Abstract
Background:
The identification of differences in protein expression resulting from methodical variations is an essential component to the interpretation of true, biologically significant results.
Aims:
We used the Lowry and Bradford methods- two most commonly used methods for protein quantification, to assess whether differential protein expressions are a result of true biological or methodical variations.
Material & Methods:
Differential protein expression patterns was assessed by western blot following protein quantification by the Lowry and Bradford methods.
Results:
We have observed significant variations in protein concentrations following assessment with the Lowry versus Bradford methods, using identical samples. Greater variations in protein concentration readings were observed over time and in samples with higher concentrations, with the Bradford method. Identical samples quantified using both methods yielded significantly different expression patterns on Western blot.
Conclusions:
We show for the first time that methodical variations observed in these protein assay techniques, can potentially translate into differential protein expression patterns, that can be falsely taken to be biologically significant. Our study therefore highlights the pivotal need to carefully consider methodical approaches to protein quantification in techniques that report quantitative differences.
Keywords: Protein assay, protein quantification, Lowry method, Bradford method
Introduction
Protein quantification is an essential component in many biological studies, particularly those evaluating quantitative protein expression. However, studies scrutinizing these techniques are limited and variations resulting from different methodologies have largely been unreported. This raises the fundamental question of whether differential protein expressions reported are a result of true biological or methodical variations[1,2].
Many diverse absorbance based colorimetric protein assays have been developed in an attempt to increase sensitivity and accuracy[3]. The Lowry and Bradford methods are the most widely used dye-binding chromogenic protein assays[4]. The Bradford assay is based on the association of specific amino acid residues, arginine, lysine, and histidine, with non-conjugated groups of Coomassie brilliant blue G-250 dye (CBB) in an acidic environment[5]. The bindings of proteins with CBB result in a color change leading to a spectral shift from 465 nm to 610 nm, and the blue color from the CBB-protein complex is generally measured at 595 nm for its maximal yield. However, variations resulting from the Bradford method may be due to 1) amino acid composition of each target protein, i.e. percentage of arginine in the target protein; 2) sample concentrations beyond 100μg/ml - 1000μg/ml[6] ; and 3) pH of buffers or types of detergents used. The Lowry method[7] is a colorimetric assay based on the interaction of protein with an alkaline copper tartrate solution and Folin reagent. The color is generated by two steps: 1) formation of protein and copper complex in an alkaline buffer, and 2) reduction reaction of Folin reagent causing a spectral shift from 405 nm to 750 nm, with maximal yield at 750 nm. Ethylenediaminetetraacetic acid (EDTA) can interfere with chromophore production with the Lowry method.
In this study, we used the Lowry and Bradford methods - two most commonly used methods for protein quantification, to assess whether differential protein expressions are a result of true biological or methodical variations.
Materials and Methods
Protein assay
We quantified protein concentration by either the Bradford or Lowry methods. Bradford method (Bio-Rad Protein Assay; cat no. 500-0006) and Lowry method (Bio-Rad DC Protein Assay; cat no. 500-0116) were used in this study. The measurements were carried out according to the manufactures instructions (Bio-Rad, Hercules, CA).
Human umbilical vein endothelial cell (HUVEC) cultures
Commercially available human umbilical vein endothelial cells (HUVECs) were obtained from Lonza (cat no. cc-2517; Allendale, NJ, USA) . HUVECs were cultured in 5% CO2/37°C incubator and grown in microvascular endothelial cell growth medium-2 (EGM-2 MV Bullet kit, CC-3202, Lonza, Allendale, NJ, USA).
HUVECs were grown to 80% confluence before commencing heat shock treatment (H) to assess the inducible protein HSP72. Heat shock treatment was performed as previously described[8]. Briefly, cells were placed into a pre-heated incubator at a temperature of 43°C for 30 minutes. Cells were harvested 24 hours following heat shock treatment. Non-heat shock treated HUVECs (NH) at the same confluency were used as a control.
