Abstract
The loss of wild type p53 tumor suppressive function and oncogenic gain-of-function of p53 mutants have been showing important implications in tumorigenesis. The p53N236S (p53N239S in human, p53S) mutation has been shown to lose wild type p53 function by yeast assay. However, its gain of function is still not clear. By gel shift assay, we showed that mutant p53S had lost its DNA binding ability to its target promoters. Further real-time PCR data confirmed that p53S had lost the function of regulating the transcription of p21 Cip1/Waf1, cyclin G, PUMA, and Bax in response to 10Gy irradiation. These data confirmed the loss of function of p53S in mammalian cells. By xenograft assay, we showed that the p53S per se was not oncogenic enough to form tumor, however, cooperating with H-RasV12, p53S could dramatically promote tumorigenesis in p53 null MEFs. Further study showed that co-expression of p53S and H-RasV12 could increase the expression level of H-RasV12 and partially eliminate the elevation of stress response proteins such as Chk2, γ-H2AX, Hsp70, Rb, p16Ink4a caused by either p53S or H-RasV12. These data suggested that p53S cross-talked with H-RasV12 and reduced the cellular stress response to oncogenic signals, which facilitated the cell growth and tumorigenesis. Together these data provided the molecular basis for the cooperation of p53S and H-RasV12 and revealed the gain of function of p53S in cross-talking with H-RasV12. This study revealed an important aspect of gain of function for p53 mutant, therefore might shed light on the clinical strategy in targeting p53 mutant.
Keywords: p53 mutant, gain of function, Ras, tumorigenesis, DNA damage response, cross-talk.
Introduction
TP53 mutation is one of the most frequent mutations occurred in human tumorigenesis. Interestingly, more than 80% of 26,608 tumor related mutations are point mutations (IARC TP53 database, version R15, November 2010) 1. Among those, seventy-three amino acids of p53 have been identified as hotspots for mutations found in human tumors by comparing the observed mutation distribution with a random multinomial distribution 2. However, it has been shown that p53 mutant proteins resulted from different site of p53 point mutations vary dramatically in terms of their loss of function and gain of function, as well as their oncogenic effects 3, 4.
One of the current strategies in cancer treatment is trying to apply personalized treatment aiming different p53 mutants. Thus, understanding of the loss and gain of function of different p53 mutants will provide the basis for personalized treatments of cancer patients.
A previous study showed that mouse embryonic fibroblasts (MEFs) from the late generation of Werner Syndrome mice (G5mTR-/-Wrn-/-) senesced very rapidly, interestingly, the senescent MEF cells are prone to escape from senescence and spontaneously immortalize. Injection of some of the immortalized cell lines into SCID mice result in tumorigenesis 5.
When we compare the molecular differences between these tumorigenic and non-tumorigenic cell lines, we found a single point mutation of p53 in all three independent tumorigenic cell lines, which cause one single amino acid change of N236S in mouse p53. The corresponding mutation in human p53 is N239S. The p53N239S mutation occurred at the L3 loop of DNA binding domain. This region is highly conserved among different species, thus might play an important role in p53 function 2, 6. IARC TP53 database (version R14, November 2009) 1 shows that human p53N239S (or p.N239S ) has been reported as a somatic mutation in 32 tumor cases, tumor origin tissues including breast, colon, stomach, hematopoietic and reticuloendothelial systems, liver and intrahepatic bile ducts, bronchus and lung, and brain etc. The widespread tumor spectrum harboring p53N239S suggested its importance in tumorigenesis. Functional assays in yeast 7 and structure-function predictions 8 indicate that p53N239S is a non-functional mutation.
However, very few studies have been done in mammalian cells for p53N239S function. p53N239S has been reported as one of the p53 mutations occurred in rheumatoid arthritis (RA) 9. When p53N239S was introduced into HS68 dermal fibroblasts together with wild type p53, it was able to up-regulate IL-6 expression 10. This result provided the evidence for the dominant negative function of p53N239S (Since there is endogenous wild type p53). However, the gain-of-function of mutant p53N239S is not well understood. The better understanding of gain of function of p53N239S mutant will benefit the cure of human diseases bearing this mutation.
Here by using non-tumorigenic p53-/- MEFs as background, we characterized the loss and gain of function of p53S. Furthermore, H-RasV12 were introduced into p53-/- MEFs with or without p53S to investigate the gain of function of p53S in cooperating with oncogenes.
