SUMMARY
Personalized medicine is expected to benefit from combining genomic information with regular monitoring of physiological states by multiple high-throughput methods. Here we present an integrative Personal Omics Profile (iPOP), an analysis that combines genomic, transcriptomic, proteomic, metabolomic, and autoantibody profiles from a single individual over a 14-month period. Our iPOP analysis revealed various medical risks, including Type II diabetes. It also uncovered extensive, dynamic changes in diverse molecular components and biological pathways across healthy and diseased conditions. Extremely high coverage genomic and transcriptomic data, which provide the basis of our iPOP, discovered extensive heteroallelic changes during healthy and diseased states and an unexpected RNA editing mechanism. This study demonstrates that longitudinal iPOP can be used to interpret healthy and disease states by connecting genomic information with additional dynamic omics activity.
INTRODUCTION
Personalized medicine aims to assess medical risks, monitor, diagnose and treat patients according to their specific genetic composition and molecular phenotype. The advent of genome sequencing and the analysis of disease states has proven to be powerful (Cancer Genome Atlas Research Network, 2011). However, its implementation for the analysis of otherwise healthy individuals for estimation of disease risk and medical interpretation is less clear. Much of the genome difficult is to interpret and many complex diseases, such as diabetes, neurological disorders and cancer, likely involve a large number of different genes and biological pathways (Ashley et al., 2010; Grayson et al., 2011; Li et al., 2011), as well as environmental contributors which can be difficult to assess. As such, the combination of genomic information along with a detailed molecular analysis of samples will be important for predicting, diagnosing and treating diseases as well as for understanding the onset, progression, and prevalence of disease states (Snyder et al., 2009).
Presently, healthy and diseased states are typically followed using a limited number of assays that analyze a small number of markers of distinct types. With the advancement of many new technologies, it is now possible to analyze upwards of 105 molecular constituents. For example, DNA microarrays have allowed the subcategorization of lymphomas and gliomas (Mischel et al., 2003), and RNA sequencing has identified breast cancer transcript isoforms (Li et al., 2011; van der Werf et al., 2007; Wu et al., 2010); (Lapuk et al., 2010). Although transcriptome and RNA splicing profiling are powerful and convenient, they provide a partial portrait of an organism's physiological state. Transcriptomic data, when combined with genomic, proteomic, and metabolomic data are expected to provide a much deeper understanding of normal and diseased states (Snyder et al., 2010). To date, comprehensive integrative omics profiling have been limited and have not been applied to the analysis of generally healthy individuals.
To obtain a better understanding of 1) how to generate an integrative Personal Omics Profile (iPOP) and examine as many biological components as possible, 2) how these components change during healthy and disease states and 3) how this information can be combined with genomic information to estimate disease risk and gain new insights into disease states, we performed extensive omics profiling of blood components from a generally healthy individual over a 14-month period (24 months total when including time points with other molecular analyses). We determined the whole genome sequence (WGS) of the subject, and together with transcriptomic, proteomic, metabolomic, and autoantibody profiles, used this information to generate an iPOP. We analyzed the iPOP of individual over the course of healthy states and two viral infections (Figure 1A). Our results indicate that disease risk can be estimated by a whole genome sequence, and by regularly monitoring health states with iPOP disease onset may also be observed. The wealth of information provided by detailed longitudinal iPOP revealed unexpected molecular complexity, which exhibited dynamic changes during healthy and diseased states, and provided insight into multiple biological processes. Detailed omics profiling coupled with genome sequencing can provide molecular and physiological information of medical significance. This approach can be generalized for personalized health monitoring and medicine.
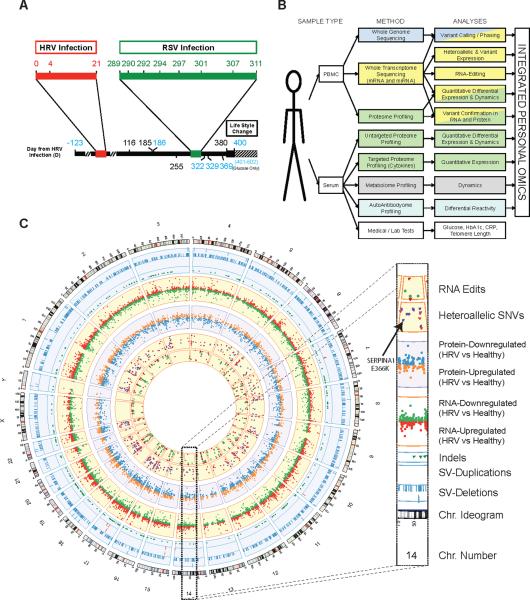
Figure 1. Summary of study.
(A) Time course summary. The subject was monitored for a total of 523 days, during which there were two infections (red bar, HRV; green bar, RSV). The black bar indicates the period when the subject 1) increased exercise; 2) ingested 81 mg of Acetylsalicylic Acid and Ibuprofen tablets each day (the latter only during the first 6 weeks of this period); and 3) substantially reduced sugar intake. Blue numbers indicated fasted time points. (B) iPOP experimental design indicating the tissues analyses involved in this study. (C) Circos (Krzywinski et al., 2009) plot summarizing iPOP. From outer to inner rings: chromosome ideogram; genomic data (pale blue ring) - structural variants > 50 bp [deletions (blue tiles), duplications (red tiles)], indels (green triangles); transcriptomic data (yellow ring) – expression ratio of HRV infection to healthy states; proteomic data (light purple ring) - ratio of protein levels during HRV infection to healthy states; transcriptomic data (yellow ring) – differential heteroallelic expression ratio of alternative allele to reference allele for missense and synonymous variants (purple dots) and candidate RNA missense and synonymous edits (red triangles, purple dots, orange triangles and green triangles, respectively).
RESULTS
Overview of Personal Omics Profiling
Our overall iPOP strategy was to: 1) determine the genome sequence at high accuracy and evaluate disease risks, 2) monitor omics components over time and integrate the relevant omics information to assess the variation of physiological states, and 3) examine in detail the expression of personal variants at the level of RNA protein to study molecular complexity and dynamic changes in disease states.
We performed iPOP on blood components [Peripheral Blood Mononuclear Cells (PBMCs), plasma and sera which are highly accessible] from a 54 year-old male volunteer over the course of 14 months. Samples used for iPOP were taken over an interval of 401 days (Days 0–400). In addition, a complete medical exam plus laboratory and additional tests were performed before the study officially launched (Day -123) and blood glucose was sampled multiple times after the comprehensive omics profiling (Days 401–602) (Figure 1A). Extensive sampling was performed during two viral infections that occurred during this period: a human rhinovirus (HRV) infection beginning on Day 0 and a respiratory syncytial virus (RSV) infection starting on Day 289. A total of 20 time points were extensively analyzed and a summary of the time course is indicated in Figure 1A. The different types of analyses performed are summarized in Figure 1B–C. These analyses, performed on PBMCs and/or serum components, included WGS, complete transcriptome analysis (providing information about the abundance of alternative spliced isoforms, heteroalleleic expression and RNA edits, as well as expression of miRNAs at selected time points), proteomic and metabolomic analyses, and autoantibody profiles. An integrative analysis of these data highlights dynamic omics changes and provides rich information about healthy and diseased phenotypes.
