Abstract
Myosins are molecular motors that convert chemical energy into mechanical work. Allosterically coupling ATP-binding, hydrolysis, and binding/dissociation to actin filaments requires precise and coordinated structural changes that are achieved by the structurally complex myosin motor domain. UNC-45, a member of the UNC-45/Cro1/She4p family of proteins, acts as a chaperone for myosin and is essential for proper folding and assembly of myosin into muscle thick filaments in vivo. The molecular mechanisms by which UNC-45 interacts with myosin to promote proper folding of the myosin head domain are not known. We have devised a novel approach, to our knowledge, to analyze the interaction of UNC-45 with the myosin motor domain at the single molecule level using atomic force microscopy. By chemically coupling a titin I27 polyprotein to the motor domain of myosin, we introduced a mechanical reporter. In addition, the polyprotein provided a specific attachment point and an unambiguous mechanical fingerprint, facilitating our atomic force microscopy measurements. This approach enabled us to study UNC-45–motor domain interactions. After mechanical unfolding, the motor domain interfered with refolding of the otherwise robust I27 modules, presumably by recruiting them into a misfolded state. In the presence of UNC-45, I27 folding was restored. Our single molecule approach enables the study of UNC-45 chaperone interactions with myosin and their consequences for motor domain folding and misfolding in mechanistic detail.
Introduction
Myosins are actin-based motor proteins that convert chemical energy from ATP hydrolysis into mechanical work. They play essential roles in a wide variety of cellular motility processes, ranging from muscle contraction to cleavage furrow ingression during cytokinesis. Type II myosin heavy chains have a molecular mass of ∼225 kDa and consist of an N-terminal globular head domain and a C-terminal rod domain. The motor activity resides within the head domain that harbors the sites for actin-binding and enzymatic activity. A model for the ATP-driven movement of muscle-specific myosin along actin filaments was proposed by Huxley in 1969 (1). This swinging cross-bridge model of muscle contraction has been refined by numerous structural and functional studies (reviewed in (2)). These studies have revealed that the myosin power stroke is the result of precise and coordinated structural rearrangements within the motor domain, amplified by a rigid lever arm domain. The precise coupling of ATP-binding, hydrolysis and release, actin binding and release, and force-generating structural changes is achieved by a complex fold of the ∼100 kDa motor domain.
Due to its complex structure, the myosin motor domain cannot spontaneously adopt its native structure in in vitro refolding experiments, in contrast to the coiled coil of the C-terminal rod domain. In vivo, molecular chaperones are necessary for the proper folding and structural maintenance of the myosin head (3–7). The myosin chaperone UNC-45, a founding member of the UCS (UNC-45/Cro1/She4p) family of proteins (8), is essential for proper folding and assembly of myosin into muscle thick filaments (9). Several studies have highlighted the importance of UCS domain-containing proteins for proper myosin function. Temperature-sensitive UNC-45 mutants cause paralysis (10) and disordered assembly of muscle thick filaments (8,10) in nematodes. All metazoan genomes analyzed thus far encode an UNC-45 ortholog. Vertebrates express a specific UNC-45 isoform in striated muscle (11). Heterologous production of skeletal muscle myosin has been reportedly achieved only in muscle-derived cell lines or lysates (3,4), consistent with a requirement of this muscle-specific UNC-45 isoform for proper myosin folding and/or assembly.
Less is known about mechanistic aspects of UNC-45 function. UNC-45 is composed of three domains: An amino-terminal TPR (tetratricopeptide repeat) domain, a ∼400 residue central region, and a ∼400 residue UCS domain (8) (Fig. 1). The TPR domain stoichiometrically binds the molecular chaperone heat shock protein 90 (Hsp90) (6). UNC-45 has been proposed to function as a cochaperone for Hsp90 (12). UNC-45 from the nematode Caenorhabditis elegans prevents the aggregation of thermally denatured myosin subfragment 1 (myosin S1, comprised almost exclusively of the motor domain; we use S1 and motor domain interchangeably throughout the text) (6). The N-terminal TPR domain is dispensable for this activity.
Figure 1.
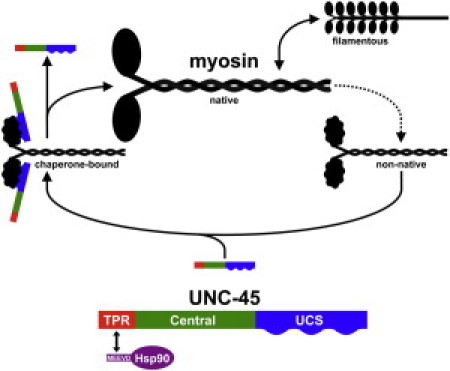
Type II myosin is composed of two dissimilar domains: An extended rod-like coiled-coil, and a structurally more complex globular motor domain. Whereas the rod folds spontaneously under various conditions, the motor domain requires the aid of molecular chaperones to acquire its functional tertiary structure. UNC-45, the UCS protein in animals, contains an amino-terminal TPR domain that mediates specific interaction with the heat shock protein Hsp90. UNC-45 is required for proper myosin folding and/or assembly. Hsp90 appears to modulate the UNC-45:myosin interaction.
