticulon-3 | gi|77567873 | 83.01 | 8.66 | 11 |
| Huntingtin-associated protein 1 | gi|30061483 | 72.34 | 4.71 | 10 |
| Proteins involved in vesicle formation | ||||
| Sec24C | gi|157822493 | 118.52 | 6.71 | 7 |
| Sec31A | gi|14717392 | 143.64 | 6.65 | 11 |
| Vesicular targeting/docking/fusion proteins | ||||
| Syntaxin 5 | gi|114152883 | 35.01 | 9.03 | 9 |
| Sec22b | gi|71043604 | 24.84 | 8.67 | 12 |
| Vesicle cargo proteins | ||||
| p58 | gi|1174158 | 57.96 | 5.92 | 8 |
| Apolipoprotein A-IV | gi|114008 | 44.45 | 5.12 | 11 |
| Proteins involved in metabolic pathways | ||||
| Phosphatidylethanolamine binding protein 1 | gi|149063506 | 10.67 | 6.26 | 9 |
| Sterol O-acyltransferase 2 | gi|40254723 | 60.49 | 8.98 | 5 |
| Lipid-binding/transport proteins | ||||
| Microsomal triglyceride transfer protein | gi|149026062 | 99.18 | 7.54 | 15 |
| Liver fatty acid binding protein p14 | gi|204074 | 13.42 | 6.74 | 7 |
| Molecular chaperones | ||||
| Heat shock protein 27 | gi|204665 | 22.90 | 6.12 | 6 |
| Heat shock cognate 71 | gi|13242237 | 70.87 | 5.37 | 14 |
| Valosin-containing protein | gi|38014694 | 89.34 | 5.14 | 11 |
| Proteins regulating cell functions | ||||
| Regucalcin | gi|13928740 | 33.40 | 5.27 | 19 |
| Annexin A5 | gi|6978505 | 35.74 | 4.91 | 8 |
| Endoplasmic reticulum regulatory proteins | ||||
| Liver microsomal Carboxylesterase | gi|562010 | 64.52 | 6.25 | 12 |
| Perchloric acid soluble protein | gi|4456766 | 10.35 | 4.93 | 6 |
| Structural proteins | ||||
| β-actin | gi|55575 | 41.73 | 5.29 | 16 |
| Tubulin, α1A | gi|50926060 | 50.13 | 4.94 | 7 |
| Miscellaneous Enzyme | ||||
| α–enolase isoform 1 | gi|158186649 | 47.13 | 6.16 | 11 |
| Glutathione S-transferase α-3 | gi|13928688 | 25.31 | 8.78 | 12 |
| Ribonuclease UK114 | gi|14269572 | 14.30 | 6.20 | 7 |
| Creatine kinase M-type | gi|6978661 | 43.01 | 6.58 | 10 |
| Other proteins | ||||
| Cell death activator Cide-B | gi|157818189 | 27.94 | 8.41 | 12 |
| Selenium binding protein 2 | gi|149030730 | 50.57 | 5.94 | 14 |
In order to identify VTV proteins with high confidence, we decided to perform LC-MS/MS mass spectrometry. The sequencing data acquired was used in the MASCOT (Matrix Science, http://www.matrixscience.com) search engine against database of NCBInr to identify proteins. Using LC-MS/MS technique, we positively identified 77 VTV proteins, which are listed in Table II. Utilizing this approach, we were able to identify more proteins that were missing in our initial proteomic analysis based on 2D-gel coupled with MALDI-TOF MS experiments. Identification of apoA-IV in VTV fractions using LC-MS/MS was significant because it indicates that this apoprotein is recruited to the nascent VLDL particle in the ER lumen. Interestingly, we identified apoB100, an essential VLDL structural protein, which is known to be present in VTVs but could not be recognized by 2D-gel and MALDI-TOF approach. We could not identify apoB48 using both approaches, which is consistent with previous published reports from Adeli’s group [41,42], however, the presence of both apoB100 and apoB48 in VTVs was confirmed by immunoblotting data (Fig. 2A). Surprisingly, we were able to identify MTP in VTVs but were unable to identify protein disulfide isomerase (PDI) utilizing both mass spectrometric approaches. Since PDI is a structural binding partner of MTP, we decided to perform immunoblotting for MTP and PDI on VTV. Figure 2A shows that both MTP and PDI are present in VTVs; however, these proteins are not concentrated in VTVs as compared to the ER. These data suggest that MTP and PDI are present in VTV but they are not the preferential cargo