Abstract
Dynamic access to genetic information is central to organismal development and environmental response. Consequently, genomic processes must be regulated by mechanisms that alter genome function relatively rapidly1-4. Conventional chromatin immunoprecipitation (ChIP) experiments measure transcription factor (TF) occupancy5, but are blind to kinetics and are poor predictors of TF function at a given locus. To measure TF binding dynamics genome-wide, we performed competition ChIP6,7 with a sequence-specific S. cerevisiae transcription factor, Rap18. Rap1 binding dynamics and Rap1 occupancy were only weakly correlated (R2 = 0.14), but binding dynamics were more strongly linked to function than occupancy. Long Rap1 residence was coupled to transcriptional activation, while fast binding turnover, which we term “treadmilling”, was linked to low transcriptional output. Thus, DNA-binding events that appear identical by conventional ChIP may have starkly different underlying modes of interaction that lead to opposing functional outcomes. We propose that TF binding turnover is a major point of regulation in determining the functional consequences of transcription factor binding, and is mediated in large part by control of competition between TFs and nucleosomes. Our model (Supplementary Fig. 1) predicts a clutch-like mechanism that rapidly engages a treadmilling transcription factor into a stable binding state, or vice-versa, to modulate TF function.
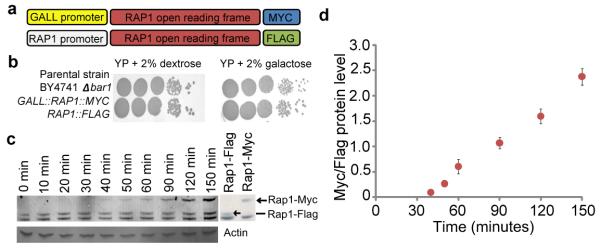
The diverse biological functions of Rap19 make it an excellent model for testing the hypothesis that binding dynamics are important for TF function. We developed a strain with two copies of RAP1. One copy of RAP1 was tagged with a 3X FLAG epitope and was constitutively expressed from the endogenous RAP1 promoter. A second copy of RAP1 was tagged with a 9X MYC epitope and was controlled by a weakened galactose-inducible promoter, GALL (Fig. 1a). This strain exhibited no growth defects in either inducing (2% Galactose) or non-inducing (2% Dextrose) conditions (Fig. 1b and Supplementary Fig. 2). To avoid cell-cycle and DNA replication effects, for the duration of the experiment the strain was arrested in G1 with alpha factor6. The induced Rap1 protein isoform could be detected as early as 30 minutes after galactose induction (Fig. 1c). The ratio of Rap1 isoforms provided an estimate of the nucleoplasmic pool of Rap1 molecules (Fig. 1d). We then performed Myc and Flag ChIP experiments independently from extract corresponding to each of 10 time points (0, 10, 20, 30, 40, 50, 60, 90, 120, 150 minutes after induction). We also performed ChIP to measure total Rap1 occupancy using a Rap1-specific antibody at 0 and 60 minutes. DNA fragments enriched in the ChIPs were detected on whole-genome tiling 12-plex microarrays containing 270,000 probes per subarray, with an average probe interval of 41 bp and an average probe length of 54 bp (Supplementary Fig. 3). The entire timecourse experiment was performed in duplicate. Procedural details can be found in Methods.
Figure 1. Development of transcription factor competition-ChIP in yeast.
(a) Schematic of Rap1 competition-ChIP yeast strain. (b) Growth comparison of competition yeast strain to wild-type in inducing (2% Galactose) and non-inducing (2% Dextrose) conditions. (c) Western blot using an antibody against Rap1 (y-300). Strains containing only a Rap1-Myc or only Rap1-Flag copy are shown to the right to indicate the size of isoform-specific bands. Actin loading control below. (d) To estimate the dynamics of induction, the ratio of induced Rap1-Myc and constitutive Rap1-Flag protein is plotted. Data is from two technical replicates of two independent time course replicates. Error bars represent standard error.