Antibodies and reagents
β-actin, heat shock protein 72 (HSP72) and albumin were our target proteins for this study. For western blot: Actin (cat no. MAB1501; Millipore, MA, USA) was used at a concentration of 1:5000; HSP72 antibody (cat no. SPA810; AssayDesigns, Michigan, USA) was used at a concentration of 1:1000. Albumin is commonly used as standard for protein measurements (cat no. B4287; Sigma, St. Louis, MO, USA).
SDS-PAGE Gel Electrophoresis and Western Blot
Sample media was mixed with 4X loading (sample) buffer containing 5% β-mercaptoethanol (Sigma, MO, USA) and Radio-Immuno Precipitation Assay (RIPA) buffer, pH 7.4 (cat no. BP-115, Boston BioProducts, MA, USA). The samples were then heated for 5 minutes at 95°C. 10-30mg of sample was loaded onto SDS-PAGE, NuPAGE Bis-Tris pre-cast polyacrylamide gels using the mini-cell system (Invitrogen, CA, USA). NuPAGE MOPS SDS running buffer was used. 500μl of antioxidant was added to the running buffer. Electrophoresis was performed at 140V-200V until adequate spread of the protein molecular marker was achieved. Following SDS-PAGE gel electrophoresis, proteins were then transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, MA, USA). Transfer was achieved using a wet-blot (Bio-Rad) transfer system. Standard Towbin transfer buffer was used containing 25mM Tris, pH 8.3, 192 mM glycine, 20% (v/v) methanol. Proteins were then visualized with an enhanced chemiluminescence detection system.
Results
Stability of the Bradford versus Lowry methods
We first assessed the stability of Bradford and Lowry methods using Albumin. Serial dilutions were made (0μg/ml, 2μg/ml, 4μg/ml, 6μg/ml, 8μg/ml and 10μg/ml), in triplicates; the measurements were carried out using a time course of 30 minutes with 5 minutes intervals. Absorbance change was calculated using the reference point of 0μg/ml at time 0, with all other samples compared to this; this value was denoted Delta. Samples measured by the Lowry method remained steady with time at all concentration preparations with Delta<0.01; while those measured by the Bradford method showed increasing Delta with time and serial concentrations. Increasing concentrations were associated with increasing Delta, from 0 to 30 minutes (2μg/ml: 0.007-0.013; 4μg/ml: 0.004-0.002; 6μg/ml: 0.012-0.04, 8μg/ml: 0.008-0.052; and 10μg/ml: 0.008-0.061) (Figure 1A). We then assessed if these differences in Delta translated into significantly different protein concentrations. As shown in Figure 1B, there is a greater variation of readings over time, with each serial dilution with the Bradford method. Therefore, we have found that the Lowry method yielded consistent absorbance readings over time with each serial dilution, while the Bradford method showed unstable results using identical preparations.
Fig. 1.