We have devised, to our knowledge, a novel approach to analyze UNC-45/myosin S1 interactions at the single molecule level using the atomic force microscope (AFM) (13). AFM is ideally suited for these studies because it mimics the directionality of the in vivo folding pathways and can capture misfolding events (14,15). However, it is difficult to observe folding with this technique in the absence of an unambiguous mechanical fingerprint. By chemically coupling a titin I27 polyprotein to the motor domain of myosin, we introduced a mechanical reporter, providing a specific attachment point and a well-characterized mechanical fingerprint in the AFM measurements. After mechanical unfolding of the chimeric protein, the otherwise robustly folding polyprotein fails to refold, presumably as a consequence of myosin S1 misfolding. UNC-45 quantitatively restores I27 folding, indicating that it efficiently prevents misfolding into a state that might represent an off-pathway folding intermediate. Thus, the polyprotein functions as a folding sensor in our measurements and allows us to directly observe the consequences of interactions between UNC-45 and myosin S1. This system should be generally applicable to study chaperone-substrate interactions at the single molecule level.
Materials and Methods
Proteins
Myosin was purified from rabbit skeletal muscle (16). We produced the myosin S1 fragment by chymotryptic digestion of myosin as described (17,18). The purity and composition of full-length myosin and the S1 subfragment proteins was confirmed by SDS-PAGE. Two highly reactive cysteine sulfhydryl groups, termed SH1 and SH2, are located close to the ATP-binding pocket in the myosin motor domain. Previous studies have shown that SH1 and SH2 exhibit differential reactivity toward a number of thiol-modifying agents (19,20). We selectively derivatized the reactive SH1 group with an environmentally sensitive fluorophore, 6-bromoacetyl-2-dimethyl-aminonaphthalene (BADAN). The excitation maximum is 387 nm and the emission is at 520 nm for the fluorescently labeled protein. The labeling stoichiometry was typically 0.85–0.95 mol BADAN/mol S1 as determined by absorption spectroscopy.
To introduce a mechanical reporter, S1 was derivatized with a tandem repeat I27 polyprotein carrying an N-terminal cysteine residue and a C-terminal His6 tag (21,22). Coupling to the S1 was achieved via the reactive cysteines (SH1/SH2) within the motor domain. For chemical coupling, the I27 tandem repeat protein (50 μM) was first incubated with a 20-fold excess of a homobifunctional thiol-reactive cross-linker (BM(PEG)3, Thermo Fisher Scientific, Rockford, IL). Excess cross-linker was removed by gel filtration on Sephadex G-25 medium. The activated I27 tandem repeat protein was incubated with a 1.5-fold excess of myosin S1 to couple the two polypeptides, resulting in a covalent linkage between the N-terminal cysteine in the I27 and the reactive thiols in the myosin motor domain. The cross-linking adduct was isolated by size exclusion chromatograpy on a Sephacryl S-300 column (GE Healthcare, Piscataway, NJ) (see protein gel in Fig. S1 in the Supporting Material).
Expression plasmids harboring mouse UNC-45b cDNA sequences (full-length) were transformed into Escherichia coli BL21 cells. The expression vectors were based on a pPROEX-HTa backbone and have an N-terminal His6-tag that can be cleaved off with TEV protease. Protein expression was induced by addition of 1 mM IPTG and shaking at 16°C overnight. The cells were resuspended in phosphate buffered saline (PBS, 10 mM sodium phosphate buffer, pH 7.4, 150 mM NaCl), and sonicated on ice in the presence of protease inhibitors. After centrifugation, the supernatant was incubated with a Ni-NTA agarose, which then was washed thoroughly with 20 mM imidazole in PBS followed by 50 mM imidazole in PBS. The protein was eluted with 250 mM imidazole in PBS and the N-terminal His6-tag was removed by digestion with TEV protease. The cleaved His6-tag, TEV-protease and imidazole were removed by size exclusion chromatograpy on a Sephacryl S-300 column (GE Healthcare).