Results
p53S mutant lost the DNA binding activity as well as the transactivation activity
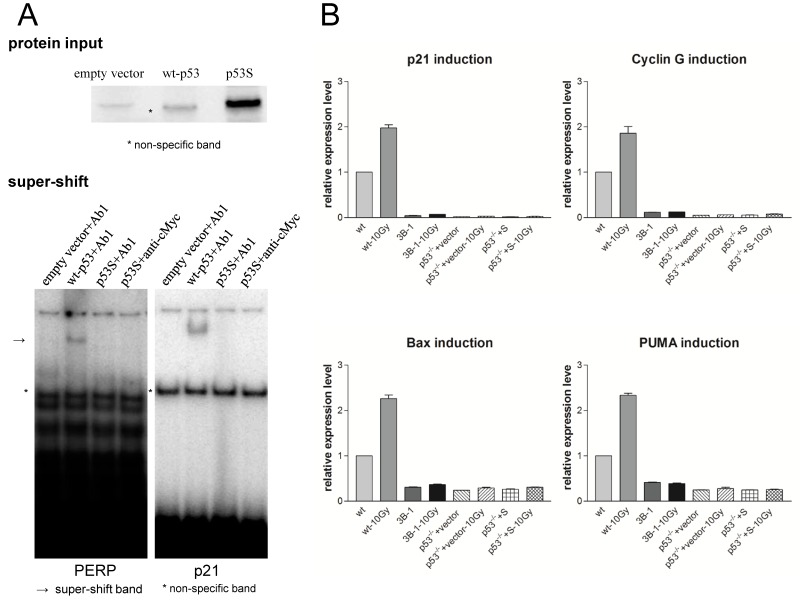
To test the DNA binding activity of p53S, gel shift assay was applied. Wild type p53 or p53S mutant proteins were obtained by in vitro translation to perform EMSA. The in vitro translated proteins were labeled with S35 and their input in EMSA assay was showed in Figure 1A, upper panel. Due to the nonspecific in vitro translated protein band showing in empty vector control (Figure 1A, upper panel), we performed super-shift assay to insure the specificity of EMSA. Specific antibodies to wild type p53 (Ab1) or p53S (Ab1 recognizing p53S and anti-cMyc recognizing Myc tag) were added to the EMSA reaction to perform super-shift assay. The results revealed that p53S lost its binding activity to both p21 and PERP promoters (Figure 1A, lower panel), suggest p53S lost the function of regulating its down stream targets.
Figure 1.
Mutant p53S lost DNA-binding and transactivation activity. A: Upper panel: The in vitro translated proteins labeled by 35S to show the input of protein amount in EMSA reactions. Empty vector was used as a control for in vitro translation. * Indicates the nonspecific protein band that appeared in the empty vector control. Lower panel: Super-shift gel images. To avoid nonspecific binding to the nonspecific band shown in the empty vector control, the super-shift in the EMSA was applied with an Ab1 antibody specific to p53 proteins and an anti-cMyc antibody specific to p53S. The p53-specific DNA-binding activity to either PERP (left) or p21 (right) promoters only occurred in the wild-type p53 control (indicated by an arrow). p53S did not show specific binding to either PERP or p21 promoter DNA tested using two specific antibodies, Ab1 and anti-cMyc. * Indicates the non-specific shifted band caused by the protein shown in the empty vector control. B: Real-time PCR analysis showed that the transactivation activity of p53S for p21, cyclin G1, puma and Bax was disrupted in response to irradiation. The relative expression levels of p21, cyclin G1, puma and Bax were measured 6 hrs after exposure to 10 Gy irradiation.
Furthermore, we introduced p53S into p53-/-MEFs and used this cell line (p53-/-+S) for testing p53S function in DNA damage response. The wild type MEFs and immortalized cell line 395-3B-1 were used as controls.
Since the control wild type MEF is not very sensitive to radiation, we use 10Gy ionized radiation (IR) to challenge the cells. After 6 hr of the IR treatment, real-time PCR was performed to analyze p53S function in cell cycle arrest or apoptotic pathways. The expression levels of the four p53 down-stream targets, p21, cyclin G1, Puma and Bax, were examined to determine the transactivation activity of p53S in response to IR. The results showed that unlike wild type p53, neither endogenous p53S from immortalized cell line 395-3B-1 nor ectopic expressed p53S in p53-/- MEF cells transactivated p21, cyclin G1, Puma and Bax in response to IR (Figure 1B).
Together these data suggested that p53S lost its transcriptional regulatory function in both cell cycle arrest and apoptotic pathways, thus lost the function of inhibiting tumorigenesis.
p53S mutant promoted cell growth and tumorigenesis in cooperating with H-RasV12
To investigate whether p53S per se is tumorigenic, using the non-tumorigenic p53-/- MEFs as the background, we introduced p53S into p53-/- MEF cells together with or without H-RasV12.