Whole Genome Sequencing
We first generated a high quality genome sequence of this individual using a variety of different technologies. Genomic DNA was subjected to deep WGS using technologies from Complete Genomics (CG, 35nt paired end) and Illumina (100nt paired end) at 150 and 120 fold total coverage, respectively, exome sequencing using three different technologies to 80–100-fold average coverage (see Extended Experimental Procedures) and analysis using genotyping arrays.
The vast majority of human sequences (91%) mapped to the hg19 (GRCh37) genome. However, because of the depth of our sequencing, we were able to identify sequences not present in the reference sequence. Assembly of the unmapped Illumina sequencing reads (60,434,531, 9% of the total) resulted in 1,425 (of 29,751) contigs (spanning 26Mb) overlapping with RefSeq gene sequences that were not annotated in the hg19 reference genome. The remaining sequences appeared unique, including 2,919 exons expressed in the RNA-Seq data (eg. Figure S1A). These results confirm that a large number of undocumented genetic regions exist in individual human genome sequences and can be identified by very deep sequencing and de novo assembly (Li et al., 2010).
Our analysis detected single nucleotide variants (SNVs), small insertions and deletions (indels) and structural variants (SVs; large insertions, deletions and inversions relative to hg19), (summarized in Table 1A and Experimental Procedures). 134,341 (4.1%) high confidence SNVs, are not present in dbSNP or the 1000G, (1000 Genomes Project Consortium, 2010) indicating that they are very rare or private to the subject. Only 302 high confidence indels reside within RefSeq protein coding exons and exhibit enrichments in multiples of 3 nucleotides (p<0.0001). In addition to indels, 2,566 high confidence SVs were identified (Experimental Procedures and Table S1) and 8,646 mobile element insertions were detected (Stewart et al., 2011).
Table 1A.
Summary and breakdown of DNA variants.
| Type | Total Variants | Total High Confidence | Heterozygous High Confidence | Homozygous High Confidence |
|---|---|---|---|---|
| TOTAL SNVs: | 3,739,701 | 3,301,521 | 1,971,629 | 1,329,892 |
| Total gene-associated SNVs: | 1,312,780 | 1,183,847 | 717,485 | 466,362 |
| TOTAL Coding/UTR: | 49,017 | 44542 | 27,383 | 17,159 |
| missense | 10,592 | 9,683 | 5,944 | 3,739 |
| nonsense | 83 | 73 | 49 | 24 |
| synonymous | 11,459 | 10,864 | 6,747 | 4,117 |
| 5'UTR | 4,085 | 2,978 | 1,802 | 1,176 |
| 3'UTR | 22,798 | 20,944 | 12,841 | 8,103 |
| intron | 1,263,763 | 1,139,305 | 690,102 | 449,203 |
| Ts/Tv | - | 2.14 | - | - |
| dbSNP | 3,493,748 | 3,167,180 | - | - |
| candidate private SNV | 245,953 | 134,341 | - | - |
| INDELS(−107~+36bp): | 1,022,901 | 216,776 | - | - |
| coding | 3,263 | 302 | ||
| Structural Variants (>50bp): | 44,781 | 2,566 | - | - |
| In 1000G project | 4,434 | 1,967 | - | - |
High confidence values are across multiple platforms (Illumina and CG) and/or Exome and RNA-Seq data. Annotations were based from variant call formatted (vcf) files for heterozygous calls: 0/1=reference (ref)/alternative (alt), 1/2=alt/alt and homozygous calls 1/1=alt/alt, 1/. (alt/alt-incomplete call). Polyphen-2 was used to identify the location of the SNVs.
Analysis of the subject's mother's genome and imputation allowed a maternal/paternal chromosomal phasing of 92.5% of the subject's SNVs and indels (see Figure S2 and Extended Experimental Procedures). 139 phased genes contain predicted compound heterozygous deleterious and/or nonsense mutations. See Table S2 for details, Data S1. Phasing enabled the assembly of a personal genome sequence of very high confidence [cf. (Rozowsky et al., 2011)],
WGS-based Disease Risk Evaluation
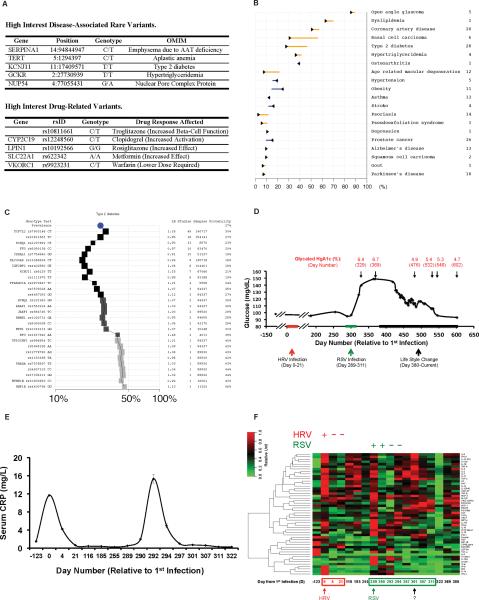
We identified variants likely to be associated with increased susceptibility to disease (Dewey et al., 2011). The list of high confidence SNVs and indels was analyzed for rare alleles (<5% of the major allele frequency in Europeans) and for changes in genes with known Mendelian disease phenotypes (data summarized in Table 1B), revealing that 51 and 4 of the rare coding SNV and indels, respectively, in genes present in OMIM are predicted to lead to loss-of-function (Table S3A). This list of genes were further examined for medical relevance (Table S3A; example alleles are summarized in Figure 2A), and 11 were validated by Sanger sequencing. High interest genes include a) a mutation (E366K) in the SERPINA1 gene previously known in the subject, b) a damaging mutation in TERT, associated with acquired aplastic anemia (Yamaguchi et al., 2005), c) variants associated with hypertriglyceridemia and diabetes, such as GCKR [homozygous] (Vaxillaire et al., 2008), and KCNJ11 [homozygous] (Hani et al., 1998) and TCF7 [heterozygous] (Erlich et al., 2009)
Table IB.
Summary of disease-related rare variants.
| Category | Count |
|---|---|
| Total Rare SNVs | 289,989 |
| Coding | 2,546 |
| Missense | 1,320 |
| Synonymous | 1,214 |
| Nonsense | 11 |
| Nonstop | 1 |
| Damaging or Possibly Damaging | 233 |
| Putative Loss-of-Function SNVs* | 51 |
|
| |
| Total Rare Indels | 51,248 |
| Coding Indels | 61 |
| Frameshift Indels | 27 |
| miRNA Indels | 3 |
| miRNA Target Sequence Indels | 5 |
| Putative Loss-of-Function Indels* | 4 |
In curated Mendelian disease genes.
Figure 2. Medical findings.
