Abstract
The objective of this work was to investigate the effect of hypertrophic cardiomyopathy-linked A8V and E134D mutations in cardiac troponin C (cTnC) on the response of reconstituted thin filaments to calcium upon phosphorylation of cardiac troponin I (cTnI) by protein kinase A. The phosphorylation of cTnI at protein kinase A sites was mimicked by S22D/S23D mutation in cTnI. Our results demonstrate that the A8V and E134D mutations had no effect on the extent of calcium desensitization of reconstituted thin filaments induced by cTnI pseudo-phosphorylation. However, the A8V mutation enhanced the effect of cTnI pseudo-phosphorylation on the rate of calcium dissociation from reconstituted thin filaments and on calcium dependence of actomyosin ATPase. Consequently, while the A8V mutation still led to a slower rate of calcium dissociation from reconstituted thin filaments upon pseudo-phosphorylation of cTnI, the ability of the A8V mutation to decrease the rate of calcium dissociation was diminished. In addition, the ability of the A8V mutation to sensitize actomyosin ATPase to calcium was diminished after cTnI was replaced by the phosphorylation mimetic of cTnI. Consistent with the hypothesis that the E134D mutation is benign, it exerted minor to no effect on the rate of calcium dissociation from reconstituted thin filaments, and on calcium sensitivity of actomyosin ATPase, regardless of cTnI phosphorylation status. In conclusion, our study enhances understanding of how cardiomyopathy-linked cTnC mutations affect the response of reconstituted thin filaments to calcium upon cTnI phosphorylation.
Familial hypertrophic cardiomyopathy (HCM)1 is an inherited cardiovascular disorder, characterized by the thickening of the heart muscle and diastolic dysfunction. HCM may lead to a variety of symptoms, such as shortness of breath, chest pain, fatigue, fainting, heart palpitations and sudden cardiac death (for review, see (1–3)). HCM has been attributed to mutations in a number of genes encoding for sarcomeric proteins, including β-myosin heavy chain, myosin binding protein C, actin, tropomyosin, cardiac troponin I (cTnI) and cardiac troponin T (cTnT) (for review, see (4–6)). Until recently, the gene encoding for cardiac troponin C (cTnC) was not considered to be associated with inherited cardiomyopathies. However, recent discoveries linked a number of mutations in cTnC to both HCM and dilated cardiomyopathy (DCM) (for review, see (5, 7)).
The Ca2+ sensor subunit of the cTn complex, cTnC, is a member of the EF-hand (helix-loop-helix motif) family of Ca2+ binding proteins. CTnC consists of the N- and C-terminal globular domains connected by an α-helical linker (for review, see (8, 9)). Each domain of cTnC contains a pair of EF-hand motifs numbered I–IV, but the first EF-hand of cTnC is unable to bind Ca2+ due to several loop residue substitutions (10). Therefore, exchange of Ca2+ with the second EF-hand of cTnC plays a direct role in the regulation of muscle contraction and relaxation. The third and fourth C-domain EF-hands are believed to be play a structural role of anchoring cTnC into the cTn complex (for review, see (11)). The α-helices within cTnC are denoted A–H, with an additional 14-residue N-helix at the N-terminus.
Both the intrinsic Ca2+ binding properties of cTnC and its interactions with other regulatory muscle proteins play an important role in controlling Ca2+ binding and exchange with myofilaments. Numerous studies have focused on elucidating the interactions between cTnC and cTnI, which play a crucial role in the regulation of cardiac muscle contractility. CTnI, an inhibitory subunit of the cTn complex, is a rod-like flexible molecule that contains an ~ 30-residue N-terminal extension region, which is absent in both skeletal and slow skeletal isoforms of TnI. The N-extension region of cTnI includes Ser22 and Ser23 residues that are targets of phosphorylation by protein kinase A (PKA). A number of studies demonstrated that phosphorylation of cTnI at PKA sites during β-adrenergic stimulation induces myofilament Ca2+ desensitization and accelerates cardiac relaxation (for review, see (12, 13)). While the additional phosphorylation sites are present in cTnI, their functional significance remains unclear and controversial (for review, see (14)).
Recently, a number of HCM- and DCM-linked cTnC mutations were shown to blunt or abolish myofilament Ca2+ desensitization induced by phosphorylation of cTnI by PKA (15–17). We wanted to determine whether blunting of the myofilament Ca2+ desensitization induced by cTnI phosphorylation was a common mechanism among cardiomyopathy-linked cTnC mutations. The objective of this study was to examine whether recently discovered HCM-linked A8V and E134D cTnC mutations (18) affect the response of reconstituted thin filaments to Ca2+ upon cTnI phosphorylation by PKA.
EXPERIMENTAL PROCEDURES
Materials
Phenyl-Sepharose CL-4B, CaCl2 and EGTA were purchased from Sigma-Aldrich (St. Louis, MO). IAANS and phalloidin were purchased from Invitrogen (Carlsbad, CA). Affi-Gel 15 affinity media were purchased from Bio-Rad (Hercules, CA). Malachite Green Oxalate and poly(vinyl alcohol) were purchased from Fisher Scientific (Pittsburgh, PA)
Protein Mutagenesis and Purification
The pET3a plasmid encoding human cTnC was a generous gift from Dr. Lawrence B. Smillie (University of Alberta, Edmonton, AB). The cTnC construct used in this work (with an exception of actomyosin ATP assays) contained C35S, T53C and C84S substitutions, to enable fluorescent labeling of cTnC on Cys53 (19, 20). The HCM-linked cTnC mutants were generated as previously described, and confirmed by DNA sequencing (19, 20). Expression and purification of cTnC and its mutants was carried out as previously described (19–21). The pET3d plasmids encoding human cTnI and human cTnT were generated by GenScript USA (Piscataway, NJ). The cTnI and cTnT subunits were bacterially expressed, purified and quantified as described (20). The cTnIS22D/S23D mutant was generated from the pET3d cTnI plasmid by primer-based site-directed mutagenesis and verified by DNA sequencing. Rabbit fast skeletal actin and myosin S1, and bovine cTm were isolated, purified, and quantified as described (19).
Labeling of cTnC and its Mutants
CTnC and its mutants were labeled with the environmentally sensitive thiol-reactive fluorescent probe IAANS on Cys53 as previously described (19, 20).
Reconstitution of the cTn Complexes
The cTn complexes were prepared and reconstituted as previously described (19, 20).