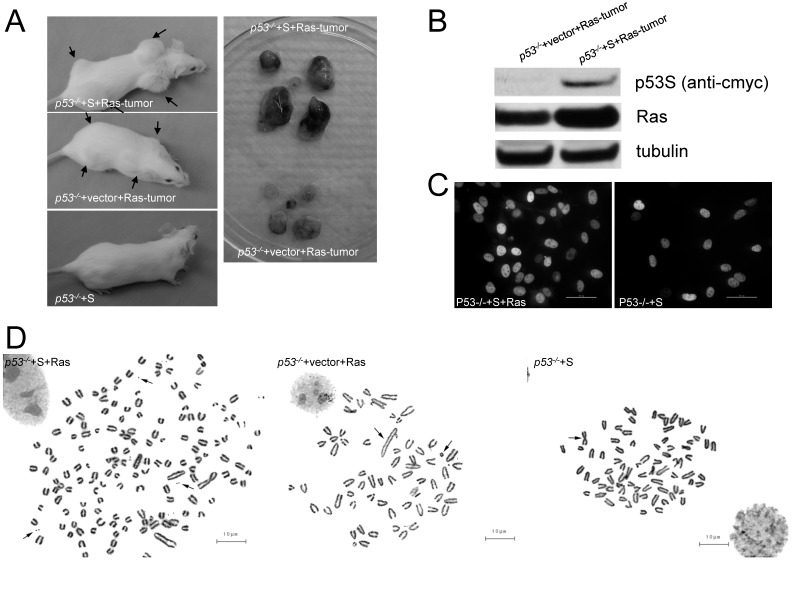
We found that, as previously reported 11, p53-/- MEFs did not form tumors by subcutaneously injecting into SCID mice (data not shown), while overexpressing H-RasV12 in p53-/- MEFs (p53-/-+Ras) resulted in tumorigenesis in 2-3 weeks (Figure 2A, left middle panel). To our surprise, introducing of p53S into p53-/- MEFs (p53-/-+S) did not generate tumors either (Figure 2A, left lower panel), suggesting that p53S per se is not enough to be tumorigenic. However, when we introduced p53S together with H-RasV12 in p53-/- MEFs (p53-/-+S+Ras), fast growing tumors were generated in 1-2 weeks (Figure 2A, left upper panel). The tumors harvested from p53-/-+S+Ras cells weighted 0.6500±0.1323g, which is significantly bigger than the tumors harvested from p53-/-+vector+Ras cells (0.1650±0.05315g). The P value is 0.014. These data strongly suggests a gain of function of p53S cooperating with H-RasV12 in tumorigenesis. Western blot revealed an elevated H-RasV12 expression in the SCID tumors derived from p53-/-+S+Ras cells comparing with tumors derived from p53-/-+vector+Ras cells (Figure 2B), which suggested a possible regulation of Ras expression by p53S. It has been shown that mutant p53 could alter its nuclear localization, and thus altered its function 12. To understand whether Ras could also regulate p53S localization, immunostaining of p53 was performed. The results showed that p53S localized in the nuclei of both p53-/-+S and p53-/-+S+Ras cells (Figure 2C), the coexpression of H-RasV12 did not change the nuclear localization of p53S.
Figure 2.
p53S mutant promoted tumorigenesis cooperating with H-RasV12. A: In vivo tumorigenesis test by subcutaneously injecting cells into SCID mice. Left upper panel: p53-/-+S+Ras cells formed fast growing tumors in 1-2 weeks. Right upper panel: p53-/-+S+Ras tumors dissected from a SCID mouse. Left lower panel: p53-/-+vector+Ras cells formed tumors in 2-3 weeks. Right lower panel: p53-/-+vector+Ras tumors dissected from a SCID mouse. B: Western blotting analysis of p53S and Ras in the tumors in panel A. Ras expression was elevated in the SCID tumors derived from p53-/-+S+Ras cells comparing with the tumors derived from p53-/-+vector+Ras cells. C: Immunostaining of exogenous p53S in p53-/- MEFs with or without H-RasV12. The bar length is 50µm. D: Cytogenetic analysis revealed multiple chromosome aberrations in p53-/-+S+Ras, p53-/-+vector+Ras tumor cells and p53-/-+S MEFs. Left, arrows pointed to double minutes; middle & right, arrows pointed to chromosome fusions. The bar length is 10µm.
It has been shown that p53 mutant gained the function of promoting both chromosome instability and cell survival 13. Furthermore, the amplification of oncogenes has been found correlated with chromosome instability, especially the increase of a specific structure: double minute chromosome 14, 15. Thus we performed cytogenetic analysis to investigate the effect of p53S on chromosome instability. Chromosome analysis revealed multiple chromosome aberrations in p53-/-+S+Ras, p53-/-+vector+Ras tumor cells and p53-/-+S MEFs in contrast with p53-/- MEFs and wild type MEFs (Figure 2D, left, arrows pointed to double minutes. middle & right, arrows pointed to chromosome fusions). Among these, dramatically increased double minutes were found in p53-/-+S+Ras cells, which again implied the oncogenic characteristics of p53S as well as the cross-talk between p53S and Ras.
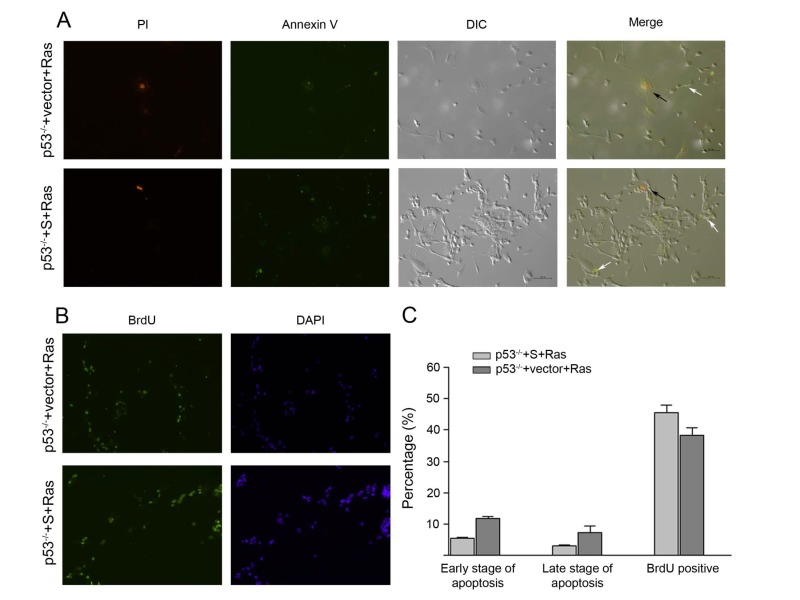
Furthermore, to understand the regulation of p53S-Ras in cell proliferation or apoptosis, the p53-/-+S+Ras, p53-/-+vector+Ras tumor cells were measured for their proliferation and apoptosis status. Annexin V staining showed that both tumor cells had small percentage of cells in apoptosis, and p53-/-+S+Ras had less apoptotic cells than p53-/-+vector+Ras tumor cells (Figure 3A, C). Brdu incorporation assay revealed that both p53-/-+S+Ras and p53-/-+vector+Ras had good percentage of proliferative cells, and p53-/-+S+Ras had a bit more proliferative cells than p53-/-+vector+Ras tumor cells (Figure 3B, C). These data suggested that p53S and H-RasV12 cooperated to increase cellular proliferation and decrease apoptosis, thus facilitated the tumor growth. These data also explained the fast growing tumors occurred in the SCID mouse injected with p53-/-+S+Ras cells.