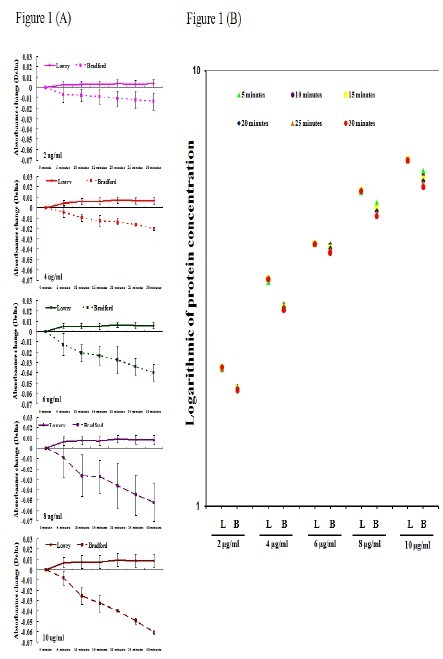
The protein concentration measurements using Lowry is more consistent than Bradford methods. The albumin, commonly used as standard for protein measurements, are prepared in serial dilutions (0μg/ml, 2μg/ml, 4μg/ml, 6μg/ml, 8μg/ml and 10μg/ml), in triplicates. The measurements were carried out using a time course of 30 minutes with 5 minutes intervals. (A) Absorbance change was calculated using the reference point of 0μg/ml at time 0, with all other samples compared to this; this value was denoted Delta. Delta is < 0.01 at every sample preparation using Lowry method; while the Delta is increasing in using Bradford method (2μg/ml: 0.007-0.013; 4μg/ml: 0.004-0.002; 6μg/ml: 0.012-0.04, 8μg/ml: 0.008-0.052; and 10μg/ml: 0.008-0.061). The X-axis in A represents 30-minutes time course on serial sample preparations; Lowry method (solid line) and Bradford method (dotted line). The Y-axis represents the Delta. (B) The Y-axis in figure 1B is protein concentration expressed in logarithmic 10. L: Lowry Method; B: Bradford Method.
Differential protein expression patterns using Bradford and Lowry methods
Our next goal was to assess whether the differential readings observed with the Lowry and Bradford methods reflected their measurable differential expression patterns, reported using protein-based techniques. Total proteins from identical samples were used with the Lowry and Bradford method. Differential protein expression patterns were assessed using HSP72 on a background of β-actin as our internal control, in human umbilical vein endothelial cells (HUVECs). β-actin is an abundant cytoskeletal protein in all cells and is the most commonly used internal control protein; HSP72 is universally induced in all organisms by stress (8). Non-heat shock treated (NH) and heat-shock treated (H) cells were subjected to western blot analysis. Protein lysates were taken from a single source from each group. For each sample, protein quantification was performed using both Lowry and Bradford methods in triplicates, followed by western blot analysis on three separate occasions.
Protein quantification results demonstrated observable variations with the Bradford method compared with the Lowry method for each sample. This confirms our observations in Figure 1 (Fig. 2A). The coefficient of variation (CV) in individual sample ranges from 0.029 to 0.09 in Lowry while 0.185 to 0.29 in Bradford.
Fig. 2.
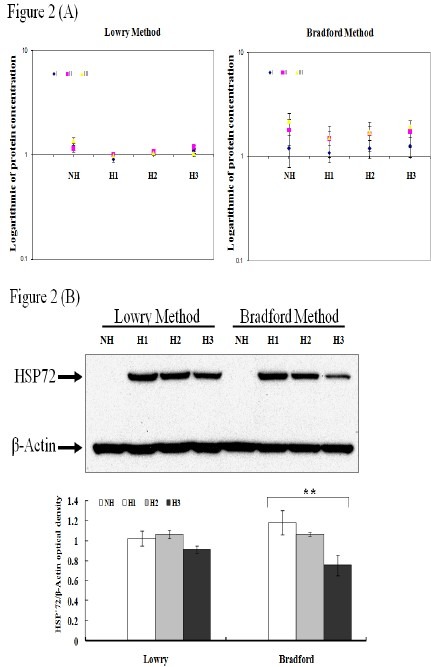
The protein concentration measurements using Lowry and Bradford resulted in protein differential expression. Total proteins are obtained from cell lysates of untreated HUVEC (NH) or 6 hours after heat shock treated HUVEC (H1-H3) from a single source from each group. (A) Protein concentrations from identical samples are measured using either Lowry or Bradford method. The measurements of protein concentration on all samples show more consistent using Lowry method while highly variable in Bradford method. The X-axis in A represents each sample; I, II, and III indicate each experiment. The Y-axis represents the protein concentration expressed in logarithmic 10. (B) The expression patterns of Heat shock protein 72 (HSP72) and β-actin from each group are examined using Western Blot. Identical samples show differential expression patterns on Western blot when concentration measurements are performed using Lowry vs. Bradford methods (triplicates) (upper panel). Protein expressions from Western blots are quantitated using Image J program (lower panel). The Y-axis represents protein expression level. The X-axis represents the method used for protein concentration measurements. ANOVA is used with P < 0.01 (**).