Reconstitution of Thin Filaments
After exhaustive dialysis against reconstitution buffer (10 mM MOPS, 150 mM KCl, 3 mM MgCl2, and 1 mM DTT, pH 7.0.), actin was mixed with an equal molar ratio of phalloidin to stabilize actin filaments. Thin filaments were reconstituted as previously described (19, 20). Briefly, actin-phalloidin (4 μM) and cTm (0.57μM) were mixed in the reconstitution buffer and kept on ice for ~ 15 minutes. The cTn complexes (0.5 μM) were subsequently added, and reconstituted thin filaments were kept on ice for ~ 15 minutes prior to use. Therefore, the stoichiometry of reconstituted thin filaments was 7:1:0.88 (actin:cTm:cTn).
Determination of Ca2+ Binding Sensitivities
All steady-state fluorescence measurements were performed using a Perkin-Elmer LS55 fluorescence spectrometer at 15°C. IAANS fluorescence was excited at 330 nm and monitored at 450 nm as μL amounts of CaCl2 were added to 2 mL of reconstituted thin filaments in titration buffer (200 mM MOPS (to prevent pH changes upon addition of Ca2+), 150 mM KCl, 2 mM EGTA, 1 mM DTT, 3 mM MgCl2, pH 7.0) at 15°C with constant stirring. The [Ca2+]free was calculated using the computer program EGCA02 developed by Robertson and Potter (22). The Ca2+ sensitivities of conformational changes were reported as a dissociation constant Kd, representing a mean of at least three titrations ± S.E. The data were fit with a logistic sigmoid function (mathematically equivalent to the Hill equation), as previously described (23).
Determination of Ca2+ Dissociation Kinetics
All kinetic measurements were performed utilizing an Applied Photophysics Ltd. (Leatherhead, UK) model SX.18 MV stopped-flow instrument with a dead time of ~1.4 ms at 15°C. The rates of conformational changes induced by EGTA removal of Ca2+ from reconstituted thin filaments were measured following IAANS fluorescence. The IAANS fluorescence was excited at 330 nm. The IAANS emission was monitored through a 510 nm BrightLine Basic™ filter from Semrock (Rochester, NY). Stopped-flow buffer consisted of 10 mM MOPS, 150 mM KCl, 3mM MgCl2 and 1 mM DTT, pH 7.0. The data were corrected for scattering artifacts as described previously (19, 20). The data were fit using a program (by P. J. King, Applied Photophysics Ltd) that utilizes the nonlinear Levenberg-Marquardt algorithm. Each koff represents an average of at least three separate experiments ±S.E., each averaging at least five traces fit with a single exponential equation.
Actomyosin S1 ATPase Assay
Reconstituted thin filaments (5μM actin, 1.0 μM cTm, 1.5 μM cTn, and 0.3 μM myosin S1) were formed at 25 °C in a buffer consisting of 50 mM MOPS, and 5 mM MgCl2, pH 7.0. EGTA (to a final concentration of 0.5 mM) and various amounts of CaCl2 were added to the 100 μL reaction mixture aliquots to achieve the desired pCa values. The ATPase reaction was initiated by addition of ATP (to a final concentration of 1 mM), and 10 μL aliquots were removed into 90 μL of 0.2M ice-cold perchloric acid in order to terminate the reaction. For determination of the Ca2+ dependence of actomyosin ATPase, the ATPase rates were measured at a single time point at which the reaction was still linear with time. In order to measure minimal and maximal specific actomyosin ATPase activities, 10 μL aliquots were terminated at 3 minute intervals (up to 12 minutes time course) by 90 μL of 0.2 M ice-cold perchloric acid. Actomyosin ATPase activity was determined by the amount of phosphate released. The amount of phosphate released was quantified using malachite green method, as previously described (20).
Statistical Analysis
Statistical significance was determined by an unpaired two-sample t-test using the statistical analysis software Minitab (State College, PA). The two means were considered to be significantly different when the p value was < 0.05. All data is shown as a mean value ± SE.
RESULTS
Location of the A8V and E134D mutations within cTnC
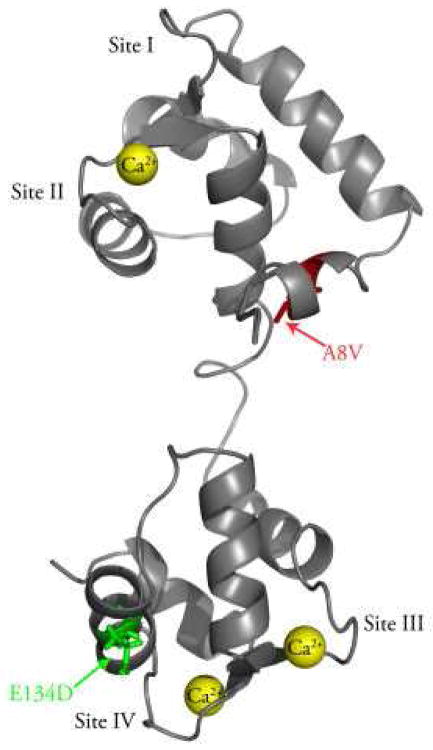
Figure 1 shows that the A8V mutation is located in the N-helix of the N-domain of cTnC, while the E134D mutation is located in the C-domain of cTnC, between Ca2+ binding sites III and IV.
Figure 1. Location of the A8V and E134D mutations in the N- and C-domains of cTnC.
The figure shows a ribbon representation of cTnC in the Ca2+ bound state (Protein Data Bank entry 1AJ4 (37)). The A8V mutation (shown in red) is located in the N-helix of the N-domain, while the E134D mutation (shown in green) is located between Ca2+ binding sites III and IV. This figure was generated using PyMOL (www.pymol.org).