Figure 3.
The apoptosis and proliferation rate in p53-/-+S+Ras and p53-/-+vector+Ras tumor cells. A: The cells were stained with Annexin-V and PI. The black arrows point to late stage apoptotic cells and the white arrows point to early stage apoptotic cells. DIC, differential interference contrast microscopic visual field. The bar length is 100µm. B: Cell proliferative status was measured by BrdU incorporation. C: The percentage of early stage apoptotic cells, late stage apoptotic cells and the BrdU-labeled proliferative cells.
The molecular basis of the cooperation between p53S and H-RasV12 in promoting tumorigenesis
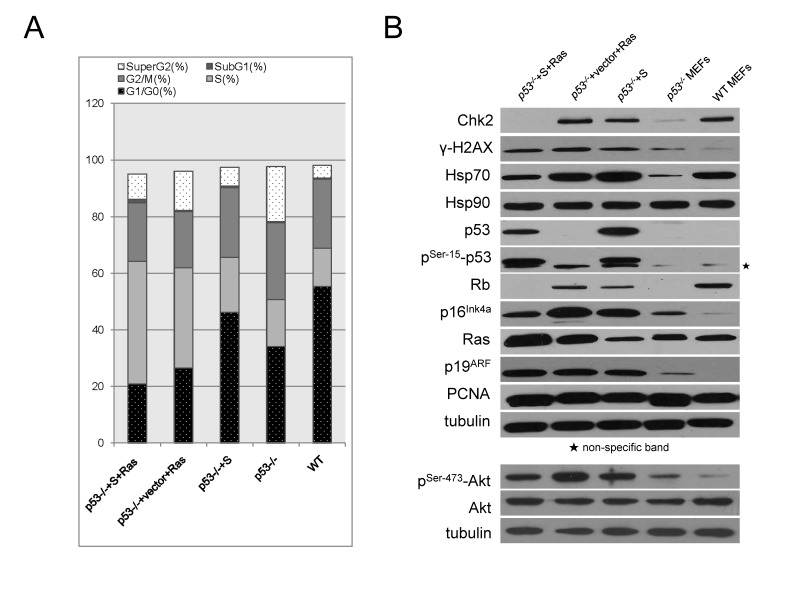
To understand the cell cycle regulation by the p53S or/and Ras, flow cytometry analysis was performed. The data showed that p53-/-+S MEFs had similar cell cycle pattern with wild type MEFs. However, both p53-/-+vector+Ras and p53-/-+S+Ras cells showed significant increase in S phase and decrease in G1/G0 phase comparing with p53-/- cells (Figure 4A). Furthermore, less p53-/-+S+Ras cells were located in G1/G0 phase and more in S phase comparing with p53-/-+vector+Ras cells (Figure 4A). This result further suggested that p53S per se is not advanced in promoting cell growth, however, cooperating with H-RasV12, they would promote cell growth greatly and facilitate tumorigenesis
Figure 4.
The regulation of cell cycle and stress responses by p53S or/and H-RasV12. A. Cell cycle analysis by FACS revealed growth advantage from cooperation of p53S and H-RasV12. The percentage of sub G1, G1/G0, S, G2/M and super G2 were shown in sections of stacked bars. B. Western blotting analysis of protein expression profile in five cell lines: p53-/- MEFs, p53-/-+S MEFs, p53-/-+vector+Ras tumor cells, p53-/-+S+Ras tumor cells and wild type MEFs (WT). Star showed the non-specific band for phosphorylated p53 blotting.
To further dissect the molecular basis of the cooperation between p53S and H-RasV12, the downstream proteins involved in gain of function of p53 mutants were analyzed in four MEF cell lines: p53-/- MEFs, p53-/-+S MEFs, p53-/-+vector+Ras tumor cells, p53-/-+S+Ras tumor cells. The wild type MEFs (WT) were used as the control.