(A) High interest disease and drug related variants in the subject's genome. (B) RiskGraph of the top 20 diseases with the highest post-test probabilities. For each disease, the arrow represents the pretest probability according to the subject's age, gender, and ethnicity. The line represents the post-test probability after incorporating the subject's genome sequence. Listed to the right are the numbers of independent disease-associated SNVs used to calculate the subject's post-test probability. (C) RiskOGram of Type 2 Diabetes. The RiskOGram illustrates how the subject's post-test probability of T2D was calculated using 28 independent SNVs. The middle graph displays the post-test probability. The left side shows the associated genes, SNVs, and the subject's genotypes. The right side shows the likelihood ratio (LR), number of studies, cohort sizes, and the post-test probability. (D) Blood glucose trend. Measurements were taken from samples analyzed at either non-fasted or fasted states; the non-fasted states (all but Days 186, 322, 329 and 369 and after Day 400) were at a fixed time after a constant meal and differed from the fasted states. Data was presented as moving average with a window of 15 days. Red and green arrows and bars indicate the times of the HRV and RSV infections, respectively. Black arrows and bars indicate the period with life style changes. (E) C-Reactive Protein trendline. (F) Serum cytokine profiles. Red box and day number, HRV infection; green box and day number, RSV infection; question mark, elevated cytokine levels indicating an unknown event at Day 301. Red is increased cytokine levels.
Genetic disease risks were also assessed by the RiskOGram algorithm, which integrates information from multiple alleles associated with disease risk (Ashley et al., 2010) (Figure 2B). This analysis revealed a modest elevated risk for coronary artery disease and significantly elevated risk levels of basal cell carcinoma (Figure 2B), hypertriglyceridemia, and Type 2 Diabetes (T2D) (Figure 2B–C).
In addition to coding region variants we also analyzed genomic variants that may affect regulatory elements (Transcription Factors, TF), which had not been attempted previously (Data S2). 14,922 (of 234,980) SNVs lie in the motifs of 36 TFs known to be associated with the binding data (see Experimental Procedures), indicating that these are likely having a direct effect on TF binding. Comparison of SNPs that alter binding patterns of NFκB and PolII sites (Kasowski et al., 2010), also revealed a number of other interesting regulatory variants, some of which are associated with human disease [eg. EDIL, (Sun et al., 2010), Figure S1B].
Medical Phenotypes Monitoring
Based on the above analysis of medically relevant variants and the RiskOGram, we monitored markers associated with high-risk disease phenotypes and performed additional medically relevant assays. These studies indicated that:
-
a)
Monitoring of glucose levels and HbA1c revealed the onset of T2D as diagnosed by the subject's physician [Day 369, Figures 2A,C (Pruitt et al., 2009)]. The subject lacked many known factors associated with diabetes (nonsmoker; BMI = 23.9 and 21.7 on Day 0 and Day 511 respectively) and glucose levels were normal for the first part of the study. However, glucose levels elevated shortly after the RSV infection (Day 301) extending for several months (Figure 2D). High levels of glucose were further confirmed using Glycated HbA1c measurements at two time points (Days 329, 369) during this period (6.4 and 6.7%, respectively). After a dramatic change in diet, exercise and ingestion of low doses of acetylsalicylic acid a gradual decrease in glucose (to approximately 93 mg/dL at Day 602) and HbA1c levels to 4.7% was observed. Insulin resistance was not evident at Day 322. The patient was negative for anti-GAD and anti-islet antibodies, and insulin levels correlated well with the fasted and non-fasted states (Figure S3C), consistent with T2D. These results indicate that a genome sequence can be used to estimate disease risk in a healthy individual, and by monitoring traits associated with that disease, disease markers can be detected and the phenotype treated.
-
b)
Although the subject contained a TERT mutation previously associated with aplastic anemia, measurements of telomere length suggested little or no decrease in telomere length and modest increase in numbers of cells with short telomeres relative to age-matched controls (Figure S3A–B). Importantly, the patient and his 83 year-old mother share the same mutation but neither exhibit symptoms of aplastic anemia, a disease previously linked to TERT mutations/dysfunction (Yamaguchi et al., 2005).
-
c)
Consistent with the elevated hyperlipidemia risk, high trigyclerides were found to be high (321 mg/dL) at the onset of the study. These levels were reduced to (81–116 mg/dL) after regularly taking simvastatin.
-
d)
We also examined the variants for their potential effects on drug response (see Extended Experimental Procedures; Figure 2A and Table S3B), including LPIN1 and SLC22A1 genotypes associated with favorable (glucose lowering) responses to two diabetic drugs, rosiglitazone and metformin, respectively.
-
e)
We followed the levels of 51 cytokines along with the C-Reactive Protein (CRP) using ELISA assays, which revealed strong induction of proinflammatory cytokines and CRP during infection (Figure 2E and 2F). We also observed a spike of many cytokines at Day 12 after the RSV infection (Day 301 overall). These data define the physiological states and serve as a valuable reference for the omic profiles integrated into a longitudinal map of health and disease states described in the next sections.
-
f)
We also profiled autoantibodies during the HRV infection. Plasma and serum samples from the first four time points (Days −123, 0, 4 and 21), along with plasma samples from 34 healthy controls were used to probe a protein microarray containing 9,483 unique human proteins spotted in duplicate. 884 antigens with increased reactivity (Data S3) in the candidate plasma relative to healthy controls were found (p< 0.01, Benjamini-Hochberg p<0.01). Among the potentially interesting results was reactivity with DOK6, an insulin receptor binding protein (NCBI Gene database). These results demonstrate that autoantibodies can be monitored and that information relevant to disease conditions can be found.
Dynamic Omics Analysis: Integrative Omics Profiling of Molecular Responses
We profiled the levels of transcripts, protein, and metabolites across the HSV and RSV infections and healthy states using a variety of approaches. RNA-Seq of 20 time points generated over 2.67 billion uniquely mapped 101b paired-end reads (123 million reads average per time point) and allowed for an analysis of the molecular complexity of the transcriptome in normal cells (PBMCs) at an unprecedented level. The relative levels of 6,280 proteins were also measured at 14 time points through differentially labeling samples using isobaric tandem mass tags (TMT), followed by liquid chromatography and mass spectrometry (LC-MS/MS) (Cox and Mann, 2010; Theodoridis et al., 2011). 3,731 PBMC proteins could be consistently monitored across most of the 14 time points (see Figure S4A, Data S4). In addition, 6,862 and 4,228 metabolite peaks were identified for the HRV and RSV infection and a total of 1,020 metabolites were tracked for both infections (see Figure S5 and Data S5(3)). Finally, as described below we also analyzed miRNAs during the HRV infection.