Fluorescence measurements
We took advantage of the environmentally sensitive fluorophore BADAN attached to myosin S1 to study its interaction with UNC-45b. 250 nM BADAN-S1 were mixed with UNC-45b at concentrations ranging from 0.5 to 10 μM and incubated for 30 min in 25 mM HEPES-KOH pH 8.0 at 25°C, 30 mM KCl, 2 mM MgCl2. Fluorescence measurements were performed using a Horiba Jobin Yvon Fluorolog-3 model FL3 Fluorescent spectrometer. Fluorescence emission of BADAN-S1 at 520 nm was measured after excitation at 387 nm relative to a control sample that contained buffer instead of UNC-45b. The fluorescence increase was analyzed assuming equilibrium between UNC-45b, BADAN-S1, and a complex of the two:
where [UNC-45b]eq, [BADAN-S1]eq, and [UNC-45b BADAN-S1]eq are the equilibrium concentrations of free UNC-45b, free BADAN-S1, and the complex of the two, respectively.
Single molecule AFM
The mechanical properties of single proteins were studied using a home-built single molecule AFM as previously described (22–26). The spring constant of each individual cantilever (MLCT or Olympus OBL, Veeco Metrology Group, Santa Barbara, CA) was calculated using the equipartition theorem (27). In a typical experiment, a small aliquot of the purified proteins (∼1–5 μl, 10–100 μg/ml) was allowed to adsorb onto either a clean glass coverslip (for myosin and S1) or a Ni-NTA-coated glass coverslip (for the S1-(I27)8 and (I27)8 constructs) (28). We incubated myosin for ∼5–15 min and then the coverslip was rinsed with myosin buffer (60 mM sodium acetate, 600 mM KCl, 25 mM imidazole, 4 mM MgCl2, and 1 mM dithiothreitol, pH 7.4). The other proteins were incubated for ∼15 min and then rinsed with PBS pH 7.4. Proteins were picked up randomly by adsorption to the cantilever tip, which was pressed down onto the sample for 1–2 s at forces of several nano-Newtons and then stretched for several hundred nm. We found that the probability of picking up a protein and stretching it from end to end was extremely low. This is because some proteins may attach to the coverslip surface nonspecifically via I27 domains. For example, in the case of the S1-I27 chimera we found that the number of I27 unfolding events observed in the force-extension curves was typically less than the total number of I27 domains (i.e., eight). All experiments were performed at room temperature (∼25°C). The pulling speed of the different unfolding and refolding experiments was in the range of 0.5–0.7 nm/ms.
Measuring the fraction of refolded I27 domains
We used a two-pulse unfolding/refolding protocol to estimate the fraction of refolded domains as described previously in (22,29). After the first stretch, a single polyprotein is allowed to relax for different time intervals. We then count the refolded domains in the second pulling. The waiting time between refolding cycles was 10 s. In a typical experiment after picking up a protein, the AFM tip is moved away from the surface (∼30–50nm) to prevent the tip picking new proteins due to cantilever drift.
Analysis of force extension curves
The elasticity of the stretched proteins was analyzed using the worm-like chain model of polymer elasticity (22,23):
where F is force, p is the persistence length, x is the end-to-end length, and Lc is the contour length of the stretched protein. The adjustable parameters are p and Lc.
Results and Discussion
Environmentally sensitive fluorescence as a reporter for UNC-45b – myosin S1 interaction
Both UNC-45b and striated muscle myosin are highly conserved among mammals: the amino acid sequences for the myosin-2 heavy chain and UNC-45b are 95% and 96% identical among the rabbit and mouse orthologs, respectively. To study the effects of UNC-45b binding to the myosin motor domain, we used subfragment 1 from rabbit skeletal muscle myosin and recombinantly expressed mouse striated muscle UNC-45b. To confirm that the purified UNC-45b interacted with myosin subfragment 1, we took advantage of the environmentally sensitive fluorophore BADAN (see Fig. 2 A) (30). The myosin motor domain harbors two reactive thiol groups, termed SH1 and SH2 (amino acid residues Cys-707 and Cys-697, respectively, in rabbit skeletal muscle myosin 2 heavy chain), located in the catalytic domain (Fig. 2 A). The BADAN fluorophore has been shown to react preferentially with the SH1 group (19), yielding fluorescently labeled myosin S1 (BADAN-S1) with a reporter fluorophore at a specific site within the catalytic domain.
Figure 2.
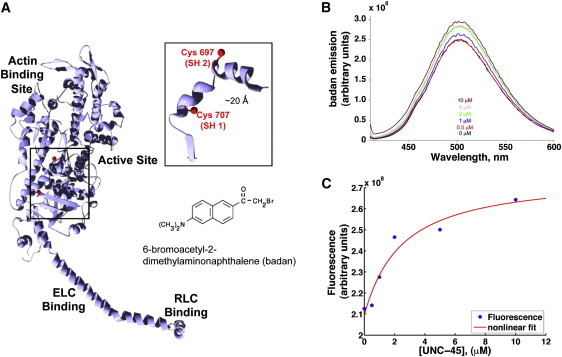
Tracking UNC-45 binding to the motor domain using environmental sensitive fluorescent probes. (A) We selectively derivatized the reactive Cys-707 group with an environmentally sensitive fluorophore, BADAN. The molecular model corresponds to the three-dimensional structure of myosin subfragment-1 (pdb 2mys, (43)). (B) Fluorescence emission spectra for BADAN-labeled S1 (BADAN-S1) as a function of UNC-45b concentration. (C) BADAN fluorescence emission (blue circles) as a function of the total UNC-45b concentration. The red line shows a fit obtained assuming complex formation with a 1:1 stoichiometry of UNC-45b and BADAN-S1, yielding an apparent KD of 1.3 μM.