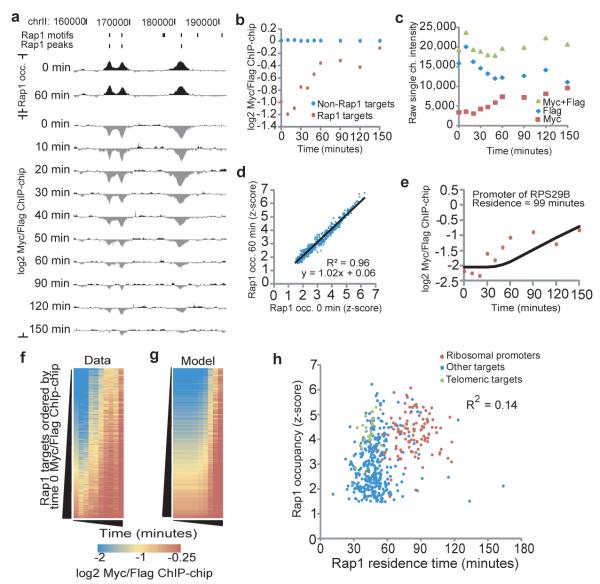
Following induction, Rap1-Myc was incorporated at targets where Rap1 had previously been shown to bind8,10 (Fig. 2a,b), indicating that the system was functioning as designed. The increase in Rap1 protein caused by the induction of the competitor did not cause an increase in the overall occupancy at the measured Rap1 site (Fig. 2c,d and Supplementary Fig. 4+5). As Rap1-Myc ChIP occupancy increased at sites of Rap1 binding, Rap1-Flag occupancy decreased coordinately (Fig. 2c,d and Supplementary Fig. 4). Thus, Rap1-Myc is competing specifically with Rap1-Flag at each locus, and Rap1-Myc binding is not the result of cooperativity or additional Rap1 binding locations.
Figure 2. Rap1-bound sites exhibit distinct replacement dynamics.
(a) A Rap1 turnover experiment over a 30-kb region of chromosome II. Rap1 motifs and peaks are indicated. (b) Average log2 Myc/Flag values for all Rap1 targets (red) increase relative to non-Rap1 targets (blue). (c) Rap1-Myc competes with Rap1-Flag for binding. Average single channel intensity for Rap1-Myc and Rap1-Flag for a single probe (id:CHR15FS000978891) in the promoter of TYE7/YOR344C shows the increase in Rap1-Myc is coincident with the loss of Rap1-Flag. (d) Total Rap1 occupancy does not change during the time-course. Average total Rap1 occupancy (log2 Rap1 IP (y-300)/input z-score) at Rap1 targets at time 0 versus 60 minutes is plotted. (e) Average log2 Myc/Flag values for the promoter of ribosomal protein gene RPL29B (red points). The model fit for the residence time parameter that best fits this data is shown (black line). (f) Colorimetric representation of log2 Myc/Flag values for all 465 Rap1 targets, sorted by the initial (normalized) log2 Myc/Flag value. (g) For each site in (f), the log2 Myc/Flag value predicted by our residence model based on the calculated residence time. (h) Rap1 occupancy (time 0 z-score) vs. Rap1 residence for 465 Rap1 targets (R2= 0.14, 0.37 spearman rank correlation).
To interpret our data, we developed a model to determine turnover rates of Rap1 by modifying a fitting algorithm used previously to measure histone H3 turnover6. Under our experimental conditions, the extracted turnover rate for a transcription factor at a binding site is equivalent to its dissociation rate, which allows us to measure residence time (Supplementary Fig. 6-8 and Supplementary Text). Our experimental system can quantify binding events that have an apparent duration of 500 seconds or longer (Supplementary Fig. 6,7 and Supplementary Text). Using our ChIP data and model we measured residence time of Rap1 at 439 peaks of Rap1 enrichment genome-wide, plus the 26 uniquely mappable telomeres (Fig. 2e-h, Supplementary Fig. 9 and Supplementary Text). Rap1 occupancy correlated only modestly with Rap1 residence (R2 = 0.14, Spearman Rank Correlation = 0.37) (Fig. 2h), and distinct dynamics of Rap1-Myc incorporation were observed at different genomic loci (Fig. 2e-h). Thus, residence times and occupancy are distinct measurements, and our system was capable of distinguishing Rap1 turnover kinetics at different loci in the same experiment.