Effect of the A8V and E134D cTnC mutations on the Ca2+ sensitivities of reconstituted thin filaments
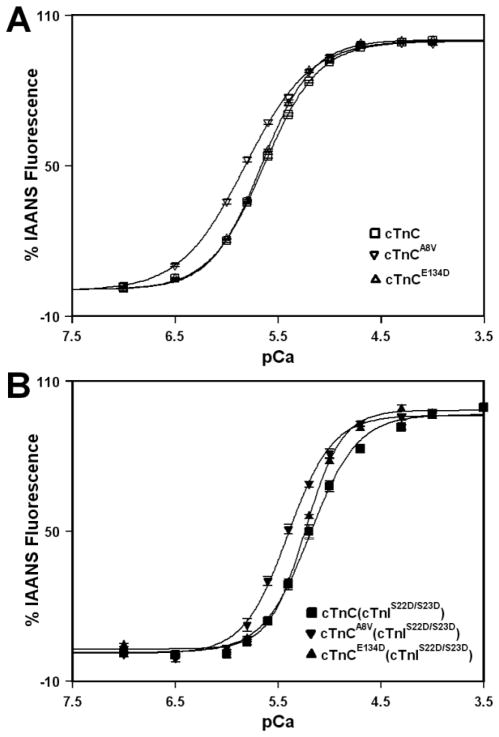
The Ca2+ induced increases in IAANS fluorescence, occurring when Ca2+ binds to the regulatory N-domain of the cTn, cTnA8V, or cTnE134D complexes reconstituted into thin filaments, are shown in Figure 2A and summarized in Table I. Thin filaments reconstituted with the cTn complex exhibited a half-maximal Ca2+ dependent increase in IAANS fluorescence with a pCa50 of 5.64 ± 0.02. Thin filaments reconstituted with the cTnA8V complex exhibited a half-maximal increase in IAANS fluorescence with a pCa50 of 5.82 ± 0.02, while thin filaments reconstituted with cTnE134D complex exhibited a half-maximal increase in IAANS fluorescence with a pCa50 of 5.660 ± 0.007. The A8V mutation affected the steepness of the curve, indicated by a slightly decreased Hill coefficient (nH), while the E134D mutation had no significant effect on nH (Table I). Thus, our results demonstrate that the A8V mutation led to a statistically significant increase in the Ca2+ sensitivity of reconstituted thin filaments, while the E134D mutation did not significantly affect the Ca2+ sensitivity of reconstituted thin filaments. Furthermore, the A8V mutation decreased the cooperativity of Ca2+ binding to reconstituted thin filaments (indicated by lower nH), while the E134D mutation had no effect on cooperativity of Ca2+ binding to reconstituted thin filaments.
Figure 2. Effect of the A8V and E134D cTnC mutations on the Ca2+ sensitivities of reconstituted thin filaments in the absence and presence of cTnI pseudo-phosphorylation.
Panel A shows increases in IAANS fluorescence, which occur as Ca2+ binds to the regulatory N-domain of the cTn (□), cTnA8V (▽) or cTnE134D (△) complexes reconstituted into thin filaments in the absence of cTnI pseudo-phosphorylation. Panel B shows increases in IAANS fluorescence, which occur as Ca2+ binds to the regulatory N-domain of cTn (■), cTnA8V (▼) or cTnE134D (▲) complexes, containing phosphomimetic of cTnI, reconstituted into thin filaments. Each data point represents the mean ± S.E. of at least three titrations fit with a logistic sigmoid function. The IAANS fluorescence was excited at 330 nm and monitored at 450 nm.
Table 1.
Effect of cTnC mutations on the Ca2+ binding properties of reconstituted thin filaments in the absence and presence of cTnI pseudo-phosphorylation.
| Protein | pCa50 | nH | Ca2+ koff (/s) |
|---|---|---|---|
| cTnC | 5.64 ± 0.02 | 1.63 ± 0.05 | 93 ± 1 |
| cTnCA8V | 5.82 ± 0.02a | 1.38 ± 0.03a | 42.0 ± 0.4a |
| cTnCE134D | 5.660 ± 0.007 | 1.76 ± 0.02 | 96 ± 1 |
| cTnC + cTnIS22D/S23D | 5.18 ± 0.04 | 2.0 ± 0.1 | 310 ± 5 |
| cTnCA8V+ cTnIS22D/S23D | 5.41 ± 0.02b | 2.0 ± 0.2 | 237 ± 8b |
| cTnCE134D+ cTnIS22D/S23D | 5.23 ± 0.01 | 2.4 ± 0.1 | 280 ± 6b |
Significantly different from their respective cTnC values (p<0.05)
Significantly different from their respective cTnC + cTnIS22D/S23D values (p<0.05)
Effect of the A8V and E134D cTnC mutations on the Ca2+ sensitivities of reconstituted thin filaments in the presence of cTnI pseudo-phosphorylation
The Ca2+ induced increases in IAANS fluorescence, occurring when Ca2+ binds to the regulatory N-domain of the cTn, cTnA8V, or cTnE134D complexes, containing phosphomimetic of cTnI (cTnIS22D/S23D), reconstituted into thin filaments, are shown in Figure 2B and summarized in Table I. Thin filaments reconstituted with the cTn complex, containing phosphomimetic of cTnI, exhibited a half-maximal Ca2+ dependent increase in IAANS fluorescence with a pCa50 of 5.18 ± 0.04. Thin filaments reconstituted with the cTnA8V complex, containing phosphomimetic of cTnI, exhibited a half-maximal increase in IAANS fluorescence with pCa50 of 5.41 ± 0.02, while thin filaments reconstituted with the cTnE134D complex, containing phosphomimetic of cTnI, exhibited a half-maximal increase in IAANS fluorescence with pCa50 of 5.23 ± 0.01. In the presence of cTnI pseudo-phosphorylation, the nH values for thin filaments reconstituted with the cTn complexes were not significantly affected by either the A8V or E134D mutation (Table I). Our results show that thin filaments reconstituted with the cTn complex underwent a substantial Ca2+ desensitization upon cTnI pseudo-phosphorylation (ΔpCa50 = −0.46 ± 0.04). The A8V mutation did not significantly affect the extent of Ca2+ desensitization induced by cTnI pseudo-phosphorylation (ΔpCa50 = −0.41 ± 0.02). The E134D mutation also did not significantly affect the extent of Ca2+ desensitization (ΔpCa50 = −0.43 ± 0.01). Therefore, our results demonstrate that neither the A8V nor E134D mutation significantly blunted the extent of the decrease in the Ca2+ sensitivity of reconstituted thin filaments induced by cTnI pseudo-phosphorylation. In addition, our results show that in the presence of cTnI pseudo-phosphorylation, the A8V mutation led to a statistically significant increase in the Ca2+ sensitivity of reconstituted thin filaments, while the E134D mutation exerted no significant effect on the Ca2+ sensitivity of reconstituted thin filaments. Furthermore, our results show that neither the A8V nor E134D mutation significantly affected the cooperativity of Ca2+ binding to reconstituted thin filaments containing phosphomimetic of cTnI.