We recorded fluorescence emission spectra of 0.5 μM BADAN-S1 alone and in the presence of increasing concentrations of UNC-45b (Fig. 2 B). We observed increased emission intensities of the reporter fluorophore with increasing concentrations of UNC-45b, indicating that the local environment of the fluorescent probe changes due to interaction of BADAN-S1 with UNC-45b. This change could be the consequence of either direct binding of the chaperone near the catalytic site, or of allosteric changes in the motor domain upon chaperone binding at a different site. The increase in fluorescence intensity approaches a plateau at UNC-45b concentrations above 5 μM (Fig. 2 C, blue circles). The red line shows a fit obtained assuming complex formation with a 1:1 stoichiometry of UNC-45b and BADAN-S1, yielding an apparent KD of 1.3 μM. We compared our data to recent pull-down experiments done for UNC-45b binding to myosin (31). Fig. S2 shows that both data sets follow a similar trend. These independent experimental techniques confirm that the recombinantly produced UNC-45b interacts robustly with the myosin motor domain. The agreement among the KD values obtained indicates that the introduction of a reporter moiety (in this case, an environmentally sensitive fluorophore) near the ATP-binding pocket in the myosin motor domain does not interfere with UNC-45b binding.
Mechanical fingerprint of myosin unfolding
Single molecule AFM has proven to be a powerful tool for studying protein folding (13). Stretching a folded protein between a surface and the tip of an AFM cantilever permits the application of force to single protein molecules, allowing the direct observation of folding and unfolding. These measurements often reveal a mechanical fingerprint that reflects the specific unfolding and refolding properties of a given protein under force. In principle, it should be possible to extend this methodology to study the consequences of chaperone binding on client protein folding. Before applying this approach to study UNC-45b – myosin interactions, we first aimed to obtain the characteristic mechanical fingerprint of myosin unfolding.
We deposited full-length myosin onto a glass coverslip and stretched single protein molecules using a silicon nitrite tip. A representative example of the resulting force extension curves is shown in Fig. 3 A. The force extension curves exhibit two features: A plateau at ∼30 pN, over which the force does not increase with extension, and poorly defined transitions at lower and higher forces, before and after the plateau, respectively. The plateau at 30 pN has previously been observed: It can be attributed to the rod domain and constitutes a fingerprint for the full-length myosin heavy chain (32–34) (see also Fig. S3). In the case of the myosin head subfragment 1 we were unable to observe characteristic transitions that could be attributed to the unfolding of the motor domain or myosin S1 (Fig. 3 C, Fig. S4). The data show that the average contour length for the full-length myosin is 316 ± 83 nm (n = 40), whereas the average contour for the S1 is much shorter, 171 ± 42 nm, n = 109. These values are consistent with the respective size of the proteins. The S1 subfragment is 845 amino acids long; the unfolded length of fully extended S1 is predicted to be around 300 nm (assuming that each amino acid contributes 0.36 nm to the contour length of the protein). In the case of native myosin, the predicted unfolded length of the coiled-coil domain (∼540 aa) plus the motor domain (845 aa), should be ∼500 nm. However, because the interaction between the AFM tip and protein occurs randomly the mean unfolded lengths should be much shorter than the predicted contour lengths. Our results indicate that the motor domain unfolds without a defined mechanical fingerprint in our experiments, perhaps because of the absence of well-defined barriers for mechanical unfolding or refolding.
Figure 3.
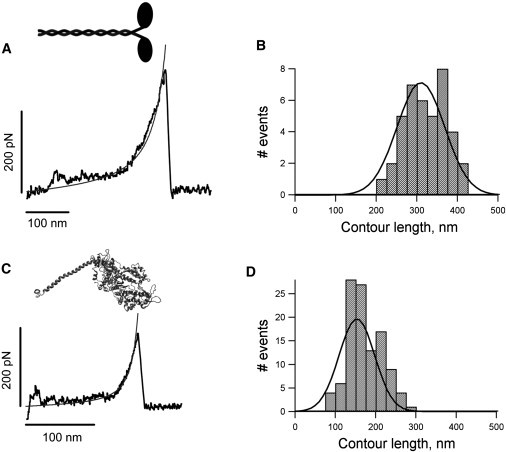
Mechanical fingerprints of the myosin domain unfolding. (A) Force-extension pattern of a single full-length myosin molecule. Full-length myosin was nonspecifically attached to glass surfaces and subject to mechanical unfolding. (B) Histogram for contour lengths obtained from 40 different force-extension profiles; the mean value is ∼315 nm (316 ± 83 nm). (C) Force-extension pattern of a single myosin head subfragment 1 (S1). (D) Histograms for contour lengths obtained for unfolded S1; the mean value is 171 ± 42 nm (n = 109). The thin lines in A and C correspond to fits to the worm-like chain equation using a persistence length, p, of 0.26 nm and an Lc of 383 nm (for the myosin trace), and p = 0.39 nm and an Lc of 183 nm (for the S1 trace).