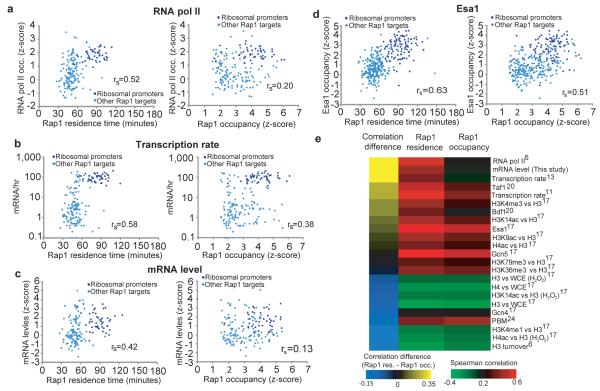
We found that efficient transcriptional activation was associated with stable Rap1 binding, while lower transcript production was associated with treadmilling, despite similar levels of Rap1 occupancy. Long Rap1 residence times occurred at ribosomal protein gene promoters, which are very highly transcribed and strongly activated by Rap16,11,12 (Fig. 2e,h). In contrast, Rap1 binding to non-ribosomal protein targets and to the infrequently transcribed telomeric and subtelomeric Rap1 sites were characterized by fast turnover (Fig. 2h, and Supplementary Fig. 10,11). Stable Rap1 binding appears to support higher mRNA production through more efficient recruitment of the RNA Polymerase II (RNA Pol II) machinery12 (Fig. 3a). Genes with stable Rap1 binding at their promoters indeed exhibited high levels of RNA Pol II association6 (Fig. 3a), high transcription initiation rates11,13 (Fig. 3b) and high mRNA levels (Fig. 3c). Rap1 occupancy does not correspond as strongly (Fig. 3a-c right). TBP turnover7 is also slow at ribosomal protein genes, suggesting that slow TF binding dynamics may be a hallmark of efficient transcription initiation11,13.
Figure 3. RNA Pol II recruitment, mRNA production, and histone acetyltransferase recruitment is associated with long Rap1 residence.
(a-d) In the left panel, Rap1 residence time is plotted on the x-axis. In the right panel, Rap1 occupancy is plotted on the x-axis. In both panels, the following is plotted on the y-axis: (a) RNA Pol II occupancy6, (b) mRNA/hr11, (c) mRNA levels at time 0 and (d) Histone acetyltransferase Esa1 occupancy z-scores17. rs is the Spearman correlation value. (e) Colorimetric representation of Spearman rank correlation between various genomic data sets and Rap1 occupancy (left) and Rap1 residence (right), ordered by the magnitude of the absolute difference between the oc cupancy and residence correlations for each comparison. WCE; Whole cell extract, PBM; Protein binding microarray. Telomeric targets excluded from analysis (Supplemental Text).
We next examined possible mechanisms for the locus-specific differences in Rap1 residence time. Nucleosomes are a major regulator of genome accessibility14, so we examined the relationship between histone modification and Rap1 binding dynamics15,16. Sites of long Rap1 residence were strongly correlated with sites of enrichment for the histone acetyltransferases Gcn5 and especially Esa117 (Fig. 3d,e). Nucleosome instability reinforced by Gcn5 and Esa1 (members of SAGA and NuA4, respectively) may stabilize Rap1 binding by reducing competition with nucleosomes18,19. Other indicators of active promoters, inclu ding H3K4me3, occupancy by the bromodomain protein Bdf1 (similar to mammalian Taf1)20, and acetylation of histone H3K9, H4, and H3K14, were also more strongly associated with Rap1 residence time than with Rap1 occupancy (Fig. 3e).