Effect of the A8V and E134D cTnC mutations on the rates of Ca2+ dissociation from reconstituted thin filaments
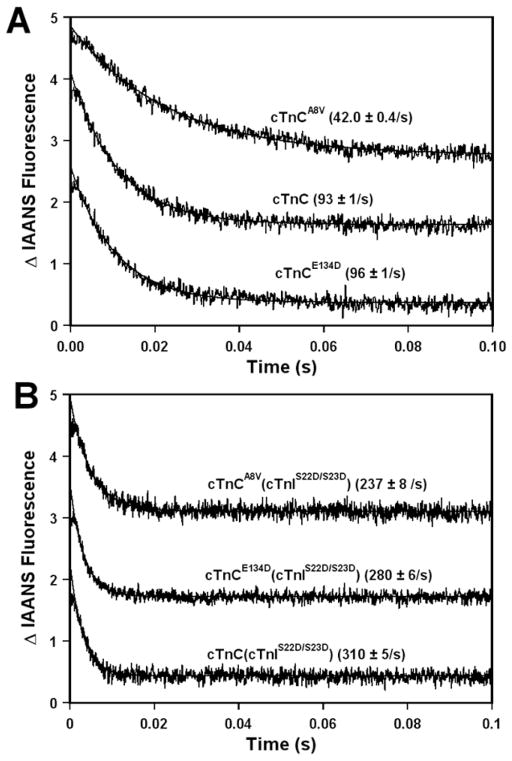
Fluorescence stopped-flow measurements, utilizing IAANS fluorescence, were conducted to determine the effect of the A8V and E134D cTnC mutations on the kinetics of Ca2+ dissociation from the regulatory N-domain of the cTn complex reconstituted into thin filaments. The results are summarized in Table I. Figure 3A shows that excess EGTA removed Ca2+ from the regulatory N-domain of the cTn, cTnA8V, or cTnE134D complexes reconstituted into thin filaments at 93 ± 1, 42.0 ± 0.4, or 96 ± 1/s, respectively. Therefore, our results show that the A8V mutation led to ~2.2-fold slower rate of Ca2+ dissociation from the regulatory N-domain of the cTn complex reconstituted into thin filaments, while the E134D mutation had no significant effect on the rate of Ca2+ dissociation.
Figure 3. Effect of the A8V and E134D cTnC mutations on the rates of Ca2+ dissociation from reconstituted thin filaments in the absence and presence of cTnI pseudo-phosphorylation.
Panel A shows the time course of decreases in IAANS fluorescence as Ca2+ was removed by excess EGTA from the regulatory N-domain of the cTn, cTnA8V or cTnE134D complexes reconstituted into thin filaments in the absence of cTnI pseudo-phosphorylation. Panel B shows the time course of decreases in IAANS fluorescence as Ca2+ was removed by excess EGTA from the regulatory N-domain of the cTn, cTnA8V or cTnE134D complexes, containing phosphomimetic of cTnI, reconstituted into thin filaments. The data traces have been normalized and staggered for clarity. Each trace is an average of at least five traces fit with a single exponential equation. The IAANS fluorescence was excited at 330 nm and monitored through a 510 nm bandpass filter.
Effect of the A8V and E134D cTnC mutations on the rates of Ca2+ dissociation from reconstituted thin filaments in the presence of cTnI pseudo-phosphorylation
Fluorescence stopped-flow measurements, utilizing IAANS fluorescence, were conducted to determine the effect of the A8V and E134D cTnC mutations on the kinetics of Ca2+ dissociation from the regulatory N-domain site of the cTn complex, containing phosphomimetic of cTnI, reconstituted into thin filaments. The results are summarized in Table I. Figure 3B shows that excess EGTA removed Ca2+ from the regulatory N-domain of the cTn, cTnA8V, or cTnE134D complexes, containing phosphomimetic of cTnI, reconstituted into thin filaments at 310 ± 5, 237 ± 8, or 280 ± 6/s, respectively. Thus, replacement of cTnI by the phosphomimetic of cTnI led to ~3.3-, 5.6- or 2.9-fold acceleration in the rate of Ca2+ dissociation from thin filaments reconstituted with the cTn, cTnA8V or cTnE134D complexes, respectively. These results indicate that the A8V mutation enhanced the extent of acceleration in the rate of Ca2+ dissociation from reconstituted thin filaments upon cTnI pseudo-phosphorylation. On the other hand, the E134D mutation had only a minor effect on the extent of acceleration in the Ca2+ dissociation rate associated with pseudo-phosphorylation of cTnI by PKA. These results also show that after cTnI was replaced by the phosphomimetic of cTnI, the A8V mutation led to ~1.3-fold slower rate of Ca2+ dissociation from the N-domain site of cTnC reconstituted into thin filaments. In addition, the E134D mutation exerted statistically significant, albeit minor, effect on the rate of Ca2+ dissociation from reconstituted into thin filaments in the presence of cTnI pseudo-phosphorylation.
Effect of the A8V and E134D cTnC mutations on the Ca2+ sensitivities of actomyosin ATPase in the absence and presence of cTnI pseudo-phosphorylation
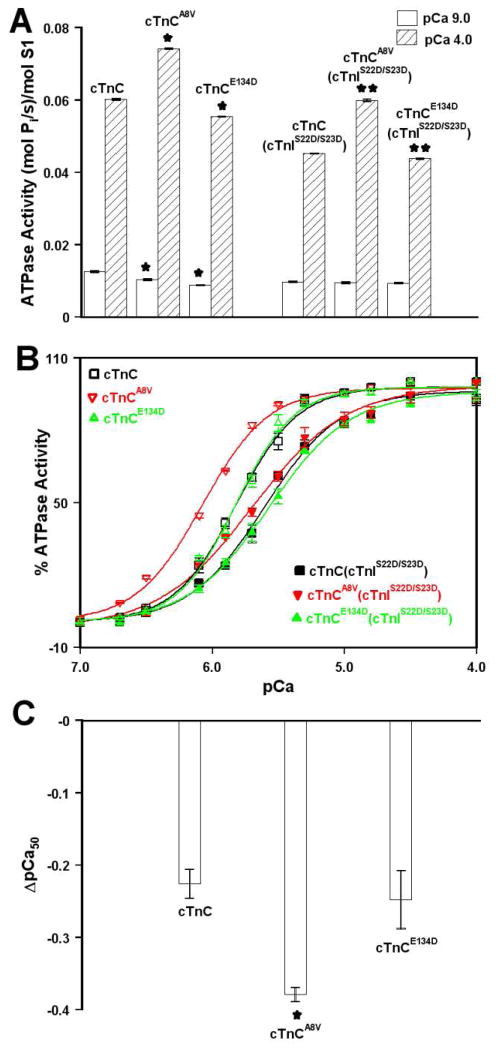
To examine the functional effect of the A8V and E134D mutations, the Ca2+ dependence of actomyosin ATPase activity was measured after reconstitution of thin filaments with the cTn, cTnA8V or cTnCE13D complexes. CTnC proteins used for the actomyosin ATPase assay were unlabeled and did not carry the C35S, T53C, and C84S mutations. First, we evaluated the effect of the A8V and E134D mutations on the specific activities of actomyosin ATPase at pCa 9.0 and pCa 4.0. The results are summarized in Table II and are shown in Figure 4A. Our results demonstrate that at pCa 9.0, neither the A8V nor E134D mutation impaired the ability of the cTn complex to inhibit actomyosin ATPase, regardless of cTnI phosphorylation status. Our results also show that in the presence of saturating Ca2+ (pCa 4.0), the A8V mutation substantially increased the ability of the cTn complex to activate actomyosin ATPase, regardless of cTnI phosphorylation status. In addition, the E134D mutation led to a significant, albeit minor, decrease in the ability of the cTn complex to activate actomyosin ATPase in the presence of saturating Ca2+, regardless of cTnI phosphorylation status.