Construction of a S1-I27 protein chimera
Because we were unable to obtain an informative fingerprint of myosin motor domain unfolding, we sought to use the well-characterized titin I27 domain as a mechanical reporter in our AFM experiments (22,35). Polyproteins composed of I27 tandem repeats unfold with a characteristic fingerprint and readily refold through many repetitions without any signs of fatigue (22). Tandem fusion constructs with I27 domains have previously been used in AFM studies to unambiguously identify proteins without knowing their fingerprint a priori and to distinguish them from nonspecific attachment events between the cantilever and the surface (see, e.g. (36–39). The introduction of I27 domains as reporters typically requires the generation of recombinant tandem fusion proteins. Recombinant expression of the myosin motor domain, however, is challenging, presumably because the complexity of its structure imposes a requirement for specific molecular chaperones. We developed a protocol to chemically couple an octameric I27 polyprotein to a specific site within the myosin motor domain of natively folded full-length myosin or myosin S1 (Fig. 4). The polyprotein harbored an N-terminal cysteine residue and a C-terminal hexahistidine (His6) tag. We chemically linked the engineered cysteine near the N-terminus of the I27 polyprotein to the reactive cysteine in the motor domain using a homobifunctional bis-maleimide cross-linker.
Figure 4.
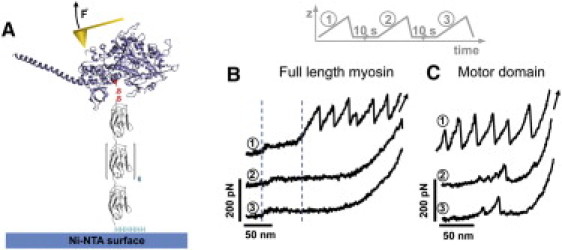
Mechanical fingerprint of derivatized full-length myosin and S1 fragment (motor domain). (A) The motor domain (blue) was derivatized with a mechanical reporter, a tandem repeat I27 polyprotein (gray) carrying an N-terminal cysteine residue and a C-terminal His6 tag. Note that the (I27)8 is both a handle reacting at the reactive cysteines in S1 and a reporter of unfolding and refolding. Coupling was achieved via the reactive cysteines (SH1/SH2) within the motor domain. The handle introduces a means of site-specific attachment via the His6 tag. (B and C) Single molecules of derivatized full-length myosin (B) or S1 (C) were subjected to repeated cycles of mechanical stretching, separated by a waiting time of 10 s at zero force (inset). The I27 sawtooth pattern is apparent in the initial unfolding trace (1), but absent in the second (2) and third (3) traces, indicating that the unfolded motor domain interferes with refolding of the I27 modules. The characteristic low-force plateau stemming from the rod (between dashed lines) is visible in all traces in A, indicating the rod folds as an independent entity and is not affected by the motor domain.
The unfolded myosin motor domain interferes with I27 refolding
We deposited the chimeric myosin – I27 constructs on a Ni2+-charged NTA surface and subjected molecules to repeated mechanical stretching in the AFM (Fig. 4, B and C), separated by pause intervals of 10 s at zero force (see inset). In the force-extension curves of both the full-length myosin and the motor domain constructs, unfolding of the I27 reporter domains is apparent during the initial stretching (Fig. 4, B and C, trace 1). In addition, the construct containing the full-length myosin (Fig. 4 B) also exhibits the plateau at 30 pN corresponding to the unraveling of the rod domain (see above). This demonstrates that in this geometry, force is applied across the I27 polyprotein, the motor domain, and the rod, validating our approach. It is possible that the sample preparation protocol also yields I278-I278 adducts, in addition to the desired I278-S1 species. However, we did not observe force extension curves with more than eight consecutive I27 unfolding events, indicating that—although these species may be present—we do not observe them in our experiments. A possible explanation could be the dual tethering of the I278-I278 to the Ni-NTA substrate; because this species has two His6-tags, it will attach to the surface on both ends, perhaps making it unlikely to be picked up by the cantilever during our AFM measurements.