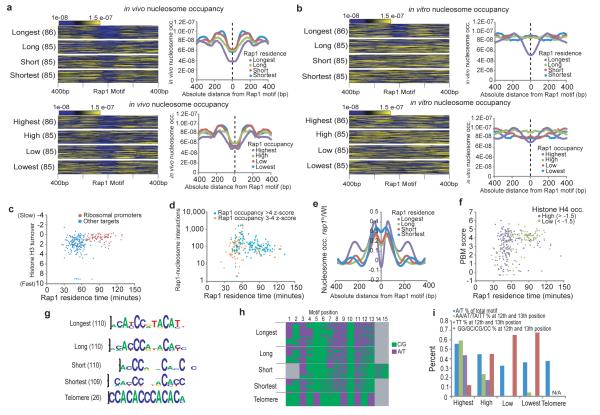
In general, sites bound by Rap1 are strongly depleted of nucleosomes21. However, the binding dynamics data allowed us to appreciate a more complex relationship. We grouped Rap1-bound loci into four categories based on their measured Rap1 residence time: Longest, Long, Short, and Shortest. We then aligned the Rap1 motifs in each category and plotted nucleosome occupancy relative to the motif position, reasoning that nucleosomes in direct proximity to the DNA motif bound by Rap1 would have a strong influence on Rap1 residence21. As expected, strong nucleosome depletion was centered on the Rap1 motif (Fig. 4a). However, as Rap1 binding turnover increased, nucleosome depletion was correspondingly less pronounced. Thus, not all highly occupied Rap1 sites are equally depleted of nucleosomes in vivo. Instead, a subset of loci at which Rap1 occupancy is high but binding turnover is also high (treadmilling), are associated with higher nucleosome occupancy (Supplementary Fig. 11b). No consistent relationship is apparent when Rap1 targets are grouped by occupancy as measured by traditional ChIP (Fig. 4a).
Figure 4. Evidence for competition between Rap1 and nucleosomes.
(a) Colorimetric representation of in vivo nucleosome occupancy centered on Rap1 binding motifs. Loci ordered by Rap1 residence time (top) or Rap1 occupancy (bottom). The total number of Rap1 targets in each group is shown in parentheses. To the right are plots of the average nucleosome occupancy for each group centered on the Rap1 motif. Targets with multiple Rap1 motifs are represented by one randomly chosen motif. (b) Same as (a) for in vitro nucleosome occupancy. (c) Histone H3 turnover vs. Rap1 residence for ribosomal protein genes (red) and other targets (blue). (d) The number of Rap1-nucleosome interactions22 detected within each Rap1 target peak boundary on a log10 scale. (e) Relative change in nucleosome occupancy following Rap1 depletion23 centered on Rap1 motifs grouped residence. A value of zero represents no relative change in nucleosome occupancy. (f) in vitro Rap1 affinity for its cognate target as measured by Protein Binding Microarray (PBM)24 compared to Rap1 residence. Colors represent histone H4 occupancy z-scores (> −1.5 purple (high),< −1.5 green (low)17. (g) Top position weight matrix motifs discovered for Rap1 targets grouped by residence. The number of targets for each group is in parentheses. (h) All motifs from the top position weight matrix for each residence group are colored by their A/T (purple) or G/C (green) content at each motif base position. (i) Percentage of A/T content for the entire motif (blue), AA/AT/TA/TT at the 12th and 13th motif position (green), TT at the 12th and 13th position (purple) and GG/GC/CG/CC at the 12th and 13th position (red) for Rap1 targets grouped by residence and telomeric regions.
We next examined nucleosome occupancy on naked DNA, in the absence of Rap1 or any protein cofactors21. Strikingly, DNA-encoded nucleosome occupancy measured in vitro is low only for the class of Rap1 targets with the most stable binding (Fig. 4b and Supplementary Fig. 11b). This pattern was not recapitulated when Rap1 targets were sorted by occupancy (Fig. 4b). This suggests that the nucleosome behavior surrounding TF motifs is at least partially encoded in DNA15, and that this DNA-encoded nucleosome occupancy can influence the binding dynamics of transcription factors, and thereby functional outputs (Supplementary Fig. 11a-c).
We sought further evidence supporting direct competition between nucleosomes and Rap1. We compared histone H3 turnover6 to Rap1 residence times and found that loci with long Rap1 residence times also had relatively slow H3 turnover. Likewise, histone H3 molecules that treadmill are found almost exclusively at sites of Rap1 treadmilling (Fig. 4c). Rap1-nucleosome interactions isolated by immunoprecipitating Rap1 following MNase digestion22 were also detected more often at treadmilling sites (Fig. 4d). Further evidence for competition is supported by a marked increase in nucleosome occupancy directly over Rap1 motifs following Rap1 depletion23 at treadmilling loci but not at loci with stable Rap1 binding (Fig. 4e). These relationships provide evidence for direct competition between Rap1 and nucleosomes.