for the VTV. The inability to identify the PDI using mass spectrometric approach raises two possibilities: (i) since immunoblotting data indicate no enrichment of PDI in the VTV; therefore it is possible that detection may be less sensitive; (ii) secondly, since there are several isoforms of PDI, it is very likely that information of a particular isoform of PDI (which is present in VTVs) is not available in the database, which is consistent with other published reports in which the identification of apoB48, VAMP7 Sar1 could not be accomplished using LC-MS/MS but these proteins were detected by immunoblotting [41,42]. Identification of rab1 and Sec31A by 2D-gel and MALDI-TOF approach was interesting because these two proteins play an important role in ER-to-Golgi transport of nascent proteins. These two proteins, however, were not found in VTVs by LC-MS/MS analysis. One possibility is that both rab1 and Sec31A are transiently associated with VTVs, which makes their detection less sensitive. As illustrated in Table II, we were able to identify proteins to be specifically associated with ER-derived vesicles and VLDL. For the sake of clarity, we have categorized the identified proteins according to their functionality (Table II).
Table II.
VTV-proteins identified with high confidence employing nano LC-MS/MS
| VTV Proteins | NCBI database Accession # | Mr (kDa) | Peptides matched |
|---|---|---|---|
| Intracellular Trafficking Proteins | |||
| Reticulon-1 | gi|16758732 | 83.01 | 7 |
| Reticulon-3 | gi|77567873 | 25.43 | 18 |
| Reticulon-3 isoform A1 | gi|158261988 | 25.43 | 16 |
| Rab6 interacting protein 2, isoform CRA_a | gi|149049596 | 108.45 | 8 |
| Rabaptin-5 | gi|2329847 | 99.13 | 11 |
| Huntingtin-associated protein 1 | gi|30061483 | 72.34 | 17 |
| Sterol carrier protein | gi|206870 | 10.47 | 10 |
| Small VCP/p97-interacting protein | gi|73919470 | 8.25 | 12 |
| Ras-related protein | gi|498257 | 20.48 | 5 |
| Ras p21-like small GTP-binding protein | gi|206567 | 24.39 | 7 |
| Proteins involved in vesicle formation | |||
| SEC23-interacting protein | gi|198442887 | 110.94 | 9 |
| Sec24C | gi|157822493 | 118.52 | 13 |
| Sec31B | gi|208973278 | 135.59 | 8 |
| Vesicular targeting/docking/fusion proteins | |||
| Membrin | gi|1907388 | 24.61 | 6 |
| Syntaxin 5 | gi|114152883 | 35.01 | 15 |
| Syntaxin 17 | gi|9297065 | 36.16 | 8 |
| Vesicle-associated membrane protein-associated protein A | gi|118142811 | 27.84 | 7 |
| Sec22b | gi|71043604 | 24.74 | 12 |
| Vesicle cargo proteins | |||
| p58 | gi|1174158 | 57.96 | 7 |
| Apolipoprotein B100 | gi|81894378 | 538.69 | 17 |
| Apolipoprotein A-IV | gi|114008 | 44.45 | 6 |
| Apolipoprotein C-I | gi|71051103 | 9.86 | 9 |
| Proteins involved in metabolic pathways | |||
| Phosphatidylethanolamine binding protein 1, isoform | gi|149063506 | 10.67 | 11 |
| Rat Phosphatidylethanolamine-Binding Protein, Chain A | gi|158428854 | 20.84 | 4 |
| Cu-Zn superoxide dismutase (EC 1.15.1.1) | gi|203658 | 15.71 | 12 |
| Sorbitol dehydrogenase | gi|77404286 | 38.23 | 7 |
| Sterol O-acyltransferase 2 | gi|40254723 | 60.49 | 5 |
| Lipid-binding/transport proteins | |||
| Fatty acid translocase/CD36 | gi|3273897 | 52.73 | 6 |
| Microsomal triglyceride transfer protein | gi|149026062 | 99.18 | 14 |
| Fatty acid binding protein 2, intestinal | gi|149025867 | 15.12 | 5 |
| Liver fatty acid binding protein p14 | gi|204074 | 13.42 | 7 |
| Cellular retinol binding protein | gi|206591 | 15.83 | 9 |
| Chain A, Liver Fatty Acid Binding Protein-Oleate | gi|157831801 | 14.28 | 11 |
| Molecular chaperones | |||
| Heat shock protein 27 | gi|204665 | 22.90 | 9 |