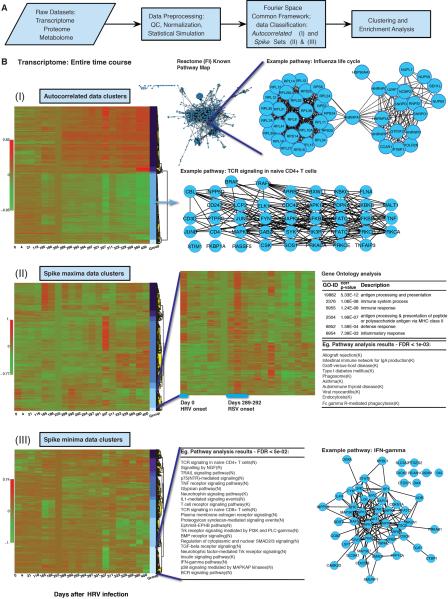
This wealth of omics information allowed us to examine detailed dynamic trends related directly to the physiological states of the individual and revealed enormous changes in biological processes that occurred during healthy and disease states. For each profile (transcriptome, proteome, metabolome), we systematically searched for two types of non-random patterns: 1) correlated patterns over time, and 2) single unusual events i.e. spikes that may occur at any given time-point defined as statistically significantly high or low signal instances compared to what would be expected by chance. To perform this analysis, we developed a novel general scheme for integrated analysis of data (see Figure S6 & Extended Experimental Procedures for further details). We used a Fourier spectral analysis approach that both normalizes the various omics data on equal basis for identifying the common trends and features, and, also accounts for dataset variability, uneven sampling and data gaps, in order to detect real-time changes in any kind of omics activity at the differential time points (See Supplementary Information). Autocorrelations were calculated to assess non-randomness of the time-series (p<0.05 one-tailed based on simulated bootstrap non-parametric distribution by sampling with replacement of the original data, n>100,000), with significant signals classified as autocorrelated (I). The remaining data was searched for spike events, which were classified as spike maxima (II) or spike minima (III) (p<0.05 one-tailed based on differences from simulated, n>100,000 random distribution of the time-series). After classification, the data were agglomerated into hierarchical clusters (using correlation distance and average linkage) of common patterns and biological relevance was assessed through GO (Ashburner et al., 2000) Analysis [Cytoscape (Smoot et al., 2011), BiNGO (Maere et al., 2005) p<0.05, Benjamini-Hochberg (Benjamini and Hochberg, 1995) adjusted p<0.05] and pathway analysis [Reactome (Croft et al., 2011) Functional Interaction, FI, networks including KEGG (Kanehisa and Goto, 2000; Smoot et al., 2011), p<0.05, FDR < 0.05]. The unified framework approach was implemented on all the different datasets both individually and in combination, and our results revealed a number of differential changes that occurred both during infectious states and the varying glucose states.
We first analyzed the different individual transcriptome, proteome (serum and PBMC) and metabolome datasets; the proteome and metabolome results are presented in the supplement (Figures S4, S5, S7 and Data S4–S7). 19,714 distinct transcript isoforms (Wang et al., 2008) corresponding to 12,659 genes (Figure S1C) were tracked for the entire time course, and their dynamic expression response was classified into either autocorrelated (I) and spike sets, further subdivided as displaying maxima (II) or minima (III) (Figure 3). The clustering and enrichment analysis displayed a number of interesting pathways in each class: In the autocorrelated group [Figure 4B (I), see also Figure S7A and Data S7(1–2) ], we found two main trends: an upward trend (2,023 genes), following the onset of the RSV infection, and a similar coincidental downward trend (2,207 genes). The upward autocorrelated trend revealed a number of pathways as enriched (p<0.002, FDR<0.05), including Protein Metabolism and Influenza Life Cycle. Additionally, the downward autocorrelation cluster showed a multitude of enriched pathways (p<0.008, FDR<0.05), such as TCR Signaling in Naïve CD4+ T cells, Lysosome, B Cell signaling, Androgen regulation and of particular interest, Insulin signaling/response pathways. These different pathways, which are activated as a response to an immune infection, often share common genes and additionally we observe many genes hitherto unknown to be involved in these pathways but displaying the same trend. Furthermore, we observed that the downward trend, that began with the onset of the RSV infection and appeared to accelerate after Day 307, coincided with the beginning of the observed elevated glucose levels in the subject.
Figure 3. Transcriptome time course analysis.
(A) Summary of approach for identification of differentially expressed components. The various omics sets were processed through a common framework involving spectral analysis, clustering and pathway enrichment analysis. (B) Pattern Classification. The different emergent patterns from the analysis of the transcriptome for the entire time course are displayed for the autocorrelation (I), spike maxima (II) and spike minima (III) classes. For different clusters, examples of gene connections in selected pathways based on Reactome (Croft et al., 2011) FI [Cytoscape (Smoot et al., 2011) plugin] are shown as networks. Example GO (Ashburner et al., 2000) enrichment analysis results from Cytoscape (Smoot et al., 2011) BiNGO (Maere et al., 2005) plugin and pathway enrichment results [Reactome (Croft et al., 2011) FI] are included.
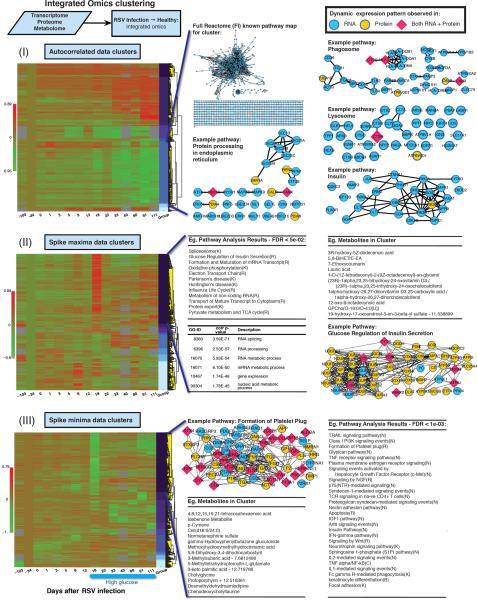
Figure 4. Integrated omics analysis.
For Days 186–400, the different emergent patterns from an integrated analysis of the transcriptome, proteome and metabolome data are displayed for autocorrelation (I), spike maxima (II) and spike minima (III) classes. For different clusters, examples of gene connections in selected pathways based on Reactome (Croft et al., 2011) and FI Cytoscape (Smoot et al., 2011) plugin are shown as networks, with constituents marked as assessed from protein data, transcriptome data or both. Example GO (Ashburner et al., 2000) enrichment analysis results from Cytoscape (Smoot et al., 2011) BiNGO (Maere et al., 2005) plugin and pathway enrichment results [Reactome (Croft et al., 2011) FI] are included.
In the dynamic spike class we again saw patterns that were concordant with phenotypes [Figure 3B, (II) and (III), see also Figure S7A, Data S7(3–14)]. A set of expression spikes displaying maxima (547 genes), that are common to the onset of both the RSV and HRV infections are associated with Phagosome, Immune processes and Phagocytosis, (p<1×10−4, FDR<6×10−3). Furthermore, a cluster that exhibits an elevated spike at the onset of the RSV infection involves the Major HistoCompatibility genes (p<7×10−4, Benjamini-Hochberg adjusted p<0.03). A large number of genes with a coexpression pattern common to both infections in the time course have yet to be implicated in known pathways and provide possible novel connections related to immune response. Finally, our spike class displaying minima showed a distinct cluster (1,535 genes) singular to day 307 (Day 14 of the RSV infection), associated with TCR signaling again, TGF receptors and T Cell, and Insulin Signaling pathways (p<0.02, FDR<0.03). Overall, the transcriptome analysis captures the dynamic response of the body responding to infection as also evidenced by our cytokine measurements, and also can monitor health changes over long periods of time, with various trends.