Given that high DNA-encoded nucleosome occupancy is associated with rapid Rap1 turnover (Fig. 4b), it stands to reason that differences in the strength of the DNA motif bound by the TF would also influence turnover. To test this, at each locus we examined the relationship between Rap1 turnover and experimentally measured in vitro Rap1 affinity24. For sites with longer Rap1 residence, Rap1’s affinity for DNA was generally high, while Rap1 sites with the fastest turnover had lower experimentally measured Rap1 affinity (Fig. 4f). Despite this relationship, among sites with strong Rap1 motifs, nucleosome occupancy was still the major factor distinguishing sites with long Rap1 residence times from those with higher turnover (Fig. 4f).
Longer in vivo Rap1 residence times at sites of high Rap1 affinity is consistent with control of the Rap1-nucleosome competition being encoded directly in DNA sequence to a substantial degree15,16. We reasoned that this would be reflected in the sequence of the DNA motifs bound by Rap1. Indeed, we found differences in the composition of the Rap1 motifs for each of the turnover categories, with the longest residence Rap1 sites preferentially containing “A” or “T” at positions 4, 8, 12, and 13 (Fig. 4g-i). These associations were not as strong when Rap1 targets were ordered by occupancy (Supplementary Fig. 12). Sites at which residence was shortest tended to contain a degenerate Rap1 binding motif (Fig. 4g).
For several other transcription factors, microscopy-based measurements at individual loci point to much shorter residence times than we measured for Rap11,3,4,25-27. For example, despite an in vitro residence time similar to Rap1 (~90 minutes9,28), Glucocorticoid receptor (GR) binding appears to be exceptionally short-lived at individual loci26,27. Nonetheless, the overall positive relation between residence time and transcriptional output appear to be held in common by Rap1 and GR3,29. The differences in Rap1 and GR binding dynamics, and the disparity between GR residence time in vitro and in vivo may reflect different modes of interactions with nucleosomes. The binding affinity of GR binding may be especially sensitive to nucleosome packaging, or may be regulated by the availability of DNA that is transiently accessible from the nucleosome surface18. This type of accessibility on the nucleosome itself could be regulated, and would not rely on the complete loss of a nucleosome14. Rap1 itself exhibits such properties, with its binding progressively inhibited as the motif recognized by Rap1 is moved closer to the nucleosome dyad30. Our data do not exclude a model in which TF binding occurs adjacent to a nucleosome, and competition occurs without complete nucleosome eviction14,22,30.
In this study, we determined Rap1 binding dynamics genomewide using competition ChIP. Rap1 occupancy was only weakly correlated with Rap1 binding turnover, showing that these are independently measurable properties. Binding turnover correlates more strongly than occupancy with many aspects of genomic function, most predominantly RNA Pol II recruitment and transcript levels. Stable Rap1 binding is associated with activation, while Rap1 treadmilling is associated with higher nucleosome occupancy, nucleosomal treadmilling, and a lack of transcription. Our work provides the basis for a model in which transcription factor binding dynamics is a major point of regulation in determining the functional consequences of transcription factor binding. Importantly, this model provides a plausible mechanism for a locus-specific switch between inactive and active TF states, or even for a rapid switch from an activator (stable binding) to a repressor (treadmilling), or vice versa. This could be achieved at any given locus through a “clutch” that alters the balance of the continual competition between transcription factors and nucleosomes (Supplementary Fig. 1). This clutch could operate through histone modification, histone variant incorporation, ATP-dependent chromatin remodeling, or any other site-directed chromatin altering activity.
Methods Summary
Strain construction
The RAP1 gene and promoter was cloned into the pRS403 plasmid and integrated by homologous recombination into the HIS3 locus of BY4741. The two copies of RAP1 were then sequentially tagged using the 9X MYC epitope from pYM20:hphNT1 at the HIS3 copy of RAP1 and the 3X FLAG tag from p3FLAG-KanMX at the endogenous RAP1 copy. The HIS3 copy of the RAP1 promoter was replaced using homologous recombination by amplifying the GALL:natNT2 promoter from the pYM-N27 plasmid. Integrations were confirmed using PCR and Western blots. The BAR1 gene was knocked out by homologous recombination using a LEU2 gene amplified from pRS405.