Although the characteristic sawtooth pattern of I27 domain unfolding is observed during the initial stretching, force extension curves collected after quenching the force and stretching the molecule again do not exhibit this sawtooth feature (Fig. 4, traces 2 and 3). Therefore, the I27 domains do not refold during the 10 s pause at zero force, in contrast to the I27 polyprotein by itself, which readily refolds within seconds ((22); see also Fig. 5). This phenomenon is observed for both full-length myosin and myosin S1, indicating that the effect is mediated by the unfolded motor domain. Thus, folding of the I27 domain is sensitive to the presence of the unfolded motor domain, perhaps because of aberrant interactions between the covalently linked unfolded polypeptides after unfolding. This interpretation is consistent with the previously reported inability of the myosin motor domain to spontaneously refold after denaturation in the absence of chaperones (4). Interestingly, the presence of the plateau at 30 pN (between dashed lines in Fig. 4 B) in repeated stretching cycles indicates that the rod domain folds as an independent entity and is not affected by interactions with the unfolded motor domain.
Figure 5.
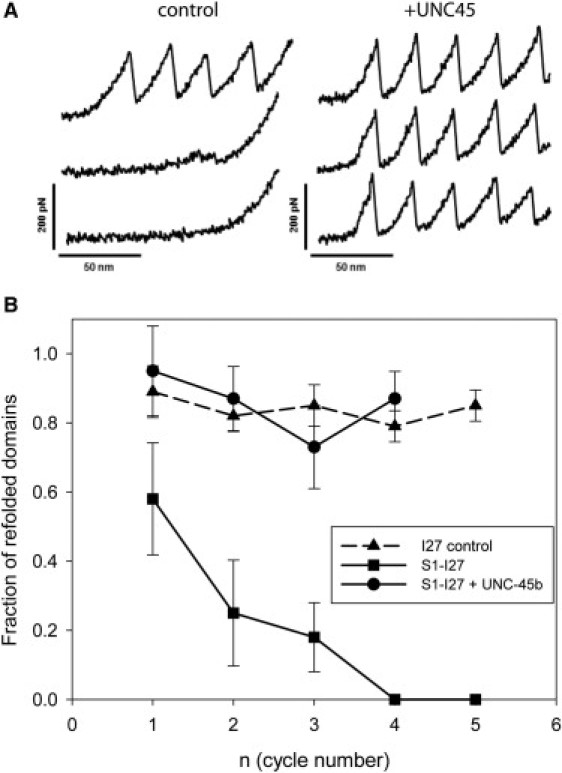
UNC-45b facilitates refolding of the titin mechanical reporter. (A) Single molecules of derivatized S1 were subjected to repeated cycles of unfolding and relaxation to zero force. In the absence of UNC-45b (control), the I27 sawtooth pattern is observed only in the initial unfolding cycle, indicating that the unfolded motor domain interferes with the refolding of the I27 modules. In the presence of 1 μM UNC-45b (right panel), full recovery of folded I27 domains is observed. This result indicates that the I27 domains function as a sensor for motor domain proper folding. UNC-45b prevents the interference between the motor domain and the I27 units, presumably by binding to the myosin motor domain. (B) Plot of the fraction of titin I27 domains as a function of refolding cycles. Single molecules of either the I27 polyprotein (triangles; 18 molecules) or derivatized S1 in the absence of chaperone (squares; 8 molecules) and in the presence of 1 μM UNC-45b (circles; 12 molecules) were subjected to repeat cycles of unfolding. The error bars correspond to the standard deviations.
UNC-45b prevents aberrant interactions of the unfolded motor domain
The observation that I27 folding is impaired in the context of the unfolded myosin motor domain indicates aberrant interactions between nonnative polypeptides. In vivo, these interactions are counteracted by molecular chaperones that prevent the formation of nonnative interactions that would otherwise interfere with proper folding. We wondered if this effect can be recapitulated in our AFM assay. We repeated the experiment described previously with the I27 – S1 construct in the presence of 1 μM UNC-45b. Strikingly, refolding of the I27 reporter domains is fully restored under these conditions (Fig. 5). Whereas in the absence of the chaperone, the I27 domains do no refold after the initial stretching cycle, indistinguishable sawtooth patterns are observed over three repeated cycles when UNC-45b is present (Fig. 5 A). Control experiments show that UNC-45 does not affect the refolding efficiency of the I27 domain alone. We also found that proteins other than UNC-45 such as bovine serum albumin do not have a significant effect on the reversible AFM-induced unfolding/refolding of I27–S1. The AFM measurements thus reveal directly an effect of the UNC-45b chaperone. Our finding suggests that UNC-45b prevents aberrant interactions between the unfolded polypeptides within the chimeric construct, presumably through interactions with the unfolded motor domain.