To further leverage the transcriptome and genome data, we performed an integrated analysis of transcriptome, proteomic and metabolomics data for each time point, observing how this corresponded to the varying physiological states monitored as described in the above sections. Because of the availability of many time points through the course of infection, we examined in detail the onset of the RSV infection, as well as extended our complete dynamics omics profile during the times that our subject began exhibiting high glucose levels. Figure 4 shows an integrated interpretation of omics data (see also Figure S7B, Data S8), where all trends are combined for each omics dataset and the common patterns emerge providing complementary information. In addition to the common patterns observed in our transcriptome analysis, new patterns emerged, some unique to protein data, some to metabolite, and some common to all. In particular we found the following interesting results: for autocorrelated clusters we found the same trends as observed in the transcriptome, additionally augmented with concordant protein expressions. Pathways such as the Phagosome, Lysosome, Protein Processing in Endoplasmic Reticulum and Insulin pathways emerged as significantly enriched (p<0.002, FDR<0.0075), and showed a downward trend post-infection, and further accelerated after about 3 weeks post the infection (this cluster comprised of 1,452 transcriptomic and 69 proteomic components, corresponding to 1,444 genes). The elevated spike class showed a maxima cluster on Day 18 post RSV infection (one time point after the cytokine maximum), with enrichment in pathways such as the Spliceosome, Glucose Regulation of Insulin Secretion and various pathways related to a stress response (p<1×10−4, FDR <0.02) - this cluster included 1,956 transcriptomic, 571 proteomic and 23 metabolomic components, corresponding to 2,344 genes. Even though current proteomic information is more limited than the full transcriptome because it follows fewer components, as evidenced in Figure 4 (II) several pathways, including the Glucose Regulation of Insulin Secretion pathway, clearly emerge from the proteomic information and would not have been observed by monitoring the transcriptome only. Additionally, in this cluster we find significant GO enrichment in splicing and metabolic processes (p<6×10−47, Benjamini-Hochberg adjusted p<10−45). Furthermore, inspection of metabolites reveals 23 that show the same exact trend (i.e. spikes at Day 18 post RSV infection); at least one, lauric acid has been implicated in fatty acid metabolism and insulin regulatory pathways (Kusunoki et al., 2007). Finally, we observe minima spikes as well, with yet another interesting group on Day 18, which showed down-regulation in several pathways (p<0.003, FDR<0.05), such as the Formation of Platelet Plug. This cluster displayed a high degree of synergy between the various omics data, comprised of 3,237 transcriptomic and 761 proteomic components corresponding 3,400 genes and 83 metabolomic components.
In summary our integrated approach revealed a clear systemic response to the RSV infection following its onset and post infection response, including a pronounced response evident at Day 18 post RSV infection. A variety of infection/stress response relative pathways were activated along with those related to the high glucose levels in the later time points. Insulin response pathways exhibit decreased signaling post infection.
Dynamic Omics Analysis: Extensive Heteroallelic Variation and RNA Editing
The considerable amount of transcriptome and proteome data allowed us to analyze and follow changes in allelic specific expression (ASE) splicing and editing at the RNA and protein levels, during healthy and disease states.
Of the 49,017 genomic variants associated with coding or UTR regions (Table 1A), 12,785 (26%) were expressed in PBMCs (≥40 read coverage; Table S4). 8,509 of the variants are heterozygous (1,113 missense) and the remainder (4,686; 684 missense) are homozygous. 8 of the 83 nonsense mutations were expressed indicating that not all nonsense mutations result in transcript loss.
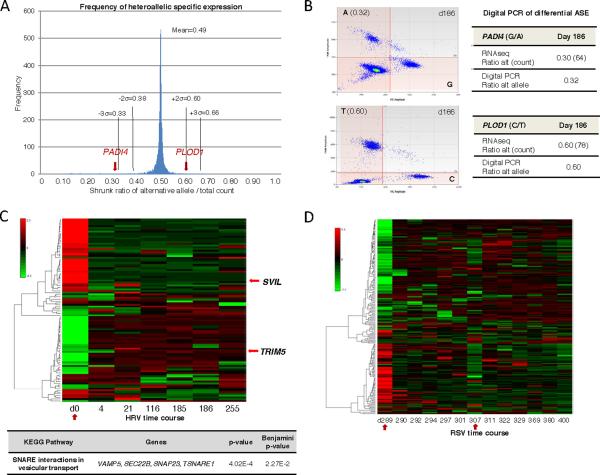
The numerous heterozygous variants allowed an analysis of the dynamics of differential ASE, (Experimental Procedures; Figure 5A, Figure S2B) in PBMCs during healthy and disease states. We found 497 and 1,047 genes that exhibited differential ASE during HRV and RSV infection, respectively (posterior probability ≥ 0.75, beta-binomial model; ≥40 reads, ≥7 time points); many of these are immune response genes, eg. PADI4 and PLOD1 (Figure 5B). Amongst the differential ASE sites 100 and 218 were specific to HRV and RSV infected states, respectively (Figure 5C–D). Differential ASE genes in the HRV compared to healthy phase were enriched for those encoding SNARE vesicular transport proteins (DAVID analysis; Benjamini p<0.05). Summing over all computed ASE alternative to total ratios revealed that non-reference heteroallelic variants were expressed at 98% of reference variants. The expression of over 50 heterozygous variants, including rare/private SNVs (0.72% of the genomic total), and differentially expressed variants (SVIL and TRIM5), was confirmed by Sanger cDNA sequencing and/or digital PCR (Hindson et al., 2011) of cDNA (Figures 5B, S2). Overall, these results demonstrate that differential ASE is pervasive in humans, particularly distinct during healthy and infected states, with many of these changes residing in immune response genes.
Figure 5. Heteroallelic expression study of PBMCs.
(A) Frequency of allelic specific expression (ASE) based on shrunk alternative/total ratios of RNA-Seq data. 143 positions fall outside the 3 standard deviations (σ) range (see Figure S2B) <0.33; >0.66), suggesting that certain heterozygous alleles (DNA level) are preferentially expressed in PBMCs. Standard deviations (σ) are denoted with dotted lines and the average ratio overlapping across all time-points is 0.49. (B) Digital droplet PCR validation of two heteroallelic expressed genes PADI4 and PLOD (relative to alternative allele). (C) Heatmap of the HRV infection time course (7 time points) showing differential ASE during HRV infection Day 0 (red arrow) relative to average shrunk ratios of healthy states (Days 116–255). (D) Heatmap of the RSV infection time course (13 time points) showing differential ASE specific to RSV infection Day 289 (red arrow) relative to average shrunk ratios of healthy states (Days 311–400), onset of T2D on Day 307 is also shown (red arrow). Heatmap ratios are relative to the alternative allele (alternative/total, posterior probability >0.75). Example of enriched KEGG pathway gene cluster (Huang et al., 2009; Benjamini p<0.05) shown below Figure 5C. See also Figures S2, S8.