Western blot analysis of identical samples demonstrated consistent results with those assessed by the Lowry method compared with the Bradford method (Fig. 2B). In addition, our results suggest that the variations in protein expression patterns observed are not due to mechanical error concluded by the analysis of all samples on three separate occasions.
Discussion
In this paper, we highlight the importance of carefully considering methodologies used for protein quantification which can potentially yield false biologically significant results. We show for the first time that methodical variations observed in these protein assay techniques, can potentially translate into differential protein expression patterns that can be falsely taken to be biologically significant. Current literature using protein-based techniques rarely report methods used for protein quantification and our study underlines the need to report this information to allow for accurate data interpretation.
Our data suggests that (1) the Bradford method results in greater variations in protein concentrations with time and in a random fashion; the differences are more profound in samples with higher concentrations; (2) using identical protein samples, Lowry and Bradford methods yield different concentration readings, subsequently leading to different expression patterns on Western blots.
No significant differences in the expression patterns of β-actin were detected between the Lowry and Bradford methods. This may be due to its high abundance in the cell and therefore variations in its expression may not be significantly detected. However, we show that methodologies chosen for protein quantification become significantly important particularly when low abundant proteins are the interest of study. Therefore, we caution the use of β-actin in isolation as an internal control for equal electrophoretic gel loading to interpret differential expressions of a target protein without considering methodologies used in protein quantification.
Unfortunately, methods used for protein quantification are commonly chosen based on individual preferences rather than taking in consideration the composition of the target protein, sample concentrations, pH of buffers or types of detergents used. As mentioned above, the Bradford assay is based on the association of specific amino acid residues, arginine, lysine, and histidine, while the Lowry method is a colorimetric assay based on the interaction of protein with an alkaline copper tartrate solution and Folin reagent. We suggest that when selecting a particular technique for protein quantification, the target protein's composition and buffers should be taken into account.
Acknowledgement
The funding support of completion of this work is in part provided by SDSC global foundation.
References
- 1.Ng V, Cho P. The relationship between total tear protein concentrations determined by different methods and standards. Graefes Arch Clin Exp Ophthalmol. 2000;238(7):571–576. doi: 10.1007/s004170000147. [DOI] [PubMed] [Google Scholar]
- 2.Okutucu B, Dinçer A, Habib O, Zihnioglu F. Comparison of five methods for determination of total plasma protein concentration. J Biochem Biophys Methods. 2007;70(5):709–711. doi: 10.1016/j.jbbm.2007.05.009. 1. [DOI] [PubMed] [Google Scholar]
- 3.Kirazov LP, Venkov LG, Kirazov EP. Comparison of the Lowry and the Bradford protein assays as applied for protein estimation of membrane-containing fractions. Anal Biochem. 1993;208(1):44–48. doi: 10.1006/abio.1993.1006. [DOI] [PubMed] [Google Scholar]
- 4.Sapan CV, Lundblad RL, Price NC. Colorimetric protein assay techniques. Biotechnol Appl Biochem. 1999;29(Pt 2):99–108. [PubMed] [Google Scholar]
- 5.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Lozzi I, Pucci A, Pantani OL, D’Acqui LP, Calamai L. Interferences of suspended clay fraction in protein quantitation by several determination methods. Anal Biochem. 2008;376(1):108–114. doi: 10.1016/j.ab.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 8.Lu TS, Chen HW, Huang MH, Wang SJ, Yang RC. Heat shock treatment protects osmotic stress-induced dysfunction of the blood-brain barrier through preservation of tight junction proteins. Cell Stress Chaperones. 2004;9(4):369–377. doi: 10.1379/CSC-45R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]