Time course
Yeast were grown overnight in YPD (Yeast Extract 1%, Peptone 2%, Dextrose 2%) and used to inoculate 800 ml of YPR (Yeast Extract 1%, Peptone 2%, Raffinose 2%) to an OD600 of 0.2 (Genesys 20 Spectrophotometer) in a 4L Erlenmeyer flask. These cells were grown to an OD600 of 0.4 and subsequently arrested using 5 μM alpha factor (400 μL of 10 mM, GenScript) until 95% of the yeast cells were unbudded (~3hrs). Cells were then induced by adding 40% galactose to a final concentration of 2%. At this time additional alpha factor was added (400 μL of 10 mM, GenScript). Samples were collected at time points 0, 10, 20, 30, 40, 50, 60, 90, 120, and 150 minutes following galactose induction. At each time point, 35ml of culture was taken and added immediately to 37% formaldehyde to a final concentration of 1% for 20 minutes. 13 ml were taken for subsequent RNA preparation. 2 ml were taken for protein preparation by pelleting cells and heating at 95°C for 5 min in 0.06 M Tris-HCL, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.0025% bromophe nol blue. All samples were frozen immediately in liquid nitrogen.
Methods
Turnover model
A mathematical model is required to interpret the data, and to obtain relative binding turnover rates. We used a modified version of a histone H3 turnover model6. The original H3 turnover model assumed that there was no competitor protein present prior to its induction6. By Western blots, we were also unable to detect the presence of the Rap1 competitor protein prior to induction. Nevertheless, at each locus we consistently measured a non-zero competitor signal from the microarray even before the competitor was induced. This likely reflects non-specific background from our microarrays. Most of the steps that could contribute to this noise, e.g. non-specific pull down from the beads, site-specific variations in the DNA amplification, or non-specific binding bias in hybridization, would affect the constitutive and competitor signal equally, and therefore we assume for simplicity that the total non-specific background signal is approximately the same for the constitutive signal and for the competitor signal in our modified turnover model. We assume that at each binding site the measured IP signal is the true IP signal plus the background:
| [1] |
We assume that at the beginning of the experiment (prior to induction) the true IP signal of the competitor is zero. The background signal at the start of the experiment is therefore the signal measured for the competitor protein A at time 0:
| [2] |
The measured background signal will generally be time-dependent because our data showed that the measured raw intensities of the IP signals for the constitutive and competitor Rap1 proteins fluctuated from one time point to the next, even though their relative proportion remained roughly the same. This suggests that there are systematic variations in either the ChIP conditions or the microarray imaging conditions from one time point to the next which would also likely influence the background signal.
The systematic changes in either the ChIP or imaging conditions can be quantified by comparing the total signal of constitutive plus competitor at each binding site as a function of time. We assume that the addition of competitor does not change total occupancy6 (Supplementary Fig. 3+4). Thus, at each binding site, the ratio of the total signal (constitutive plus competitor) at time t versus time 0 generates a scaling factor to account for systematic fluctuations over time. This scaling factor (the brackets in Eq. (3) below) can be used to calculate the background at time t based on the background at time 0:
| [3] |
With this formula, we can calculate an occupancy ratio in the presence of background. First note that the occupancy ratio R(t) in the absence of background is defined as the ratio of the IPs of the competitor and constitutive signals:
| [4] |
We define a measured occupancy ratio mR(t) that includes the background signal:
| [5] |
where the second equality arises by substitution from Eq. (1) assuming that the background is the same in the competitor and constitutive signals. Using Eqs. (2)(3)(4), Eq. (5) can be rewritten as:
| [6] |
where C0 = mIPA(0)/(IPA(0) + IPB (0)). This constant can be expressed in terms of measurable quantities by using Eq. (1) and t he previously stated assumption IPA(0) = 0 to yield:
| [7] |
where mR(0) is the measured occupancy ratio at time 0. In practice, we calculated C0 by averaging over the first 3 time points which all showed no detectable competitor signal. With this estimate of C0, Eq. (6) enables calculation of an occupancy ratio in the presence of a microarray background signal by using the occupancy ratio R(t) calculated in the absence of background6.