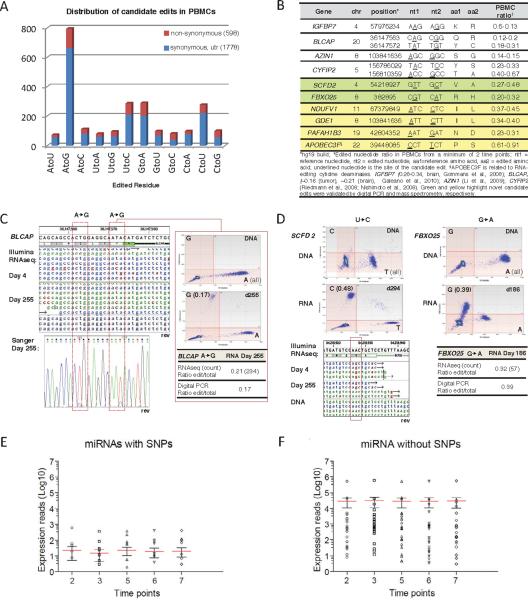
The depth of our RNA-Seq data enabled us to re-evaluate the extent of RNA editing, typically an adenosine to inosine (A-to-I) conversion (Li et al., 2009b) or cytidine to uridine (C-to-U), in normal human cells. We found 2,376 high confidence RNA edits, including 795 A-to-I (A-to-G) and 277 C-to-U deamination-like edits (Figures 6A, S8). 587 edits in 175 genes were predicted to cause amino acid substitutions [Polyphen-2 (Adzhubei et al., 2010)]; the remainder were nonsense (11), synonymous (435) or located in 5'/3' UTRs (103/1,240). 10 edited bases causing amino acid substitutions were validated by Sanger cDNA sequencing and/or digital droplet PCR, as well as by identification of their peptide counterparts by mass spectrometry (Figure 6B). Interestingly, we identified A-to-G edits (Figure 6B), eg. IGFBP7, BLCAP and AZIN1 in PBMCs that were known to occur in other tissues (Gommans et al., 2008; Levanon et al., 2005), indicating that the same RNA can be edited in other cell types. BLCAP exhibited two edited changes, Figure 6C, with edited/total ratios of 0.12–0.2 and 0.18–0.31, respectively, comparable to the 0.21 ratio previously observed in the brain (Galeano et al., 2010).
Figure 6. RNA editing and miRNA expression of PBMCs.
(A) Distribution of RNA editing types in missense (red) and synonymous and UTRs (blue), based on seven or more time points (total 20 time points). (B) Selected summary of known and novel RNA edits expressed in PBMCs. RNA edits were validated by digital PCR (green) and proteomic mass spectrometry (yellow). (C) Detail of 2 missense causing edit sites in BLCAP. Selected data from RNA-Seq at Day 4 and Day 255 (top left), Sanger sequencing of Day 255 cDNA (bottom left) and digital PCR (right panel) are shown. (D) Digital droplet PCR analysis of novel edit sites in SCFD2 (left) and FBXO25 (right) genes show no variants in DNA, while in RNA, editing is evident (top left quadrant). (E and F) Expression of SNV-containing and SNV-free miRNA, respectively, for Days 4, 21, 116, 185 and 186. Red lines: mean; error bars: standard error of the mean. Genome browsers, chromatograms and digital PCR data were analyzed with software from DNAnexus Inc., Chromas Ltd. and Quantalife™, respectively. See related Figures S2, S8 and Supplementary Data.
Furthermore, we found and validated two new missense causing edits, U-to-C in SCFD2 and G-to-A in FBXO25 (Figure 6D), indicating an amination-like RNA-editing mechanism, previously not observed in human cells. Our results reveal that a large number of edits occur and exhibit dynamic changes in populations of PBMCs. The total number of edited RNAs, while extensive, is significantly lower than that reported in human lymphoblastoid lines and very different in its distribution (Li et al., 2011). We believe that in addition to tissue-specific variation, the observed differences are also likely due to overcalling of false-positive SNVs, a problem we corrected with deep exome sequencing, removal of repeat regions and strings of close-proximity variants (Figure 6).
Finally, to determine whether the non-reference allele and edited RNAs serve as templates for protein synthesis, we generated proteome databases for 4,586 missense SNVs and all 30,385 edits and used them to search our mass spectra from the untargeted protein profiling experiments as well as in a targeted approach to directly search for 500 edited proteins (see Extended Experimental Procedures). Peptides for 48 SNVs and 51 edits were identified (FDR <0.01 and requiring one unique peptide per protein; Data S9). 17/17 selected SNVs (100%) validated by Sanger sequencing. 7 and 6 of peptides derived from the SNV and edited transcripts, respectively, were unique to a single protein in the IPI database (Kersey et al., 2004) and classified as high confidence. These results indicate that a large fraction of personal variants are expressed as transcripts and a number of these are also translated as proteins.
miRNA Variant Analysis
In addition to the omics profiling above, we identified 619–681 known miRNAs from PBMCs per time point (>10 reads, Days 4, 21, 116, 185 and 186), 106 of which showed dynamic changes (eg. Figure S3D–E). Examination of miRNA editing revealed 50 edited miRNAs (C-to-U or A-to-I) with stringent criteria (edited reads >5% of total reads or >399 modified reads) indicating that ~4% of expressed miRNAs are potentially edited. 18 miRNAs contain edits located within the functionally critical `seed sequences', potentially affecting their mRNA targets. Interestingly, expression of SNV-containing miRNAs was generally higher compared to SNV-free miRNA (Figure 6E–F). In addition to edits, analysis of the SNVs located in miRNAs revealed that most (25 of 31) SNVcontaining miRNAs were not expressed. These miRNAs were among those discovered in cancer cell lines (Jima et al., 2010) and may not normally be highly expressed in PBMCs from healthy individuals.
DISCUSSION
Our study is the first to perform extensive personal iPOP of an individual through healthy and disease states. It reveals extensive complex and dynamic changes in the omics profiles, especially in the transcriptomes, between healthy states and viral infections, and between non-diabetic and diabetic states. iPOP provides a multi-dimensional view of medical states, including healthy states, response to viral infection, recovery, and T2D onset. Our study indicates that disease risk can be assessed from a genome sequence and illustrates how traits associated with disease can be monitored to identify varying physiological stages. We show that large numbers of molecular components are present in blood samples and can be measured (>3 billion measurements taken over 20 time points). For the transcriptome many of these arise from differential splicing, ASE and editing events. By observing dynamic molecular changes that correspond to physiological states, this proof-of-principle study offers a pilot implementation of personalized medicine. The information obtained may greatly help in the design and application of personalized health monitoring, diagnosis, prognosis and treatment.