Table 2.
Effect of cTnC mutations on the properties of actomyosin ATPase in the absence and presence of cTnI pseudo-phosphorylation.
| Protein | Activity at pCa 9.0 (mol Pi/s)/mol S1 | Activity at pCa 4.0 (mol Pi/s)/mol S1 | pCa50 | nH |
|---|---|---|---|---|
| cTnC | 0.0125 ± 0.0003 | 0.0601 ± 0.0003 | 5.83 ± 0.01 | 2.0 ± 0.1 |
| cTnCA8V | 0.010 ± 0.0003a | 0.0742 ± 0.0002a | 6.074 ± 0.006a | 1.6 ± 0.1 |
| cTnCE134D | 0.0088 ± 0.0001a | 0.0553 ± 0.001a | 5.82 ± 0.02 | 1.98 ± 0.04 |
| cTnC + cTnIS22D/S23D | 0.0097 ± 0.0002 | 0.0451 ± 0.0001 | 5.60 ± 0.02 | 1.5 ± 0.1 |
| cTnCA8V+ cTnIS22D/S23D | 0.0095 ± 0.0003 | 0.0599 ± 0.0003b | 5.70 ± 0.01b | 1.24 ± 0.08 |
| cTnCE134D+ cTnIS22D/S23D | 0.0093 ± 0.0002 | 0.0438 ± 0.0001b | 5.57 ± 0.04 | 1.42 ± 0.04 |
Significantly different from their respective cTnC values (p<0.05)
Significantly different from their respective cTnC + cTnIS22D/S23D values (p<0.05)
Figure 4. Effect of the A8V and E134D cTnC mutations on the Ca2+ sensitivity of actomyosin ATPase in the absence and presence of cTnI pseudo-phosphorylation.
Panel A shows the specific actomyosin ATPase activities of thin filaments reconstituted with the cTn, cTnA8V or cTnE134D complexes at pCa 9.0 (open bars) or pCa 4.0 (bars filled with slanted lines) in the absence and presence (indicated by cTnIS22D/S23D) of cTnI pseudo-phosphorylation. Each data point represents a mean ± S.E. of three separate experiments (each carried out in triplicate). Values marked with * are significantly different from their respective cTnC values (p<0.05). Values marked with ** are significantly different from their respective cTnC + cTnIS22D/S23D values (p<0.05). Panel B shows the Ca2+ dependent activity of actomyosin ATPase in the absence of cTnI pseudo-phosphorylation for thin filaments reconstituted with the cTn (□), cTnA8V (red ▽) or cTnE134D (green △) complexes as a function of pCa. Panel B also shows the Ca2+ dependent activity of actomyosin ATPase in the presence of cTnI pseudo-phosphorylation for thin filaments reconstituted with the cTn (■), cTnA8V (red ▼) or cTnE134D (green ▲) complexes as a function of pCa. Each data point represents a mean ± S.E. of at least three separate experiments (each carried out in triplicate). Data sets were individually normalized for each mutant cTn complex, and fit with logistic sigmoid. The experimental conditions were as described under “Experimental Procedures” section. Panel C shows ΔpCa50 data for Ca2+ dependence of actomyosin ATP activity of thin filaments reconstituted with the cTn complexes (mutant cTn pCa50 (with cTnIS22D/S23D) – mutant cTn pCa50 (without cTnIS22D/S23D)). Value marked with * is significantly different from its control value (p< 0.05).
For thin filaments reconstituted with the cTn, cTnA8V or cTnE134D complexes, half-maximal Ca2+ activation occurred with a pCa50 of 5.83 ± 0.01, 6.074 ± 0.006, or 5.82 ± 0.02, respectively (Figure 4B and Table II). These results indicate that the A8V mutation led to a statistically significant increase in the Ca2+ sensitivity of actomyosin ATPase, while the E134D mutation had no significant effect on the Ca2+ sensitivity of actomyosin ATPase. The nH values for actomyosin ATPase curves were not significantly affected by either the A8V or E134D mutation (Table II). For thin filaments reconstituted with cTn, cTnA8V or cTnE134D complexes, containing phosphomimetic of cTnI, the Ca2+ half-maximal activation occurred with a pCa50 of ± 5.60 ± 0.02, 5.70 ± 0.01, or 5.57 ± 0.04, respectively (Figure 4B). Thus, actomysoin ATPase activity of thin filaments reconstituted with the cTn complex underwent a substantial Ca2+ desensitization upon cTnI pseudo-phosphorylation (ΔpCa50 = −0.23 ± 0.02). The A8V mutation significantly enhanced the extent of Ca2+ desensitization induced by cTnI pseudo-phosphorylation (ΔpCa50 = −0.38 ± 0.01), while the E134D mutation did not significantly affect the extent of Ca2+ desensitization (ΔpCa50 = −0.25 ± 0.04) (Figure 4C). In the presence of cTnI pseudo-phosphorylation, the nH values for actomyosin ATPase curves were not significantly affected by either the A8V or E134D mutation (Table II). Our results show that after cTnI was replaced by the phosphomimetic of cTnI, the A8V mutation still resulted in a statistically significant increase in the Ca2+ sensitivity of actomyosin ATPase, while the E134D mutation had no significant effect on the Ca2+ sensitivity of actomyosin ATPase. Thus, our results indicate that the A8V mutation significantly increased the Ca2+ sensitivity of actomyosin ATPase regardless of cTnI phosphorylation status. On the other hand, the E134D mutation exerted no significant effect on the Ca2+ sensitivity of actomyosin ATPase, regardless of cTnI phosphorylation status. In addition, our results show that neither the A8V nor E134D cTnC mutation significantly affected the cooperativity of Ca2+ activation of actomoysin ATPase, regardless of cTnI phosphorylation status. Furthermore, our results show that the A8V mutation enhanced the extent of Ca2+ desensitization of actomyosin ATPase induced by cTnI pseudo-phosphorylation, while the E134D mutation did not affect the extent of Ca2+ desensitization.