| Heat shock protein 70 | gi|396270 | 70.10 | 7 |
| Heat shock cognate 71 | gi|13242237 | 70.87 | 12 |
| Heat shock protein HSP 90-α | gi|28467005 | 84.81 | 7 |
| Heat-responsive protein 12, isoform CRA_b | gi|149066547 | 10.74 | 5 |
| Valosin-containing protein | gi|38014694 | 89.34 | 12 |
| UDP glucose:glycoprotein Glucosyltransferase 1 | gi|224471866 | 178.37 | 11 |
| Proteins regulating cell functions | |||
| Regucalcin | gi|13928740 | 33.40 | 17 |
| Cell cycle checkpoint protein RAD17 | gi|67078464 | 77.31 | 8 |
| Annexin A2 | gi|9845234 | 38.70 | 10 |
| Annexin A5 | gi|6978505 | 35.74 | 11 |
| Endoplasmic reticulum regulatory proteins | |||
| Liver microsomal Carboxylesterase | gi|562010 | 64.52 | 14 |
| Perchloric acid soluble protein | gi|4456766 | 10.35 | 9 |
| Peptidyl-prolyl cis-trans isomerase B | gi|11968126 | 22.81 | 10 |
| Structural proteins | |||
| β-actin | gi|55575 | 41.73 | 11 |
| Synphilin-1 | gi|157817380 | 105.82 | 7 |
| Actin-related protein 3 | gi|70912366 | 47.35 | 9 |
| Dynamin | gi|404073 | 98.14 | 12 |
| Tubulin, α1A | gi|50926060 | 50.13 | 8 |
| Tubulin, β2c | gi|38014578 | 49.83 | 11 |
| Miscellaneous Enzyme | |||
| Serine/threonine protein kinase | gi|207083 | 52.80 | 14 |
| Phosphoglycerate kinase 1 | gi|40254752 | 44.53 | 7 |
| α–enolase isoform 1 | gi|158186649 | 47.13 | 16 |
| α–enolase isoform 2 | gi|158186651 | 47.16 | 11 |
| Triosephosphate isomerase | gi|38532559 | 26.90 | 7 |
| Spermine synthase | gi|76559933 | 41.31 | 6 |
| Aldehyde dehydrogenase | gi|1403721 | 54.73 | 8 |
| Alcohol dehydrogenase | gi|202727 | 39.64 | 10 |
| Nucleoside diphosphate Kinase A | gi|19924089 | 17.20 | 9 |
| Phosphoinositide phosphatase SAC1 | gi|11095248 | 67.03 | 7 |
| Glutathione S-transferase Mu 2 | gi|28933457 | 25.70 | 11 |
| Glutathione S-transferase Yb-1 subunit (EC 2.5.1.18) | gi|204503 | 25.91 | 13 |
| Glutathione S-transferase α-3 | gi|13928688 | 25.31 | 11 |
| Lysophosphatidylcholine acyltransferase 3 | gi|58476497 | 56.01 | 14 |
| Lysophosphatidylcholine acyltransferase 1 | gi|213688411 | 59.80 | 7 |
| Glycerol-3-phosphate dehydrogenase | gi|603583 | 80.97 | 4 |
| Ribonuclease UK114 | gi|14269572 | 14.30 | 12 |
| Creatine kinase M-type | gi|6978661 | 43.01 | 10 |
| Other proteins | |||
| Cell death activator Cide-B | gi|157818189 | 27.94 | 15 |
| Epidermal growth factor receptor kinase substrate 8-like protein 3 | gi|157823675 | 68.60 | 5 |
| BCL6 co-repressor-like 1 | gi|149060097 | 144.46 | 13 |
| Selenium binding protein 2 | gi|149030730 | 50.57 | 7 |
| Cell death activator CIDE-3/fat specific protein 27 | gi|66730262 | 27.21 | 10 |
| Protein CYR61 precursor | gi|31542447 | 41.70 | 13 |
4. DISCUSSION
In present studies, we fully characterized VTVs and carried out detailed proteomic analyses to describe the VTV proteome. Using a cell free in vitro ER-budding assay and sucrose density gradients, we generated, isolated and purified VTVs to homogeneity. The purity of VTVs was determined by the enrichment of cargo proteins like apoB100 and apoA-IV and by the exclusion of ER resident proteins such as calnexin [26]. That the VTV fraction was not contaminated by other ER-derived vesicles formed during VTV-budding was established by our data that show albumin, a cargo protein of ER-derived PTV, was not present in VTV fractions which is consistent with previous published reports [26]. An important characteristic of secretory transport vesicle is that they enrich certain proteins and exclude other proteins at time of their genesis. Concentration of apoB100 or apoA-IV and exclusion of albumin in VTVs meet these criteria authenticating our VTV preparations.