We speculate that differential expression of ASE/edits may be important in monitoring and assessing disease states. In this respect the genes/proteins in which one isoform is abundant in one condition (e.g. disease or healthy state) whereas another is abundant in another (e.g. disease state) may provide unique physiological advantages to the individual in distinct environmental conditions. Since multiple genes in our study that exhibit ASE and editing changes are involved in immune function, we speculate that these components are particularly valuable for mediating immune responses to environmental conditions such as exposure to pathogens. Likewise miRNA SNVs and edits, which also undergo differential expression, may confer unique biological responses.
Although we analyzed a single individual, insights were gained by integrating the multiple omics profiles associated with distinct physiological states. Through examination of molecular patterns, clear signatures of dynamic biological processes were evident, including immune responses during infection, insulin signaling response alterations after the RSV infection. Indeed, careful monitoring of omics changes across multiple time points for the same individual revealed detailed responses, which might not have been evident due to inter-individual variability had the analyses been performed on groups. Hence, we expect that our longitudinal personalized profiling approach provides novel and valuable information on an individual basis.
We focused on a generally healthy subject who exhibited no apparent disease symptoms. This is a critical aspect of personalized medicine, which is to perform iPOP and evaluate the importance and changes of all the profiles in ordinary individuals. These results have important implications and suggest new paradigm shifts: first, genome sequencing can be used to direct the monitoring of specific diseases (in this study, aplastic anemia and diabetes) and second, by following large numbers of molecules a more comprehensive view of disease states can be analyzed to follow physiological states.
Our study revealed that many distinct molecular events and pathways are activated both through viral infection and the onset of diabetes. Indeed, the monitoring of large numbers of different components revealed a steady decrease of insulin-related responses that are associated with diabetes-insulin response pathways occurring from the early healthy state to a high glucose state. Although many of the activated and repressed pathways could be detected through transcript profiling, some were detected only with the proteomics data and some with the combined set of data. In addition a large number of novel connections with diabetes and insulin signaling using metabolites, miRNAs, and autoantibodies were observed. One particularly interesting response detected with the proteomics data was the onset of the elevated glucose response that was tightly associated with the RSV infection and a particular subclinical response at day 12/18 post-infection. It is tempting to speculate that the RSV infection and/or the associated event at day 12/18 triggered the onset of high glucose/T2D. Although viral infections have been associated with T1D (van der Werf et al., 2007), we are unaware of viral infection associated with T2D. Inflammation and activated innate immunity have been associated with T2D (Pickup, 2004), and we speculate that perhaps RSV triggered aberrant glucose metabolism through activation of a viral inflammation response in conjunction with a predisposition toward T2D. Although this cannot be proven with the analyses from a single individual, this study nonetheless serves as proof-of-principle that iPOP can be performed and provide valuable information. Because diabetes is a complex disease there may be many ways to acquire high glucose phenotype; longitudinal iPOP analysis of a large number of individuals may be extremely valuable to dissecting the disease and its various subtypes, as well providing information into the molecular mechanism of its onset.
Finally, we believe that the wealth of data generated from this study will serve as a valuable resource to the community in the developing field of personalized medicine. A large database with the complete time-dynamic profiles for more individuals that acquire infections and other types of diseases will be extremely valuable in the early diagnostics, monitoring and treatment of disease states.
EXPERIMENTAL PROCEDURES
Full methods and associated references can be found in the Extended Experimental Procedures section.
WGS was performed at Complete Genomics Inc. and Illumina Inc.. High confidence SNVs were mostly correct as evidenced by: a) Illumina Omni1-Quad genotyping arrays (99.3% sensitivity); b) a Ti/Tv ratio of 2.14 as expected (1000 Genomes Project Consortium, 2010); c) Illumina capture and DNA sequencing (92.7% accuracy) and d) Sanger sequencing of 36 randomly selected SNVs (36/36 validated, Table S1A). In contrast, the low confidence SNVs had a Ti/Tv of only 1.46 and an accuracy of 63.8% (19 of 33 confirmed by Sanger sequencing, Table S1A). Similarly, the majority of the 216,776 high confidence indels are likely to be correct as a) Sanger sequencing validated 14 of 15 (93%) tested indels, and b) exome-sequencing validated most indels (4,706, 82%); meanwhile the 806,125 low confidence indels had a low validation rate (5,225, 0.65%). SVs were called using: 1) Paired-end mapping (Chen et al., 2009), 2) Read depth (Abyzov et al., 2011), 3) Split reads (Ye et al., 2009), and 4) Junction mapping (Lam et al., 2010) to the breakpoint junction database from the 1000G (Mills et al., 2011) 2,566 were found by two different methods or platforms (CG or Illumina) and were called high confidence; >90% of these were in the database of genome variants.
Strand-specific RNA-Seq libraries were prepared as described previously (Parkhomchuk et al., 2009) and sequenced on 1–3 lanes of Illumina's HiSeq 2000 instrument. The TopHat package (Trapnell et al., 2009) was used to align the reads to the hg19 reference genome, followed by Cufflinks for transcript assembly and RNA expression analysis (Trapnell et al., 2010). The Samtools package (Li et al., 2009a) was used to identify variants including single nucleotide variants (SNV) and Indels. Small RNAs were prepared from PBMCs for the first five time points; sequencing was performed according to Illumina's Small RNA v1.5 Sample Preparation Guide.
The Luminex 51-plex Human Cytokines assay were performed at the Stanford Human Immune Monitoring Center. For mass spectrometry, proteins were prepared from PBMC cell lysates, labeled at lysine's using the TMT isobaric tags by Pierce, and digested with trypsin and analyzed using reverse phase LC coupled to a Thermo Scientific (LTQ)-Orbitrap Velos instrument. In order to profile serum, 14 major glycoproteins were first removed using the Agilent Human 14 Multiple Affinity Removal System (MARS) column) in order to analyze the less abundant constituents. Metabolites were extracted by 4x serum volume of equal mixture of methanol, acetonitrile and acetone and separated using our Agilent 1260 liquid chromatography. Hydrophobic molecules were profiled using reversed phase UPLC followed by APCI-MS and hydrophilic molecule were analyzed using HILIC UPLC followed by ESI-MS in either the positive or negative mode.
For the integrated analysis, per omics set, for each time-series curve the Lomb-Scargle transformation (Hocke and Kämpfer, 2009; Lomb, 1976; Scargle, 1982, 1989) for unevenly sampled gapped time-series data was implemented (Ahdesmaki et al., 2007; Glynn et al., 2006; Van Dongen et al., 1999; Yang et al., 2011; Zhao et al., 2008). This allowed us to obtain a periodogram, which was used to calculate then the autocorrelations and then reconstruct the time-series with even sampling, allowing standard time-series analysis and performing data clustering, while taking the time intervals into account (see Extended Experimental Procedures).
Autoantibodyome profiling was performed using the Invitrogen ProtoArray Protein Microarray v5.0 according to the manufacturer's instructions.