DISCUSSION
Recently, several mutations of cTnC, including A8V and E134D, were linked to HCM (18). Compared to wild-type cTnC, the A8V mutation led to higher force recovery and increased Ca2+ sensitivity of force development in skinned fibers (18). The E134D mutation did not affect either the extent of force recovery or the Ca2+ sensitivity of force generation (18), and was hypothesized to be a polymorphism (5). The main objective of this study was to examine whether HCM-linked A8V and E134D cTnC mutations affect the response of reconstituted thin filaments to Ca2+ upon phosphorylation of cTnI by PKA. In order to mimic phosphorylation of cTnI by PKA, Ser22 and Ser23 residues of cTnI were substituted by Asp. It is important to note that substitutions of Ser with Asp do not always recapitulate the effects of phosphorylation on the properties of the protein. Thus, use of pseudo-phosphorylation to elucidate the effects of phosphorylation should be carefully considered. However, a number of studies demonstrated that effects of cTnI pseudo-phosphorylation on properties of cTnI were similar to that of phosphorylation. For example, NMR analysis of cTnC-cTnI complexes demonstrated that pseudo-phosphorylated cTnI provided a good structural mimetic for cTnI phosphorylated by PKA (24). In addition, the effects of S22D/S23D cTnI mutation on the Ca2+ sensitivity of reconstituted thin filaments and on the Ca2+ sensitivity of myofibrillar ATPase were shown to be very similar to that of actual PKA phosphorylation (25, 26). Thus, we are confident that pseudo-phosphorylation of cTnI is able to mimic the properties of phosphorylated cTnI.
The A8V mutation is located within the 14-residue N-helix of cTnC, a region known to modulate Ca2+ binding and exchange with the regulatory N-domain site (27, 28). Consistent with previous studies (29, 30), the A8V mutation increased the Ca2+ sensitivity of reconstituted thin filaments by slowing the rate of Ca2+ dissociation. The slower rate of Ca2+ dissociation from reconstituted thin filaments could potentially result in diastolic dysfunction, a hallmark of HCM. In addition, in the absence of cTnI pseudo-phosphorylation, the A8V mutation led to a slight decrease in cooperativity of Ca2+ binding to reconstituted thin filaments, as indicated by an altered slope of IAANS fluorescence–pCa relationship. It is possible that the A8V mutation alters interactions between cTnC and other subunits of the cTn complex, ultimately affecting near-neighbor regulatory unit interactions along the thin filament.
Our results indicate that the A8V mutation did not significantly affect the extent of the decrease in the Ca2+ sensitivity of the cTn complex reconstituted into thin filaments, associated with pseudo-phosphorylation of cTnI by PKA. Furthermore, the A8V mutation did not abolish the acceleration of the Ca2+ dissociation rate from the regulatory N-domain site of the cTn complex reconstituted into thin filaments, associated with pseudo-phosphorylation of cTnI by PKA. In fact, the rate of Ca2+ dissociation from the cTnA8V complex reconstituted into thin filaments was accelerated by a greater extent upon cTnI pseudo-phosphorylation.
To examine the functional effect of the A8V mutation on the response of reconstituted thin filaments to Ca2+ induced by cTnI pseudo-phosphorylation, we measured the Ca2+ dependence of the actomyosin ATPase. Consistent with the effect of the A8V mutation on Ca2+ sensitivity of reconstituted thin filaments and with a previous report (29), the A8V mutation sensitized actomyosin ATPase to Ca2+. In addition to its Ca2+ sensitizing effect, the A8V mutation led to a substantial increase in the ability of cTnC to activate actomyosin ATPase in the presence of saturating Ca2+, regardless of cTnI phosphorylation status. However, the A8V mutation did not blunt the extent of Ca2+ desensitization of actomyosin ATPase induced by cTnI pseudo-phosphorylation. On the contrary, the Ca2+ sensitizing effect of the A8V mutation on actomyosin ATPase was diminished upon cTnI pseudo-phosphorylation due to a larger decrease in the Ca2+ sensitivity caused by cTnI pseudo-phosphorylation.
Numerous studies demonstrated that abnormal response of myofilaments to Ca2+ can lead to severe pathophysiological consequences (for review, see (5, 31)). Available experimental evidence shows that correcting the abnormal Ca2+ sensitivity can rescue hypertrophic and restrictive phenotypes in transgenic mouse models (32, 33). Since the A8V mutation did not abolish the response of reconstituted thin filaments to Ca2+ upon cTnI pseudo-phosphorylation, drugs that mimic the effects of cTnI phosphorylation could be potentially designed to correct the pathophysiological consequences due to the Ca2+ sensitizing effect of the A8V and perhaps other cardiomyopathy-linked mutations. Alternatively, regulatory proteins that desensitize reconstituted thin filaments to Ca2+, such as the N-terminal truncated cTnI (32, 34–36), might themselves be used as therapeutic tools to threat hypertrophy and diastolic dysfunction associated with the Ca2+ sensitizing HCM-linked mutations.
We also examined the effect of the E134D mutation on the response of reconstituted thin filaments to Ca2+ upon pseudo-phosphorylation of cTnI by PKA. Earlier studies demonstrated that the E134D mutation did not affect Ca2+ binding properties of reconstituted thin filaments (18, 29, 30). However, the E134D mutation could have led to alterations in Ca2+ binding properties of cTnC reconstituted into thin filaments when cTnI was phosphorylated, as was previously observed with several cardiomyopathy-linked cTnC mutations (15–17). Thus, we decided to determine whether the E134D mutation altered Ca2+ binding properties of reconstituted thin filaments upon pseudo-phosphorylation of cTnI by PKA. Our results indicate that in the absence of pseudo-phosphorylation, the E134D mutation exerted no effect on the Ca2+ sensitivity and the rate of Ca2+ dissociation from reconstituted thin filaments. However, in the presence of cTnI pseudo-phosphorylation, the E134D mutation led to statistically significant, albeit minor, decrease in the rate of Ca2+ dissociation from reconstituted thin filaments. In addition, the E134D mutation had no significant effect on the Ca2+ sensitivity of actomyosin ATPase, regardless of cTnI phosphorylation status. However, the E134D mutation led to statistically significant, albeit minor, decrease in ability of the cTn complex to activate actomyosin ATPase in the presence of saturating Ca2+, regardless of cTnI phosphorylation status. Since the effects of the E134D mutation on the Ca2+ binding properties of reconstituted thin filaments were rather modest, these results are consistent with the idea that the E134D mutation is a rare polymorphism. However, the possibility remains that the E134D mutation exerts its effect not due to changes in the Ca2+ sensitivity but through a different, yet unknown mechanism. These questions can be answered by developing an animal model bearing the E134D mutation.