We found that either introduction of p53S or H-RasV12 in p53-/- cells resulted in the up-regulation of Chk2, Hsp70 and γ-H2AX, indicated that cellular DNA damage responses (DDR) could be initiated by p53S or H-RasV12 solo overexpression (Figure 4B, lane p53-/-+S and lane p53-/-+vector+Ras, comparing with lane p53-/- ). It has been reported that oncogenic signals such as Ras could stimulate cellular DDR16. Here we showed that p53S could also stimulate DDR, suggesting that p53S gained oncogenic characteristics. At the same time, the cell cycle inhibitors p16Ink4a and Rb were also up-regulated by p53S or H-RasV12 solo overexpression (Figure 4B, lane p53-/-+S and lane p53-/-+vector+Ras, comparing with lane p53-/- ), suggesting the p16Ink4a -Rb tumor suppression axis remained intact and could be activated by either p53S or H-RasV12 independent of wild type p53 function.
Interestingly, co-expression of p53S and H-RasV12 partially eliminated the up-regulation of Chk2, Hsp70, γ-H2AX, p16Ink4a and Rb in response to solo expression of either p53S or H-RasV12 (Figure 4B, lane p53-/-+S+Ras, comparing with lane p53-/-+S and lane p53-/-+vector+Ras). These might contribute to the rapid cell cycle progression by co-expression of p53S or H-RasV12.
As we have observed in the SCID tumor tissue (Figure 2B), the co-expression of p53S and H-RasV12 in p53-/- background resulted in an enhanced expression of H-RasV12 comparing with overexpression of H-RasV12 itself, provided us a direct evidence for the cooperation between p53S and H-RasV12 (Figure 4B, Ras).
We also found that the expression of p19ARF was stimulated by either p53S or H-RasV12 overexpression, which is consistent with the stress response of p19ARF to oncogenic stimulations. However, under this genetic context (p53-/-+S or p53-/-+S+Ras), the only p53 proteins existed in the cells are mutant p53S. the stabilization mechanism of p53 by p19ARF acted on p53S mutant proteins, which further stabilized the protein level of p53S and facilitated the tumorigenesis (Figure 4B, p19 ARF).
To further understand the effect of p53S-Ras on cellular proliferation, we also checked the level of active Akt, which is known to be the down stream proliferation molecule of Ras pathway. The results revealed that total Akt level are not changed in the presence of p53S or H-RasV12 (Figure 4B, Akt). It is very interesting that p53S itself could increase the level of phosphorylated Akt: pSer-473-Akt (Figure 4B, p53-/- +S), although not as great as H-RasV12 (Figure 4B, p53-/- +Ras). Since the induction of active phosphorylated Akt is known as the downstream of Ras regulation, this data again suggested the Ras-like oncogenic property of p53S. However, when both p53S and H-RasV12 were present, the level of pSer-473-Akt decreased (Figure 4B, p53-/- +S+Ras). Giving the fact that p53-/-+S+Ras resulted in fast growing tumor (Figure 2A), we speculated that a balanced active Akt level (not too high, not too low) might be best for tumor growth.
Together these data suggested that the p53S cross-talk with H-RasV12, reduced the cellular stress response to oncogenic signals, and promoted the proliferative potential, which facilitate the cell growth and tumorigenesis.
Discussion
The current strategies in cancer treatment are trying to apply personalized treatment aiming different p53 mutants. Understanding of the loss and gain of function of different p53 mutants provides the basis for personalized treatments.
Seventy-three amino acid of p53 have been identified as hotspots for mutations found in human tumors by comparing the observed mutation distribution with a random multinomial distribution. N239 is one of the medium hot spots (p < 0.01) located at the protein surface involving in DNA interaction 2. A study in yeast classified N239S as one of the non-discriminant p53 mutants that are unable to discriminate between different p53 responsive elements 17. By assessment of its transactivation capacities in yeast assays 7, N239S was classified as a non-functional mutant. The computational analysis and yeast assays also suggested that N239S mutation located in the loop region of the DNA binding domain is a mild mutation comparing to those mutations located in the helix or β-sheet region of the DNA binding domain. Despite of these predictions, N239S somatic mutations were found in 32 cases of human tumors 1, suggesting its important function in human tumorigenesis.
In this study, we showed by EMSA that the p53S lost its DNA binding ability to promoters regulated by wild type p53. Real-time PCR analysis confirmed that p53S lost the function in regulating the transcription of p21 Cip1/Waf1, cyclin G, PUMA, and Bax in response to 10Gy irradiation. These results suggested that p53S lost the function of wild type p53 in regulating cell cycle arrest and apoptosis, thus lost the ability of tumor suppressing. Our data confirmed the loss of function of p53S in mammalian cells with p53 null background.
Previous study has showed that different p53 point mutations varied dramatically in their impacts on p53 function 4. The mutation occurred at amino acid N239 is a good example. A substitution mutation N239Y has been found to facilitate in restoring transcriptional activity in a subset of cancer mutants, including V143A, G245S and R249S 18. N239Y mutation could stabilize the p53 protein and rigidify the local structure without perturbing it 19, 20. Moreover, a very recent study showed that N239Y mutant was more active than wild-type p53 in terms of Bax transcription and apoptotic activity, which correlated very well with its structure stability 21. Thus it is very important to understand the gain of function of each mutant.