One of the major criteria for bona fide VTVs is their size that should be large enough to accommodate VLDL-sized particles. To ascertain that vesicle fraction contain bona fide VTVs, we carried out detailed electron microscopic studies. As presented in figure 2 B&C and [26], our data strongly suggest we successfully generated and purified VTVs. In this study, we show TEM electron micrographs for the first time, which allowed us to determine that one VLDL particle is present per vesicle. This observation is consistent with our previous findings pertaining to ER-to-Golgi transport of pre-chylomicrons where we reported that ER-derived PCTV contains one pre-chylomicron [34]. The size of PCTVs was considerably large (average size ~ 300 nm) which was thought to be the reason for one particle per vesicle, however, the current data acquired on VTV, which is approximately one third of PCTV in size, support the thesis that cargo determines the size of the vesicle.
That the nascent VLDL exits the ER in a specialized vesicle, the VTV, has been demonstrated by our laboratory and by Fisher’s group [26,27] and a very small number of proteins generally associated with either VLDL or intracellular trafficking were reported to be present in VTVs. Using Western blotting, we identified a limited number of proteins such as apoB100, apoB48, sec22b, syntaxin5, Sar1, rBet1, and membrin [26,28]. We reasoned that characterization of VTV proteome would provide us an important tool to study various aspects of this important intracellular compartment such as factors regulating the formation of larger vesicle, cargo-selection mechanism, etc. Since VTVs are unique transport vesicles produced specifically in liver and their full characterization has not been carried out, our first approach to study their proteome was to resolve VTV-proteins by 2D-gel followed by mass spectrometric analysis. Figure 3 shows 2D-gel pattern of VTV-proteins revealing several hundred protein-bands, each of these band were excised and analyzed by MALDI-TOF. Utilizing this strategy we positively identified 28 proteins that include cargo-proteins (i.e. VLDL-proteins), proteins associated with ER-to-Golgi transport carriers, chaperones, SNAREs, vesicular-structure and proteins involved in different metabolic pathways. Since this is the first ever proteomic study performed on VLDL-transport vesicles, which are distinct in their cargo and size, a number of the identified proteins have not been reported to be associated with other transport vesicles. Interestingly, many of the first-time identified proteins in a bona fide transport vesicle (i.e. VTV), have been associated with lipid metabolism. Even though 2D-gel coupled with MALDI-TOF analysis enabled us to identify a number of VTV-proteins, a major barrier with this approach was the resolution of very-high Mr proteins along with very-low Mr proteins by 2D-gels. Consequently, apoB100 and apoB48 (540 kDa and 250 kDa, respectively) could not be recognized simultaneously with L-FABP (14 kDa protein) using the same 2D-gel, which is consistent with other published reports [41,42]. To overcome this limitation, we decided to carry out LC-MS/MS analysis of VTV-proteins, which enabled us to identify most of the proteins associated with VTVs irrespective of their Mr values. As listed in Table II, a wide range of proteins involved in various cellular functions including intracellular protein/lipoprotein transport pathways and lipid metabolism were identified. Interestingly, we could be able to detect signature VTV-cargo (VLDL) protein apoB100 along with apoAIV and apoC-I in our LC-MS/MS analysis.