Supplementary Material
HIGHLIGHTS
Physiological states analyzed by first integrative Personal Omics Profiling
Extensive molecular changes revealed during different health states
Individual disease risk predicted from genomics data
Extensive heteroallele and RNA editing during healthy and disease states
Acknowledgements
M.S. is funded by grants from Stanford University and the NIH. M.G. is funded by grants from the NIH. G.I.M. is funded by NIH training grant. K.J.K., J.T.D. and S.H. are supported by the NIH/NLM training grant T15-LM007033. T.E.K. and R.B.A are funded by NIH/NIGMS R24-GM61374. M.A. Blasco′s laboratory is funded by the Spanish Ministry of Science and Innovation Projects SAF2008-05384 and CSD2007-00017, European Union FP7 Projects 2007-A-201630 (GENICA) and 2007-A-200950 (TELOMARKER), European Research Council Advanced Grant GA#232854, the Körber Foundation, the Fundación Marcelino Botín and Fundación Lilly (España). F.E.D. was supported by NIH/NHLBI training grant T32 HL094274. E.A.A. was supported by NIH/NHLBI KO8 HL083914, NIH New Investigator DP2 Award OD004613, and a grant from the Breetwor Family Foundation. We dedicate this manuscript to Dr. Tara A. Gianoulis, an enthusiastic advocate for genomic science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest R.B.A., E.A., A.B. and M.S. serve as founders and consultants for Personalis. R.B.A. is a consultant to 23andMe. M.S. is a member of the scientific advisory board of GenapSys and a consultant for Illumina. M.A.B. acts as consultant and holds stock in Life Length, S.L.
REFERENCES
- 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahdesmaki M, Lahdesmaki H, Gracey A, Shmulevich l., Yli-Harja O. Robust regression for periodicity detection in non-uniformly sampled time-course gene expression data. BMC Bioinformatics. 2007;8:233. doi: 10.1186/1471-2105-8-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. Quantitative, High-Resolution Proteomics for Data-Driven Systems Biology. Annu Rev Biochem. 2010 doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Research. 2011;39:D691–697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich HA, Valdes AM, Julier C, Mirel D, Noble JA. Evidence for association of the TCF7 locus with type I diabetes. Genes Immun. 2009;10(Suppl 1):S54–59. doi: 10.1038/gene.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Chen J, Mushegian AR. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb-Scargle periodograms. Bioinformatics. 2006;22:310–316. doi: 10.1093/bioinformatics/bti789. [DOI] [PubMed] [Google Scholar]
- Grayson BL, Wang L, Aune TM. Peripheral blood gene expression profiles in metabolic syndrome, coronary artery disease and type 2 diabetes. Genes Immun. 2011;12:341–351. doi: 10.1038/gene.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, Velho G, Froguel P. Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians. Diabetologia. 1998;41:1511–1515. doi: 10.1007/s001250051098. [DOI] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocke K, Kämpfer N. Gap filling and noise reduction of unevenly sampled data by means of the Lomb-Scargle periodogram. Atmos Chem Phys. 2009;9:4197–4206. [Google Scholar]
- Jima DD, Zhang J, Jacobs C, Richards KL, Dunphy CH, Choi WW, Yan Au W, Srivastava G, Czader MB, Rizzieri DA, et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood. 2010;116:e118–127. doi: 10.1182/blood-2010-05-285403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusunoki M, Tsutsumi K, Nakayama M, Kurokawa T, Nakamura T, Ogawa H, Fukuzawa Y, Morishita M, Koide T, Miyata T. Relationship between serum concentrations of saturated fatty acids and unsaturated fatty acids and the homeostasis model insulin resistance index in Japanese patients with type 2 diabetes mellitus. J Med Invest. 2007;54:243–247. doi: 10.2152/jmi.54.243. [DOI] [PubMed] [Google Scholar]
- Lam HY, Mu XJ, Stutz AM, Tanzer A, Cayting PD, Snyder M, Kim PM, Korbel JO, Gerstein MB. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nature biotechnology. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, Hu Z, Simpson K, Pachter L, Durinck S, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res. 2010;8:961–974. doi: 10.1158/1541-7786.MCR-09-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009a;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009b;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Zheng H, Luo R, Zhu H, Li Q, Qian W, Ren Y, Tian G, Li J, et al. Building the sequence map of the human pan-genome. Nature biotechnology. 2010;28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- Lomb N. Least-squares frequency analysis of unequally spaced data. Astrophysics and space science. 1976;39:447–462. [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic acids research. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup JC. Inflammation and activated innate immunity in the pathogenesis of type 2 diabetes. Diabetes Care. 2004;27:813–823. doi: 10.2337/diacare.27.3.813. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Research. 2009;37:D32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Abyzov A, Wang J, Alves P, Raha D, Harmanci A, Leng J, Bjornson R, Kong Y, Kitabayashi N, et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scargle JD. Studies in astronomical time series analysis. II-Statistical aspects of spectral analysis of unevenly spaced data. The Astrophysical Journal. 1982;263:835–853. [Google Scholar]
- Scargle JD. Studies in astronomical time series analysis. III-Fourier transforms, autocorrelation functions, and cross-correlation functions of unevenly spaced data. The Astrophysical Journal. 1989;343:874–887. [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Du J, Gerstein M. Personal genome sequencing: current approaches and challenges. Genes & development. 2010;24:423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Weissman S, Gerstein M. Personal phenotypes to go with personal genomes. Mol Syst Biol. 2009;5:273. doi: 10.1038/msb.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, Stutz AM, Urban AE, Grubert F, Lam HY, Lee WP, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS genetics. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Liang XT, Pan K, Wang H, Zhao JJ, Li JJ, Ma HQ, Chen YB, Xia JC. High expression level of EDIL3 in HCC predicts poor prognosis of HCC patients. World J Gastroenterol. 2010;16:4611–4615. doi: 10.3748/wjg.v16.i36.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoridis G, Gika HG, Wilson ID. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom Rev. 2011 doi: 10.1002/mas.20306. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf N, Kroese FG, Rozing J, Hillebrands JL. Viral infections as potential triggers of type 1 diabetes. Diabetes Metab Res Rev. 2007;23:169–183. doi: 10.1002/dmrr.695. [DOI] [PubMed] [Google Scholar]
- Van Dongen HP, Olofsen E, VanHartevelt JH, Kruyt EW. A procedure of multiple period searching in unequally spaced time-series with the Lomb-Scargle method. Biol Rhythm Res. 1999;30:149–177. doi: 10.1076/brhm.30.2.149.1424. [DOI] [PubMed] [Google Scholar]
- Vaxillaire M, Cavalcanti-Proenca C, Dechaume A, Tichet J, Marre M, Balkau B, Froguel P. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57:2253–2257. doi: 10.2337/db07-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- Yang R, Zhang C, Su Z. LSPR: an integrated periodicity detection algorithm for unevenly sampled temporal microarray data. Bioinformatics. 2011;27:1023–1025. doi: 10.1093/bioinformatics/btr041. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Agyepong K, Serpedin E, Dougherty ER. Detecting Periodic Genes from Irregularly Sampled Gene Expressions: A Comparison Study. EURASIP Journal on Bioinformatics and Systems Biology. 2008;2008 doi: 10.1155/2008/769293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.