Our data revealed that p53S cross-talked with H-RasV12 and gain new functions in promoting tumorigenesis. It has been shown that activated Ras could induce the accumulation of p53, p21, p16Ink4a, and p14/p19ARF, decrease expression of cyclin A, and reduce kinase activity of CDK2 22, 23, thus initiate signaling transduction to cellular senescence. Further study revealed that Ras-initiated DNA replication forks could serve as effective DNA damage signals and induced DNA damage response, which resulted in cellular senescence 16. These data clearly showed that the oncogenic effect of Ras is highly dependent on the dysfunction of intact cellular DNA damage response pathway, especially, p53 status. Loss of p53 function removes the barrier for oncogenic growth signals, which passively cooperates with Ras. Recent researches found that p53 inactivation induced the RasV12 pathway through diverse mechanisms involving the p53 targets BTG2 and ATF3, which in turn facilitated the malignant transformed phenotype of cells and the accentuated expression of chemokines, CXCL1; interleukins, IL-1β; ECM-related genes, MMP3 24. These data revealed the cancer-related gene signatures, which underlies the cross-talk between the p53 tumor suppressor and Ras oncogene.
It has reported that concomitant endogenous expression of Trp53(R172H) and K-Ras(G12D) to the mouse pancreas caused cooperative development of invasive and widely metastatic carcinoma 25, suggested that p53 mutant could actively cooperate with Ras in tumorigenesis. The frequent occurrence of both p53 & Ras mutation in human tumors also suggest the active interaction between them.
Here we showed that the p53N239S actively cooperated with H-RasV12 and facilitated tumorigenesis in xenografted mice. We found that in molecular level, p53S could promoted the expression of H-RasV12. This effect suggested that p53S could directly or indirectly regulate the transcription of H-RasV12. We are working on CHIP assay to reveal possible targets for p53S binding, the results might reveal the mechanism underlying Ras regulation by p53S.
Furthermore, we found that in p53-/- background, introduction of p53S could stimulate the DDR response, as suggested by the up-relation of γ-H2AX, Chk2, p19, p16Ink4a, and HSP70, similar to the responses to H-RasV12. Either H-RasV12 or p53S could stimulate the response of p16Ink4a-Rb pathway, but together they deregulated these responses, thus could reduce the inhibition of cell cycle and promote tumorigenesis. These data revealed the Ras-like oncogenic properties of p53S, it is reasonable to speculate that p53S could also stimulate the growth signals in cells, thus promoting tumorigenesis. Supporting this, we found that either p53S or H-RasV12 could induce the level of phosphorylated Akt (pSer-473-Akt), which is an important down stream proliferative signal for Ras pathway 26. Interestingly, when both p53S and H-RasV12 were present, the level of pSer-473-Akt decreased. Giving the fact that p53-/-+S+Ras resulted in fast growing tumor, we speculated that a balanced active Akt level (not too high, not too low) might be best for tumor growth. It has been found that active Akt could increase the cellular ROS level thus promoting cellular senescence in wild type MEFs 27. This finding suggests that active Akt plays an important role in homeostatic regulation of cell viability 28. Our data is consistent with this hypothesis.
Together these data explained the molecular basis for the fast tumorigenesis in the cells co-expressing p53S and H-RasV12 and strongly indicated the cross-talk between p53S and H-RasV12.
It is worth to notice that p53N239S has also been found in rheumatoid arthritis synovial tissue and synoviocytes, possibly involved in positively interfering immune system by regulating the IL-6 production, suggesting its role in RA pathogenesis 10.
To further understand the function of p53S, knock-in p53N236S mouse model will be utilized to clarify its role in aging-related mesenchymal tumorigenesis, as well as RA.
Conclusions
Our data confirmed the loss of function of p53S mutant in mammalian cells. More importantly, these data revealed the Ras-like oncogenic properties of p53S and suggested that p53S cross-talked with H-RasV12 and reduced the cellular stress response to oncogenic signals, which facilitated the cell growth and tumorigenesis. This study revealed an important aspect of gain of function for p53 mutant, therefore might shed light on the clinical strategy in targeting p53 mutant.
Materials and Methods
Cell lines and constructs
All cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (Hyclone, CA) in 3% oxygen and 5% CO2 incubator at 37°C.
p53S cDNA fragment was cloned from immortalized cell line 395-3B-1 by RT-PCR and put into retroviral vector PQCXIP and PQCXIH (Clontech, CA), tagged with cMyc peptide. The sequence was confirmed by sequencing and the expression of the constructed p53S-cmyc was confirmed by Western blotting.
Either empty vector PQCXIH or PQCXIH-p53S construct or pBabe-H-RasV12 construct (as described previously 22) were introduced into p53-/- MEF. After 1-2 months antibiotics selection, cell colonies were picked up, expression of p53S or H-RasV12 was analyzed by both Western blotting and immunostaining. By this way, p53-/- MEFs stably introduced both p53S and H-RasV12 (p53-/-+S+Ras MEFs), or H-RasV12 and empty PQCXIH vector (p53-/-+vector+Ras MEFs), or p53S only ( p53-/-+S MEFs ) were established.
To detect stress response, cells were treated with 10Gy ionized irradiation. Six hours after treatment, cells were harvested for further experiments.