The presence of coat-complex II (COPII) proteins in VTVs was interesting in two different aspects. First, COPII proteins are known to facilitate vesicle generation from the ER membranes and once the vesicle is in cytosol, most of the COPII proteins come off immediately and recycle back to ER membrane for the next cycle of vesicle biogenesis [13,14]. Since VTVs were isolated from budding-reaction mixture and purified using sucrose density gradient, the presence of COPII proteins indicates that COPII proteins remain on VTVs after their release from the ER membranes and not because of cytosolic contamination, which is consistent with our previous published work pertaining to intestinal pre-chylomicron transport vesicle, the PCTV [33–35]. Secondly, it is intriguing that the mechanisms of biogenesis of VTVs and PCTVs are different in their requirement for COPII proteins. The formation of PCTV from intestinal ER is independent of COPII proteins whereas VTV-budding from hepatic ER requires COPII proteins [26,34,43]. Moreover, it has been shown that COPII proteins play an important role for the fusion of PCTV with intestinal cis-Golgi [39], therefore, it is possible that COPII protein(s) may have similar role in VTV-Golgi fusion event.
The identification of cideB (cell death-inducing DFF45-like effector b) in VTV fraction was noteworthy because it has recently been shown to play an important role in VLDL maturation [44]. In their elegant study Ye et al. have shown that silencing of cideB gene leads to the secretion of small VLDL particles from the liver [44]. However, it is not known if cideB is involved in the biogenesis of VTVs and the absence of cideB interferes with the VTV-mediated transport of nascent VLDLs to the cis-Golgi where significant modifications to VLDL-apoB take place and apoAI is added to the VLDL [9]. Another role of cideB can be argued that it facilitates the transfer of triglycerides a role suggested by Ye et al. [44], however, the presence of microsomal triglyceride transfer protein (MTP) in VTV fractions diminishes the possibility. Other lipid interacting proteins detected in VTVs include liver-fatty acid binding protein (L-FABP), intestinal-FABP and fatty acid translocase/CD36. It is not yet known what role these lipid interacting proteins play in the biogenesis of VTVs or VTV-mediated export of VLDL to hepatic cis-Golgi. The detection of L-FABP in VTVs is of great interest because it has been shown to initiate the formation of PCTVs in small intestine [45]; however, Sar1 has been demonstrated to initiate the process of VTV-budding from hepatic ER membranes. It may be possible that L-FABP functions as a sorting protein for VLDL-packaging into VTVs because it interacts with apoB [35,45]. Another protein positively identified in VTV fraction is fatty acid translocase/CD36 that caught our attention because in previous published data, fatty acid translocase/CD36 has been demonstrated to be a major player in PCTV-formation and it is a one of the component of multi-protein complex, which is required the PCTV-budding [35].
Formation of fusogenic SNARE-complex requires several proteins other than SNAREs; these proteins facilitate SNARE pairing. Heat shock proteins specifically Hsc70, has been shown to play a role in SNARE-complex formation [46]. Identification of several heat shock proteins, hsc27, hsc71, hsp70, hsp90 in VTV fractions is interesting because any of these proteins or a complex of these proteins may be important for VTV-Golgi fusion event. Our data (unpublished) show that hsc71 interacts with Sec22b, a v-SNARE on VTV, suggesting its role in SNARE-complex formation. However, further experiments are required to substantiate the role of heat shock proteins in Fusion-complex formation.
5. CONCLUSIONS
VTVs play an important role in VLDL secretion from the liver, however these vesicles are poorly investigated and partially characterized. Although in our previous studies we identified a number of proteins using Western blotting, a detailed catalogue of VTV proteins is imperative. In this study, we have fully characterized VTVs morphologically and biochemically and carried out a detailed proteomic analysis of VTVs. We have identified a number of proteins that might be important in different aspects of VTV-mediated VLDL transport such as VLDL-selection into VTV, formation of large vesicle to accommodate VLDL particle, docking of VTV with its target, cis-Golgi etc. Further studies are required to establish the functional role of VTV proteins and the VTV proteome reported in this report can provide a basic tool to study the mechanisms underlying VLDL biogenesis, maturation, intracellular trafficking and secretion from the liver.