R(t) is the probability that a locus is occupied by the competitor protein divided by the probability that it is occupied by the constitutive protein6. If P is the probability that the competitor occupies a given locus, then the probability that the constitutive protein occupies the locus is 1– P(t), and so R(t) becomes:
| [8] |
This probability satisfies the following differential equation6:
| [9] |
Here λ is the turnover rate at each locus, and A(t) and B(t) are the cellular concentrations of the free competitor and constitutive proteins. We measured A(t) and B(t) at all time points by Western blot. To determine the turnover rateλ for each locus we tunedλ to fit the measured occupancy ratio mR(t) at that locus. Specifically, we varied λ in Eq. (9) such that the value of R(t) obtained from Eq. (8) yields the best fit to our measured occupancy ratio when R(t) is substituted into Eq. (6).
The modified turnover model (Eq. (6)) was implemented in Matlab 2009b (The MathWorks, Natick, MA) and Eq. (9) was solved numerically using the ODE45 function. The Matlab routine lsqcurvefit was used to fit the models to experimental data and extract the turnover rate λ. We sampled a range of different starting guesses to avoid the detection of local minima. The MATLAB source code for the modified turnover model is available at http://code.google.com/p/ccc-process/.
Plasmids
The following plasmids were used in generation o f the Rap1 turnover strain: pRS40331,pRS40531, pYM20:hphNT132, p3FLAG-KanMX33, and pYM-N2732.
Chromatin immunoprecipitation and DNA amplification
Chromatin immunoprecipitation was performed on whole cell extract from crosslinked cells as described previously using anti-Flag (M2, Sigma), anti-Myc (clone 9E10, Millipore), and anti-Rap1 (y-300, Santa Cruz Biotechnology)8. IP and/or Input DNA was amplified using the GenomePlex Complete Whole Genome Amplification (WGA) kit (WGA2-50RXN, Sigma) and then reamplified using GenomePlex WGA Reamplification Kit (WGA3-50RXN, Sigma) using provided protocols. DNA was purified using Zymo columns according to the manufacturer’s instructions (Zymo Research).
Hybridization and processing of data from high resolution HD4 microarrays
For Nimblegen high resolution HD4 microarrays, amplified ChIP material was sent directly to Nimblegen where it was labeled and hybridized according to protocols in chapter 3 and 4 of the NimbleGen Arrays User’s Guide ChIP-chip Analysis, Version 3.1, 27 May 2008. Biweight mean scaled ratios are used as input for lowess normalization. All HD4 array data is deposited in GEO under Accession GSE32351.
Modified lowess normalization
Standard lowess normalization results in depressed binding ratios at the most highly enriched probes in ChIP-chip experiments. We therefore implemented a modified lowess normalization designed specifically for ChIP-chip based on the method described by Van Werven et al.7. The lowess function is determined for probes that do not show enrichment by defining a subset of probes which are the most enriched features in a MA plot. This lowess normalization is then applied to the entire data set with linear extrapolation to probes outside the range of the lowess fit, which corrects for the deflated binding ratios at enriched probes.
We implemented a comparable method which only varied in that we defined the enriched group for the calculation of the lowess function based on the sites we used to define Rap1 target enrichment for our turnover time course. We then consider all probes within +/− 2000 bp of these sites to be affected by Rap1, with all other probes forming the reference group for normalization (Supplementary Fig. 8a-d). Each time-point is normalized separately but we use the same group of reference probes for the normalization. While we believe using this modified lowess normalization approach is the most appropriate way to normalize the data, we find qualitative and quantitatively similar Rap1 turnover values without normalization (Data not shown).