Conclusions
The myosin motor domain converts chemical energy into mechanical work: It allosterically couples ATP binding and hydrolysis at one site to the binding to actin filaments and transduction of force at a second site. This highly specialized function is achieved by the intricate structure of the myosin head. Perhaps as a consequence of this structural intricacy, the myosin polypeptide is unable to fold spontaneously and depends on molecular chaperones for productive folding to its native, functional conformation. UNC-45 and other members of the UCS protein family are required for the folding of the myosin motor into a mechanochemically competent protein (3,5,40–42). However, the molecular mechanisms by which the molecular chaperone UNC-45 interacts with myosin to promote proper folding of the myosin motor domain are not known.
The experimental approach described here is novel, to our knowledge, and provides a first step toward elucidating, in mechanistic detail, the effect of molecular chaperones on myosin motor domain folding. The data obtained using single-molecule AFM techniques reveal that the motor domain does not fold after mechanical unfolding. Misfolding often results in aggregation, hampering the characterization of nonnatively structured species in ensemble measurements. Our single-molecule approach circumvents this limitation and, in combination with a titin I27-based reporter, allows us to observe the effect of UNC-45 on the nonnative myosin motor domain. After mechanical unfolding, the motor domain presumably recruits the I27 domains into a misfolded state. In the presence of UNC-45, titin domain folding was restored. Thus, the approach described here is new, to our knowledge, and powerful for investigating the mechanism of action of UNC-45 on myosin function and folding.
Future experiments will pursue the unfolding and refolding of myosin in the presence of different combinations of UNC-45, its C-terminal UCS domain and other molecular chaperones, such as Hsp90. Furthermore, assembly of myosin must involve the folding of the motor domain, the association of the light chains, and dimerization of the long C-terminal α-helical segments of the two heavy chains. Hence, future experiments under more physiological conditions will help to resolve the folding pathways of the myosin motor domain and to better understand myosin assembly into functional structures.
Acknowledgments
We thank Paul Nicholls for helping with the purification of the myosin S1 fragment protein and Dr. Jose Barral for helpful discussions.
This work was funded by National Institutes of Health grants R01DK073394 (A.F.O.), by the University of Texas Medical Branch Claude D. Pepper Older Americans Independence Center NIH/NIA Grant P30 AG024832 (to A.F.O.), John Sealy Memorial Endowment Fund for Biomedical Research (A.F.O.), R01AR050051 (H.F.E.), Muscular Dystrophy Association and the Cecil and Ida Green Endowment (H.F.E.).
Footnotes
Christian M. Kaiser's present address is Department of Physics and QB3 Institute, University of California, Berkeley, CA.
Supporting Material
References
- 1.Huxley H.E. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 2.Spudich J.A. The myosin swinging cross-bridge model. Nat. Rev. Mol. Cell Biol. 2001;2:387–392. doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 3.Srikakulam R., Winkelmann D.A. Myosin II folding is mediated by a molecular chaperonin. J. Biol. Chem. 1999;274:27265–27273. doi: 10.1074/jbc.274.38.27265. [DOI] [PubMed] [Google Scholar]
- 4.Chow D., Srikakulam R., Winkelmann D.A. Folding of the striated muscle myosin motor domain. J. Biol. Chem. 2002;277:36799–36807. doi: 10.1074/jbc.M204101200. [DOI] [PubMed] [Google Scholar]
- 5.Srikakulam R., Winkelmann D.A. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- 6.Barral J.M., Hutagalung A.H., Epstein H.F. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- 7.Kachur T.M., Pilgrim D.B. Myosin assembly, maintenance and degradation in muscle: role of the chaperone UNC-45 in myosin thick filament dynamics. Int. J. Mol. Sci. 2008;9:1863–1875. doi: 10.3390/ijms9091863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barral J.M., Bauer C.C., Epstein H.F. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J. Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landsverk M.L., Li S., Epstein H.F. The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J. Cell Biol. 2007;177:205–210. doi: 10.1083/jcb.200607084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein H.F., Thomson J.N. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- 11.Price M.G., Landsverk M.L., Epstein H.F. Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J. Cell Sci. 2002;115:4013–4023. doi: 10.1242/jcs.00108. [DOI] [PubMed] [Google Scholar]
- 12.Liu L., Srikakulam R., Winkelmann D.A. Unc45 activates Hsp90-dependent folding of the myosin motor domain. J. Biol. Chem. 2008;283:13185–13193. doi: 10.1074/jbc.M800757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oberhauser A.F., Carrión-Vázquez M. Mechanical biochemistry of proteins one molecule at a time. J. Biol. Chem. 2008;283:6617–6621. doi: 10.1074/jbc.R700050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberhauser A.F., Marszalek P.E., Fernandez J.M. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 1999;6:1025–1028. doi: 10.1038/14907. [DOI] [PubMed] [Google Scholar]