In summary, we examined the effect of HCM-linked A8V and E134D cTnC mutations on the response of reconstituted thin filaments to Ca2+ upon pseudo-phosphorylation of cTnI by PKA. Our results show that neither the A8V nor E134D mutations significantly affected the extent of Ca2+ desensitization of reconstituted thin filaments induced by cTnI pseudo-phosphorylation. In fact, the A8V mutation enhanced the effect of cTnI pseudo-phosphorylation on the rate of calcium dissociation from reconstituted thin filaments. Consequently, while the A8V mutation still led to a slower rate of Ca2+ dissociation from reconstituted thin filaments upon cTnI pseudo-phosphorylation, the ability of the A8V mutation to decrease the rate of Ca2+ dissociation was diminished. In addition, the ability of the A8V mutation to sensitize actomyosin ATPase to Ca2+ was diminished after cTnI was replaced by the phosphomimetic of cTnI. Consistent with the hypothesis that E134D mutation is a rare polymorphism, it exerted minor to no effect on Ca2+ binding properties of cTnC reconstituted into thin filaments, and on the Ca2+ dependence of actomyosin ATPase, regardless of cTnI phosphorylation status. In conclusion, this study enhances understanding of how cardiomyopathy-linked cTnC mutations affect the response of reconstituted thin filaments to Ca2+ upon cTnI phosphorylation.
Acknowledgments
We thank Dr. Lawrence Smillie (University of Alberta) for the generous gift of the human cTnC plasmid. We also thank Karen Veloso (University of Houston) for technical assistance.
Footnotes
This research was funded by NIH grant R00HL087462 (to S.B.T)
Abbreviations: HCM, Hypertrophic cardiomyopathy; DCM, dilated cardiomyopathy; cTnC, cardiac troponin C; cTnCA8V, cTnC with the A8V mutation, cTnCE134D, cTnC with the E134D mutation; cTnI, cardiac troponin I; phosphomimetic of cTnI, cTnI with S22D/S23D mutation; cTnT, cardiac troponin T; cTn, cardiac troponin complex: (cTnC-cTnI-cTnT); cTnA8V, the cTn complex containing cTnCA8V: (cTnCA8V-cTnI-cTnT); cTnE134D, the cTn complex containing cTnCE134D: (cTnCE134D-cTnI-cTnT); IAANS, 2-(4′-(iodoacetamido)anilino)naphthalene-6-sulfonic acid; EGTA, ethylene glycol-bis(2-aminoethyl)-N,N,N′,N′-tetraacetic acid; DTT, dithiothreitol; MOPS, 3-(N-morpholino)propanesulfonic acid; Kd, dissociation constant; koff, dissociation rate.
References
- 1.Chung MW, Tsoutsman T, Semsarian C. Hypertrophic cardiomyopathy: from gene defect to clinical disease. Cell Res. 2003;13:9–20. doi: 10.1038/sj.cr.7290146. [DOI] [PubMed] [Google Scholar]
- 2.Bashyam MD, Savithri GR, Kumar MS, Narasimhan C, Nallari P. Molecular genetics of familial hypertrophic cardiomyopathy (FHC) J Hum Genet. 2003;48:55–64. doi: 10.1007/s100380300007. [DOI] [PubMed] [Google Scholar]
- 3.Arad M, Seidman JG, Seidman CE. Phenotypic diversity in hypertrophic cardiomyopathy. Hum Mol Genet. 2002;11:2499–2506. doi: 10.1093/hmg/11.20.2499. [DOI] [PubMed] [Google Scholar]
- 4.Tardiff JC. Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes. Heart Fail Rev. 2005;10:237–248. doi: 10.1007/s10741-005-5253-5. [DOI] [PubMed] [Google Scholar]
- 5.Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD. Mutations in Troponin that cause HCM, DCM AND RCM: what can we learn about thin filament function? J Mol Cell Cardiol. 2009;48:882–892. doi: 10.1016/j.yjmcc.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez JE, McCudden CR, Willis MS. Familial hypertrophic cardiomyopathy: basic concepts and future molecular diagnostics. Clin Biochem. 2009;42:755–765. doi: 10.1016/j.clinbiochem.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 7.Tardiff JC. Thin filament mutations: developing an integrative approach to a complex disorder. Circ Res. 2011;108:765–782. doi: 10.1161/CIRCRESAHA.110.224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farah CS, Reinach FC. The troponin complex and regulation of muscle contraction. Faseb J. 1995;9:755–767. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- 9.Filatov VL, Katrukha AG, Bulargina TV, Gusev NB. Troponin: structure, properties, and mechanism of functioning. Biochemistry (Mosc) 1999;64:969–985. [PubMed] [Google Scholar]
- 10.van Eerd JP, Takahashi K. The amino acid sequence of bovine cardiac tamponin-C. Comparison with rabbit skeletal troponin-C. Biochem Biophys Res Commun. 1975;64:122–127. doi: 10.1016/0006-291x(75)90227-2. [DOI] [PubMed] [Google Scholar]
- 11.Davis JP, Tikunova SB. Ca(2+) exchange with troponin C and cardiac muscle dynamics. Cardiovasc Res. 2008;77:619–626. doi: 10.1093/cvr/cvm098. [DOI] [PubMed] [Google Scholar]
- 12.Solaro RJ, Rosevear P, Kobayashi T. The unique functions of cardiac troponin I in the control of cardiac muscle contraction and relaxation. Biochem Biophys Res Commun. 2008;369:82–87. doi: 10.1016/j.bbrc.2007.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solaro RJ. Multiplex kinase signaling modifies cardiac function at the level of sarcomeric proteins. J Biol Chem. 2008;283:26829–26833. doi: 10.1074/jbc.R800037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solaro RJ, van der Velden J. Why does troponin I have so many phosphorylation sites? Fact and fancy. J Mol Cell Cardiol. 2010;48:810–816. doi: 10.1016/j.yjmcc.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidtmann A, Lindow C, Villard S, Heuser A, Mugge A, Gessner R, Granier C, Jaquet K. Cardiac troponin C-L29Q, related to hypertrophic cardiomyopathy, hinders the transduction of the protein kinase A dependent phosphorylation signal from cardiac troponin I to C. Febs J. 2005;272:6087–6097. doi: 10.1111/j.1742-4658.2005.05001.x. [DOI] [PubMed] [Google Scholar]