Antibodies
Antibodies used for Western blotting were anti-Chk2 (1:500, BD Transduction Laboratories, CA), anti-Rb (1:500, BD Transduction Laboratories, CA), anti-Ras (1:500, BD Transduction Laboratories, CA), anti-cMyc (9E10) (1:500, Santa Cruz, CA), anti- p16Ink4a (M-156) (1:500, Santa Cruz, CA), anti- p21 (F-5) (1:500, Santa Cruz, CA), anti-p53 Ab1(clone PAb240) (1:250, Neomarker, CA), anti-phospho-p53 (Ser15) (1:500, Cell Signaling, MA), anti-Hsp70 (1:1000, Stressgen, Canada), anti-Hsp90 (1:1000, Stressgen, Canada), anti-PCNA (1:1000, Upstate, NY), anti-p19 (1:1250, Upstate, NY), anti-γ-H2AX (1:1000, Upstate, NY), anti-γ-tubulin (1:5000, Upstate, NY).
Elecrophoretic mobility shift assay (EMSA)
The ability of p53S to bind DNA was tested using EMSA modified from a previously described protocol 29. Briefly, pcDNA3 empty vector, pcDNA3-p53 or pcDNA3-p53S-myc were used for in vitro translation (TNT® T7 Coupled Reticulocyte Lysate System, Promega, WI) to provide wild-type or mutant p53S protein for use in the EMSA reaction. The protein input level was checked by 35S labeling. p21 and PERP promoter oligonucleotides (sequences are as previously described 30) were synthesized and labeled with 32P-γ-ATP. The EMSA reaction was performed in a buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl, 0.5 mM EDTA, 1 mM DTT, 10% glycerol, and 0.45 μg/μl salmon sperm DNA. EMSA products were separated on a 4-20% 1x TBE gel (Invitrogen, CA). The super-shift was done by adding either p53 antibody Ab1 (PAb421, Calbiochem, NJ) or anti-cMyc antibody (Santa Cruz, CA).
Real-time RT-PCR
RNA was extracted from cells 6 hrs after exposure to 10 Gy irradiation and was reverse transcribed to cDNA. Real-time PCR was performed using a SYBR-Green PCR master mix according to the manufacturer's instructions (Applied Biosystems, CA). The primers used were as follows: p21 (Forward primer: 5' CCA GGC CAA GAT GGT GTC TT 3', Reverse primer: 5' TGA GAA AGG ATC AGC CAT TGC 3'), Cyclin G (Forward primer: 5' CCG GTC CGT GAC GCC 3', Reverse primer: 5' AGT TCA ACA ATC CGA AAA GCT GA 3'), Bax (Forward primer: 5' GTT TCA TCC AGG ATC GAG CAG 3', Reverse primer: 5' CCC CAG TTG AAG TTG CCA TC 3'), puma (Forward primer: 5´ GTA CGG GCG GCG GAG ACG AG 3', Reverse primer: 5´ GCA CCT AGT TGG GCT CCA TTT CTG 3'), and GAPDH (Forward primer: 5' TCA CCA CCA TGG AGA AGG C 3', Reverse primer: 5' GCT AAG CAG TTG GTG GTG CA 3') 31-34.
Injection of cells into SCID mice and tumor cells harvest
1x 106 cells were injected subcutaneously into each site of SCID mice. When the size of the largest tumor reached 1 cm, the mice were sacrificed and the tumors were collected and digested in a tumor digesting cocktail (4 mg/ml collagenase D and 4 mg/ml dispase II). Isolated tumor cells were plated and cultured in DMEM supplemented with 10% fetal bovine serum. All mouse procedures were performed with the approval of the Animal Care and Use Committee of the Kunming University of Science & Technology (approval ID: M2009-011).
Cytogenetics analysis
Metaphase chromosomes from cells were prepared as described 35 and subjected to Giemsa staining. About 30 metaphases from each sample were analyzed in detail.
Cell cycle analysis
Cell cycle analysis was done by flow cytometry (BD, FACS Vantage SE). Data were analyzed by Flowjo. Percentages of cells in G1, S, and G2 were determined using the Dean-Jett-Fox algorithm.
Annexin V staining and BrdU incorporation
Annexin V staining was performed with the Annexin V-FLUOS staining kit (Roche, Germany) to detect apoptotic cells. The stained cells were analyzed by fluorescence microscopy. The early stage apoptotic cells only showed Annexin V positive staining, and the late stage apoptotic or dead cells showed both Annexin V and propidium iodide positive staining. The apoptotic cells are counted and the apoptosis rate were calculated. BrdU incorporation was performed with the In Situ cell proliferation kit FLUOS (Roche, Germany) according to the manufacturer's instructions. Briefly, cells were labeled with BrdU (final concentration: 10 μM) for 1 hr and immunostained with anti-BrdU-FLUOS antibody. The BrdU incorporation rate was measured by counting cells labeled with anti-BrdU antibody.
Acknowledgments
This work was supported by NSFC grants (30771194, 30970598, 31170735) to Y.L, The New Century Excellent Scholarship to Y.L, NSFC grant (30960152) to W.T. Yunnan Educational Science Foundation (09J0001) to S. J. We thank Dr. Sandy Chang and Dr. Wilfredo Cosme-Blanco for kindly providing the p53-/- MEFs.
Biographies
Shuting Jia is a PhD candidate in Kunming University of Science & Technology.
Lanjun Zhao is also a PhD candidate in Kunming University of Science & Technology.