Acknowledgments
This study was supported by NIH’s DK-81413 (to SAS) from the National Institute of Diabetes And Digestive and Kidney Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes And Digestive and Kidney Diseases or the National Institutes of Health.
Abbreviations used
- VLDL
very low-density lipoprotein
- VTV
VLDL transport vesicle
- LDL
low-density lipoprotein
- ApoB
apolipoprotein B
- ER
endoplasmic reticulum
- SNARE
soluble N-ethylmaleimide-sensitive factor-attachment protein receptor
- FFA
free fatty acid
- MTP
microsomal triglyceride transfer protein
- TAG
triacylglycerol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sehayek E, Eisenberg S. The role of native apolipoprotein B-containing lipoproteins in atherosclerosis: cellular mechanisms. Curr Opin Lipidol. 1994;5:350–353. doi: 10.1097/00041433-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Ginsberg HN. New perspectives on atherogenesis: role of abnormal triglyceride-rich lipoprotein metabolism. Circulation. 2002;106:2137–2142. doi: 10.1161/01.cir.0000035280.64322.31. [DOI] [PubMed] [Google Scholar]
- 3.Swift LL. Assembly of very low density lipoproteins in rat liver: a study of nascent particles recovered from the rough endoplasmic reticulum. J Lipid Res. 1995;36:395–406. [PubMed] [Google Scholar]
- 4.Rustaeus S, Lindberg K, Stillemark P, Claesson C, Asp L, Larsson T, Boren J, Olofsson SO. Assembly of very low density lipoprotein: a two-step process of apolipoprotein B core lipidation. J Nutr. 1999;129:463S–466S. doi: 10.1093/jn/129.2.463S. [DOI] [PubMed] [Google Scholar]
- 5.Shelness GS, Ingram MF, Huang XF, DeLozier JA. Apolipoprotein B in the rough endoplasmic reticulum: translation, translocation and the initiation of lipoprotein assembly. J Nutr. 1999;129:456S–462S. doi: 10.1093/jn/129.2.456S. [DOI] [PubMed] [Google Scholar]
- 6.Bakillah A, Nayak N, Saxena U, Medford RM, Hussain MM. Decreased secretion of ApoB follows inhibition of ApoB-MTP binding by a novel antagonist. Biochemistry. 2000;39:4892–4899. doi: 10.1021/bi9924009. [DOI] [PubMed] [Google Scholar]
- 7.Olofsson SO, Asp L, Boren J. The assembly and secretion of apolipoprotein B-containing lipoproteins. Curr Opin Lipidol. 1999;10:341–346. doi: 10.1097/00041433-199908000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Rusinol A, Verkade H, Vance JE. Assembly of rat hepatic very low density lipoproteins in the endoplasmic reticulum. J Biol Chem. 1993;268:3555–3562. [PubMed] [Google Scholar]
- 9.Gusarova V, Seo J, Sullivan ML, Watkins SC, Brodsky JL, Fisher EA. Golgi-associated maturation of very low density lipoproteins involves conformational changes in apolipoprotein B, but is not dependent on apolipoprotein E. J Biol Chem. 2007;282:19453–19462. doi: 10.1074/jbc.M700475200. [DOI] [PubMed] [Google Scholar]
- 10.Macri J, Adeli K. Conformational changes in apolipoprotein B modulate intracellular assembly and degradation of ApoB-containing lipoprotein particles in HepG2 cells. Arterioscler Thromb Vasc Biol. 1997;17:2982–2994. doi: 10.1161/01.atv.17.11.2982. [DOI] [PubMed] [Google Scholar]
- 11.Tran K, Thorne-Tjomsland G, DeLong CJ, Cui Z, Shan J, Burton L, Jamieson JC, Yao Z. Intracellular assembly of very low density lipoproteins containing apolipoprotein B100 in rat hepatoma McA-RH7777 cells. J Biol Chem. 2002;277:31187–31200. doi: 10.1074/jbc.M200249200. [DOI] [PubMed] [Google Scholar]
- 12.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 13.Barlowe C. COPII and selective export from the endoplasmic reticulum. Biochim Biophys Acta. 1998;1404:67–76. doi: 10.1016/s0167-4889(98)00047-0. [DOI] [PubMed] [Google Scholar]