- 16.Pinto JR, Siegfried JD, Parvatiyar MS, Li D, Norton N, Jones MA, Liang J, Potter JD, Hershberger RE. Functional characterization of TNNC1 rare variants identified in dilated cardiomyopathy. J Biol Chem. 2011;286:34404–34412. doi: 10.1074/jbc.M111.267211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biesiadecki BJ, Kobayashi T, Walker JS, Solaro RJ, de Tombe PP. The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ Res. 2007;100:1486–1493. doi: 10.1161/01.RES.0000267744.92677.7f. [DOI] [PubMed] [Google Scholar]
- 18.Landstrom AP, Parvatiyar MS, Pinto JR, Marquardt ML, Bos JM, Tester DJ, Ommen SR, Potter JD, Ackerman MJ. Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations in TNNC1-encoded troponin C. J Mol Cell Cardiol. 2008;45:281–288. doi: 10.1016/j.yjmcc.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis JP, Norman C, Kobayashi T, Solaro RJ, Swartz DR, Tikunova SB. Effects of thin and thick filament proteins on calcium binding and exchange with cardiac troponin C. Biophys J. 2007;92:3195–3206. doi: 10.1529/biophysj.106.095406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tikunova SB, Liu B, Swindle N, Little SC, Gomes AV, Swartz DR, Davis JP. Effect of calcium-sensitizing mutations on calcium binding and exchange with troponin C in increasingly complex biochemical systems. Biochemistry. 2010;49:1975–1984. doi: 10.1021/bi901867s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swindle N, Tikunova SB. Hypertrophic cardiomyopathy-linked mutation D145E drastically alters calcium binding by the C-domain of cardiac troponin C. Biochemistry. 2010;49:4813–4820. doi: 10.1021/bi100400h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson S, Potter JD. The regulation of free Ca2+ ion concentration by metal chelators. Methods in Pharmacology. 1984;5:63–75. [Google Scholar]
- 23.Tikunova SB, Rall JA, Davis JP. Effect of hydrophobic residue substitutions with glutamine on Ca(2+) binding and exchange with the N-domain of troponin C. Biochemistry. 2002;41:6697–6705. doi: 10.1021/bi011763h. [DOI] [PubMed] [Google Scholar]
- 24.Finley N, Abbott MB, Abusamhadneh E, Gaponenko V, Dong W, Gasmi-Seabrook G, Howarth JW, Rance M, Solaro RJ, Cheung HC, Rosevear PR. NMR analysis of cardiac troponin C-troponin I complexes: effects of phosphorylation. FEBS Lett. 1999;453:107–112. doi: 10.1016/s0014-5793(99)00693-6. [DOI] [PubMed] [Google Scholar]
- 25.Lu QW, Hinken AC, Patrick SE, Solaro RJ, Kobayashi T. Phosphorylation of cardiac troponin I at protein kinase C site threonine 144 depresses cooperative activation of thin filaments. J Biol Chem. 2010;285:11810–11817. doi: 10.1074/jbc.M109.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakthivel S, Finley NL, Rosevear PR, Lorenz JN, Gulick J, Kim S, VanBuren P, Martin LA, Robbins J. In vivo and in vitro analysis of cardiac troponin I phosphorylation. J Biol Chem. 2005;280:703–714. doi: 10.1074/jbc.M409513200. [DOI] [PubMed] [Google Scholar]
- 27.Chandra M, da Silva EF, Sorenson MM, Ferro JA, Pearlstone JR, Nash BE, Borgford T, Kay CM, Smillie LB. The effects of N helix deletion and mutant F29W on the Ca2+ binding and functional properties of chicken skeletal muscle troponin. J Biol Chem. 1994;269:14988–14994. [PubMed] [Google Scholar]
- 28.Luo Y, Davis JP, Smillie LB, Rall JA. Determinants of relaxation rate in rabbit skinned skeletal muscle fibres. J Physiol. 2002;545:887–901. doi: 10.1113/jphysiol.2002.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto JR, Parvatiyar MS, Jones MA, Liang J, Ackerman MJ, Potter JD. A functional and structural study of troponin C mutations related to hypertrophic cardiomyopathy. J Biol Chem. 2009;284:19090–19100. doi: 10.1074/jbc.M109.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto JR, Reynaldo DP, Parvatiyar MS, Dweck D, Liang J, Jones MA, Sorenson MM, Potter JD. Strong cross-bridges potentiate the Ca(2+) affinity changes produced by hypertrophic cardiomyopathy cardiac troponin C mutants in myofilaments: a fast kinetic approach. J Biol Chem. 2011;286:1005–1013. doi: 10.1074/jbc.M110.168583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marston SB. How do mutations in contractile proteins cause the primary familial cardiomyopathies? J Cardiovasc Transl Res. 2011;4:245–255. doi: 10.1007/s12265-011-9266-2. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Charles PY, Nan C, Pinto JR, Wang Y, Liang J, Wu G, Tian J, Feng HZ, Potter JD, Jin JP, Huang X. Correcting diastolic dysfunction by Ca(2+) desensitizing troponin in a transgenic mouse model of restrictive cardiomyopathy. J Mol Cell Cardiol. 2010;49:402–411. doi: 10.1016/j.yjmcc.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagatheesan G, Rajan S, Petrashevskaya N, Schwartz A, Boivin G, Arteaga GM, Solaro RJ, Liggett SB, Wieczorek DF. Rescue of tropomyosin-induced familial hypertrophic cardiomyopathy mice by transgenesis. Am J Physiol Heart Circ Physiol. 2007;293:H949–958. doi: 10.1152/ajpheart.01341.2006. [DOI] [PubMed] [Google Scholar]
- 34.Yu ZB, Zhang LF, Jin JP. A proteolytic NH2-terminal truncation of cardiac troponin I that is up-regulated in simulated microgravity. J Biol Chem. 2001;276:15753–15760. doi: 10.1074/jbc.M011048200. [DOI] [PubMed] [Google Scholar]
- 35.Feng HZ, Chen M, Weinstein LS, Jin JP. Removal of the N-terminal extension of cardiac troponin I as a functional compensation for impaired myocardial beta-adrenergic signaling. J Biol Chem. 2008;283:33384–33393. doi: 10.1074/jbc.M803302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbato JC, Huang QQ, Hossain MM, Bond M, Jin JP. Proteolytic N-terminal truncation of cardiac troponin I enhances ventricular diastolic function. J Biol Chem. 2005;280:6602–6609. doi: 10.1074/jbc.M408525200. [DOI] [PubMed] [Google Scholar]
- 37.Sia SK, Li MX, Spyracopoulos L, Gagne SM, Liu W, Putkey JA, Sykes BD. Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain. J Biol Chem. 1997;272:18216–18221. doi: 10.1074/jbc.272.29.18216. [DOI] [PubMed] [Google Scholar]