- 15.Randles L.G., Rounsevell R.W., Clarke J. Spectrin domains lose cooperativity in forced unfolding. Biophys. J. 2007;92:571–577. doi: 10.1529/biophysj.106.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollard T.D. Myosin purification and characterization. Methods Cell Biol. 1982;24:333–371. doi: 10.1016/s0091-679x(08)60665-2. [DOI] [PubMed] [Google Scholar]
- 17.Weeds A.G., Pope B. Studies on the chymotryptic digestion of myosin. Effects of divalent cations on proteolytic susceptibility. J. Mol. Biol. 1977;111:129–157. doi: 10.1016/s0022-2836(77)80119-8. [DOI] [PubMed] [Google Scholar]
- 18.Mornet D., Ue K., Morales M.F. Proteolysis and the domain organization of myosin subfragment 1. Proc. Natl. Acad. Sci. USA. 1984;81:736–739. doi: 10.1073/pnas.81.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiratsuka T. ATP-induced opposite changes in the local environments around Cys(697) (SH2) and Cys(707) (SH1) of the myosin motor domain revealed by the prodan fluorescence. J. Biol. Chem. 1999;274:29156–29163. doi: 10.1074/jbc.274.41.29156. [DOI] [PubMed] [Google Scholar]
- 20.Takashi R., Tonomura Y., Morales M.F. 4,4′-Bis (1-anilinonaphthalene 8-sulfonate) (bis-ANS): a new probe of the active site of myosin. Proc. Natl. Acad. Sci. USA. 1977;74:2334–2338. doi: 10.1073/pnas.74.6.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 22.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bullard B., Linke W.A., Leonard K. Varieties of elastic protein in invertebrate muscles. J. Muscle Res. Cell Motil. 2002;23:435–447. doi: 10.1023/a:1023454305437. [DOI] [PubMed] [Google Scholar]
- 24.Miller E., Garcia T., Oberhauser A.F. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 2006;91:3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberhauser A.F., Badilla-Fernandez C., Fernandez J.M. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 26.Oberhauser A.F., Marszalek P.E., Fernandez J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 27.Florin E.L., Rief M., Gaub H.E. Sensing specific molecular interactions with the atomic force microscope. Biosens. Bioelectron. 1995;10:895–901. [Google Scholar]
- 28.Sakaki N., Shimo-Kon R., Kinosita K., Jr. One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 2005;88:2047–2056. doi: 10.1529/biophysj.104.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 30.Maier R., Eckert B., Schmid F.X. Interaction of trigger factor with the ribosome. J. Mol. Biol. 2003;326:585–592. doi: 10.1016/s0022-2836(02)01427-4. [DOI] [PubMed] [Google Scholar]
- 31.Ni W., Hutagalung A.H., Epstein H.F. The myosin-binding UCS domain but not the Hsp90-binding TPR domain of the UNC-45 chaperone is essential for function in Caenorhabditis elegans. J. Cell Sci. 2011;124:3164–3173. doi: 10.1242/jcs.087320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwaiger I., Sattler C., Rief M. The myosin coiled-coil is a truly elastic protein structure. Nat. Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi Y., Khatri B.S., Kawakami M. Dynamics of the coiled-coil unfolding transition of myosin rod probed by dissipation force spectrum. Biophys. J. 2010;99:257–262. doi: 10.1016/j.bpj.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Root D.D., Yadavalli V.K., Wang K. Coiled-coil nanomechanics and uncoiling and unfolding of the superhelix and alpha-helices of myosin. Biophys. J. 2006;90:2852–2866. doi: 10.1529/biophysj.105.071597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberhauser A.F., Hansma P.K., Fernandez J.M. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA. 2001;98:468–472. doi: 10.1073/pnas.021321798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandal M., Valle F., Samorì B. Conformational equilibria in monomeric alpha-synuclein at the single-molecule level. PLoS Biol. 2008;6:e6. doi: 10.1371/journal.pbio.0060006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dougan L., Li J., Fernandez J.M. Single homopolypeptide chains collapse into mechanically rigid conformations. Proc. Natl. Acad. Sci. USA. 2009;106:12605–12610. doi: 10.1073/pnas.0900678106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegmann S., Schöler J., Muller D.J. Competing interactions stabilize pro- and anti-aggregant conformations of human Tau. J. Biol. Chem. 2011;286:20512–20524. doi: 10.1074/jbc.M111.237875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Best R.B., Li B., Clarke J. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 2001;81:2344–2356. doi: 10.1016/S0006-3495(01)75881-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toi H., Fujimura-Kamada K., Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lord M., Pollard T.D. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J. Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lord M., Sladewski T.E., Pollard T.D. Yeast UCS proteins promote actomyosin interactions and limit myosin turnover in cells. Proc. Natl. Acad. Sci. USA. 2008;105:8014–8019. doi: 10.1073/pnas.0802874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayment I., Rypniewski W.R., Holden H.M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