- 14.Jensen D, Schekman R. COPII-mediated vesicle formation at a glance. J Cell Sci. 2011;124:1–4. doi: 10.1242/jcs.069773. [DOI] [PubMed] [Google Scholar]
- 15.Fromme JC, Orci L, Schekman R. Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 2008;18:330–336. doi: 10.1016/j.tcb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Gurkan C, Stagg SM, Lapointe P, Balch WE. The COPII cage: unifying principles of vesicle coat assembly. Nat Rev Mol Cell Biol. 2006;7:727–738. doi: 10.1038/nrm2025. [DOI] [PubMed] [Google Scholar]
- 17.Tang BL, Wang Y, Ong YS, Hong W. COPII and exit from the endoplasmic reticulum. Biochim Biophys Acta. 2005;1744:293–303. doi: 10.1016/j.bbamcr.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Hughes H, Stephens DJ. Assembly, organization, and function of the COPII coat. Histochemistry and Cell Biology. 2008;129:129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato K, Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007;581:2076–2082. doi: 10.1016/j.febslet.2007.01.091. [DOI] [PubMed] [Google Scholar]
- 20.Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoulders CC, Stephens DJ, Jones B. The intracellular transport of chylomicrons requires the small GTPase, Sar1b. Curr Opin Lipidol. 2004;15:191–197. doi: 10.1097/00041433-200404000-00012. [DOI] [PubMed] [Google Scholar]
- 22.Levy E, Harmel E, Laville M, Sanchez R, Emonnot L, Sinnett D, Ziv E, Delvin E, Couture P, Marcil V, Sane AT. Expression of Sar1b enhances chylomicron assembly and key components of the coat protein complex II system driving vesicle budding. Arterioscler Thromb Vasc Biol. 2011;31:2692–2699. doi: 10.1161/ATVBAHA.111.233908. [DOI] [PubMed] [Google Scholar]
- 23.Charcosset M, Sassolas A, Peretti N, Roy CC, Deslandres C, Sinnett D, Levy E, Lachaux A. Anderson or chylomicron retention disease: molecular impact of five mutations in the SAR1B gene on the structure and the functionality of Sar1b protein. Mol Genet Metab. 2008;93:74–84. doi: 10.1016/j.ymgme.2007.08.120. [DOI] [PubMed] [Google Scholar]
- 24.Georges A, Bonneau J, Bonnefont-Rousselot D, Champigneulle J, Rabes JP, Abifadel M, Aparicio T, Guenedet JC, Bruckert E, Boileau C, Morali A, Varret M, Aggerbeck LP, Samson-Bouma ME. Molecular analysis and intestinal expression of SAR1 genes and proteins in Anderson’s disease (Chylomicron retention disease) Orphanet J Rare Dis. 2011;6:1. doi: 10.1186/1750-1172-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada T, Miyashita M, Fukuhara J, Sugitani M, Ueno T, Samson-Bouma ME, Aggerbeck LP. Anderson’s disease/chylomicron retention disease in a Japanese patient with uniparental disomy 7 and a normal SAR1B gene protein coding sequence. Orphanet J Rare Dis. 2011;6:78. doi: 10.1186/1750-1172-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqi SA. VLDL exits from the endoplasmic reticulum in a specialized vesicle, the VLDL transport vesicle, in rat primary hepatocytes. Biochem J. 2008;413:333–342. doi: 10.1042/BJ20071469. [DOI] [PubMed] [Google Scholar]
- 27.Gusarova V, Brodsky JL, Fisher EA. Apolipoprotein B100 exit from the endoplasmic reticulum (ER) is COPII-dependent, and its lipidation to very low density lipoprotein occurs post-ER. J Biol Chem. 2003;278:48051–48058. doi: 10.1074/jbc.M306898200. [DOI] [PubMed] [Google Scholar]