Dr. Wenru Tang is an associate professor in Molecular Genetics in Kunming University of Science & Technology, and has been working in epigenetics and gene expression regulation for 8 years.
Dr. Ying Luo was an instructor in MD Anderson Cancer Center, and is now a full professor in Molecular Genetics in Kunming University of Science & Technology. Dr. Luo has been working on genetic signaling pathway for DNA damage response and carcinogenesis for 15 years.
References
- 1.Petitjean A. et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 2.Walker D.R. et al. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18(1):211–8. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 3.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9(10):701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 4.Joerger A.C, Ang H.C, Fersht A.R. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci U S A. 2006;103(41):15056–61. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laud P.R. et al. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19(21):2560–70. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin A.C. et al. Integrating mutation data and structural analysis of the TP53 tumor-suppressor protein. Hum Mutat. 2002;19(2):149–64. doi: 10.1002/humu.10032. [DOI] [PubMed] [Google Scholar]
- 7.Kato S. et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100(14):8424–9. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathe E. et al. Predicting the transactivation activity of p53 missense mutants using a four-body potential score derived from Delaunay tessellations. Hum Mutat. 2006;27(2):163–72. doi: 10.1002/humu.20284. [DOI] [PubMed] [Google Scholar]
- 9.Firestein G.S. et al. Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proc Natl Acad Sci U S A. 1997;94(20):10895–900. doi: 10.1073/pnas.94.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Z. et al. Dominant-negative p53 mutations in rheumatoid arthritis. Arthritis Rheum. 1999;42(6):1088–92. doi: 10.1002/1529-0131(199906)42:6<1088::AID-ANR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 11.Jacks T. et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4(1):1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 12.Olivier M. et al. Recent advances in p53 research: an interdisciplinary perspective. Cancer Gene Ther. 2009;16(1):1–12. doi: 10.1038/cgt.2008.69. [DOI] [PubMed] [Google Scholar]
- 13.Murphy K.L, Dennis A.P, Rosen J.M. A gain of function p53 mutant promotes both genomic instability and cell survival in a novel p53-null mammary epithelial cell model. FASEB J. 2000;14(14):2291–302. doi: 10.1096/fj.00-0128com. [DOI] [PubMed] [Google Scholar]
- 14.Trent J. et al. Evidence for rearrangement, amplification, and expression of c-myc in a human glioblastoma. Proc Natl Acad Sci U S A. 1986;83(2):470–3. doi: 10.1073/pnas.83.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka K. et al. Amplification of c- oncogene MYC and point mutation of N-RAS oncogene point mutation in acute myelocytic leukemias with double minute chromosomes. Leukemia. 1993;7(3):469–71. [PubMed] [Google Scholar]
- 16.Di Micco R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 17.Campomenosi P. et al. p53 mutants can often transactivate promoters containing a p21 but not Bax or PIG3 responsive elements. Oncogene. 2001;20(27):3573–9. doi: 10.1038/sj.onc.1204468. [DOI] [PubMed] [Google Scholar]
- 18.Brachmann R.K. et al. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J. 1998;17(7):1847–59. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolova P.V. et al. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc Natl Acad Sci U S A. 1998;95(25):14675–80. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolova P.V. et al. Mechanism of rescue of common p53 cancer mutations by second-site suppressor mutations. EMBO J. 2000;19(3):370–8. doi: 10.1093/emboj/19.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoo K.H, Mayer S, Fersht A.R. Effects of stability on the biological function of p53. J Biol Chem. 2009;284(45):30974–80. doi: 10.1074/jbc.M109.033183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serrano M. et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 23.Tremain R. et al. Defects in TGF-beta signaling overcome senescence of mouse keratinocytes expressing v-Ha-ras. Oncogene. 2000;19(13):1698–709. doi: 10.1038/sj.onc.1203471. [DOI] [PubMed] [Google Scholar]
- 24.Buganim Y. et al. p53 Regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res. 2010;70(6):2274–84. doi: 10.1158/0008-5472.CAN-09-2661. [DOI] [PubMed] [Google Scholar]
- 25.Hingorani S.R. et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 26.Vivanco I, Sawyers C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 27.Nogueira V. et al. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell. 2008;14(6):458–70. doi: 10.1016/j.ccr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolado I, Nebreda A.R. AKT and oxidative stress team up to kill cancer cells. Cancer Cell. 2008;14(6):427–9. doi: 10.1016/j.ccr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Johnson T.M. et al. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet. 2005;37(2):145–52. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- 30.Reczek E.E. et al. Multiple response elements and differential p53 binding control Perp expression during apoptosis. Mol Cancer Res. 2003;1(14):1048–57. [PubMed] [Google Scholar]
- 31.Martinez-Gac L. et al. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol Cell Biol. 2004;24(5):2181–9. doi: 10.1128/MCB.24.5.2181-2189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwakuma T. et al. Mutation at p53 serine 389 does not rescue the embryonic lethality in mdm2 or mdm4 null mice. Oncogene. 2004;23(46):7644–50. doi: 10.1038/sj.onc.1207793. [DOI] [PubMed] [Google Scholar]
- 33.Bruins W. et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol. 2004;24(20):8884–94. doi: 10.1128/MCB.24.20.8884-8894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu W.S. et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123(4):641–53. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 35.Chang S. et al. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17(1):88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]