- 28.Siddiqi S, Mani AM, Siddiqi SA. The identification of the SNARE complex required for the fusion of VLDL-transport vesicle with hepatic cis-Golgi. Biochem J. 2010;429:391–401. doi: 10.1042/BJ20100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 30.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 32.Hay JC, Scheller RH. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- 33.Mansbach CM, Siddiqi SA. The biogenesis of chylomicrons. Annu Rev Physiol. 2010;72:315–333. doi: 10.1146/annurev-physiol-021909-135801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siddiqi SA, Gorelick FS, Mahan JT, Mansbach CM., 2nd COPII proteins are required for Golgi fusion but not for endoplasmic reticulum budding of the pre-chylomicron transport vesicle. J Cell Sci. 2003;116:415–427. doi: 10.1242/jcs.00215. [DOI] [PubMed] [Google Scholar]
- 35.Siddiqi S, Saleem U, Abumrad NA, Davidson NO, Storch J, Siddiqi SA, Mansbach CM., 2nd A novel multiprotein complex is required to generate the prechylomicron transport vesicle from intestinal ER. J Lipid Res. 2010;51:1918–1928. doi: 10.1194/jlr.M005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqi SA, Mansbach CM., 2nd PKC zeta-mediated phosphorylation controls budding of the pre-chylomicron transport vesicle. J Cell Sci. 2008;121:2327–2338. doi: 10.1242/jcs.022780. [DOI] [PubMed] [Google Scholar]
- 37.Siddiqi SA, Mahan J, Siddiqi S, Gorelick FS, Mansbach CM., 2nd Vesicle-associated membrane protein 7 is expressed in intestinal ER. J Cell Sci. 2006;119:943–950. doi: 10.1242/jcs.02803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqi SA, Siddiqi S, Mahan J, Peggs K, Gorelick FS, Mansbach CM., 2nd The identification of a novel endoplasmic reticulum to Golgi SNARE complex used by the prechylomicron transport vesicle. J Biol Chem. 2006;281:20974–20982. doi: 10.1074/jbc.M601401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddiqi S, Siddiqi SA, Mansbach CM., 2nd Sec24C is required for docking the prechylomicron transport vesicle with the Golgi. J Lipid Res. 2010;51:1093–1100. doi: 10.1194/jlr.M002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto S, Nakamura M. Calnexin: its molecular cloning and expression in the liver of the frog, Rana rugosa. FEBS Lett. 1996;387:27–32. doi: 10.1016/0014-5793(96)00443-7. [DOI] [PubMed] [Google Scholar]
- 41.Wong DM, Webb JP, Malinowski PM, Macri J, Adeli K. Proteomic profiling of the prechylomicron transport vesicle involved in the assembly and secretion of apoB-48-containing chylomicrons in the intestinal enterocytes. Proteomics. 2009;9:3698–3711. doi: 10.1002/pmic.200800914. [DOI] [PubMed] [Google Scholar]
- 42.Wong DM, Webb JP, Malinowski PM, Xu E, Macri J, Adeli K. Proteomic profiling of intestinal prechylomicron transport vesicle (PCTV)-associated proteins in an animal model of insulin resistance (94 char) J Proteomics. 2010;73:1291–1305. doi: 10.1016/j.jprot.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Brodsky JL, Gusarova V, Fisher EA. Vesicular trafficking of hepatic apolipoprotein B100 and its maturation to very low-density lipoprotein particles; studies from cells and cell-free systems. Trends Cardiovasc Med. 2004;14:127–132. doi: 10.1016/j.tcm.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Ye J, Li JZ, Liu Y, Li X, Yang T, Ma X, Li Q, Yao Z, Li P. Cideb, an ER- and lipid droplet-associated protein, mediates VLDL lipidation and maturation by interacting with apolipoprotein B. Cell Metab. 2009;9:177–190. doi: 10.1016/j.cmet.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 45.Neeli I, Siddiqi SA, Siddiqi S, Mahan J, Lagakos WS, Binas B, Gheyi T, Storch J, Mansbach CM., 2nd Liver fatty acid-binding protein initiates budding of pre-chylomicron transport vesicles from intestinal endoplasmic reticulum. J Biol Chem. 2007;282:17974–17984. doi: 10.1074/jbc.M610765200. [DOI] [PubMed] [Google Scholar]
- 46.Joglekar AP, Hay JC. Evidence for regulation of ER/Golgi SNARE complex formation by hsc70 chaperones. Eur J Cell Biol. 2005;84:529–542. doi: 10.1016/j.ejcb.2004.12.028. [DOI] [PubMed] [Google Scholar]