Hybridization and processing of data from low resolution PCR based arrays
1 μg of amplified DNA was labeled with either dUTP Cy5 (PA55022, GE Healthcare) or Cy3 (PA53022, GE Healthcare) for low resolution PCR-based arrays. Purified labeled DNA was hybridized to PCR-based arrays representing the whole yeast genome and covering all coding and non-coding regions at an average resolution of approximately 800 bp10. The time course was performed in duplicate, one in each dye orientation, with the Myc and Flag samples then comparatively hybridized to an array for each time point. Arrays were scanned using an Axon 4000B scanner, and analyzed using Genepix 6.0 software (Axon). Only spots with <10% saturated input pixels and a signal intensity of greater than 500 (background-corrected sum of medians for both channels) were used for the analysis. Data was further normalized in the UNC microarray database with the normalized median log2 ratio of Rap1-Myc/Rap1-Flag being used for further analysis. All low resolution array data is deposited in GEO under Accession GSE27377. We did not use this ChIP-chip data in any of our analysis except in Supplementary Figure 3.
Reverse transcription, cDNA labeling, and expression arrays
Total RNA was extracted by the hot phenol method as previously described34. 30 μg of total RNA was reverse transcribed into cDNA using reagents and protocols provided with SuperScript II reverse transcriptase (Invitrogen; Cat. No. 18064-014) containing an amino-allyl-dDUTP mix (50x aa-dUTP mixture; 1mg amino-allyl dUTP (Sigma) dissolved with 32 μl of 100 mM dATP, dGTP, dCTP, 12.7 μl 100 mM of dTTP, and 19.3 μl of dH2O) and an anchored oligo dT primer (22mer; IDT). Reactions were incubated for 2hrs at 42°C, then heated at 95°C for 5 min and snap cooled on ice. RNA was hydrolyzed by addition of 13μl of 1 N NaOH and 1μl of 0.5M EDTA followed by incubation at 67°C. Reactions were then neutralized with 50μl of 1M HEPES pH 7.5. cDNA was purified on Zymo columns (Zymo Research; D4003) using seven volume excess of DNA binding buffer. cDNA was eluted off of columns using 5μl of 50mM sodium bicarbonate ph 9.0. cDNA was fluorescently labeled using Amersham CyDye Post-Labeling Reactive Dye Packs (#RPN5661). Each dye pack was resuspended in 11μl DMSO and 3μl of mixture was used per reaction. Cy dyes and aa-dUTP cDNAs were allowed to couple for 2 hours in the dark. Labeled cDNAs were cleaned up using Zymo columns with seven volumes excess DNA binding buffer and eluted with 10mM Tris-Cl ph 8.0 and hybridized to arrays as described previously.
For comparative hybridization, input genomic DNA from the experimental Rap1 turnover strain was extracted using phenol chloroform. 4μg of genomic DNA was denatured at 100°C with 10μg of random hexamer (IDT) then snap cooled on ice for 10 minutes. Samples were then incubated with 50 units of Klenow (Exo-) (NEB), and 1X Buffer 2 (NEB) in a total volume of 50μl at 37°C for 2hrs. Samples were cleaned up with Zymo columns, eluted in 5μl of 50mM sodium bicarbonate pH 9.0 and coupled to Cy dyes as for cDNA. Expression studies were performed on PCR-based arrays which were prepared, processed and analyzed as for the low resolution ChIP arrays10.
Defining regions of Rap1 enrichment
Rap1 ChIP-seq data from yeast strain BY4741 grown in YPD (Yeast extract 1%, Peptone 2%, and Dextrose 2%) were used to determine precise sites of Rap1 binding. Peaks and peak summits were called using MACS with a bandwidth of 300 p-value cutoff of 1e-5. Peaks in our turnover data set were called on total Rap1 occupancy at time zero using Peakpicker35 to ensure we identified all Rap1 peaks which were present in our turnover conditions. For analysis, we then used only MACS Chip-seq peak regions that had at least 1bp of overlap with our time course peaks, and had a z-score > 1.5 at time zero. Seven regions with a z-score > 1.5 at time zero that were identified at time zero of the Rap1 time course but not the Chip-seq experiment were also included to ensure full representation of Rap1 enriched regions in our experiment. Of the 457 total Rap1 peak regions identified we did not analyze 18. 15 targets had an estimated residence time of under 500 seconds, which is too short to measure with our system (Supplementary Fig. 6). 3 targets were also excluded which had residence times which exceeded 1×1010 seconds and showed exceptionally poor fits to the model. The average log2 Myc/Flag level for all probes which fell within +/− 150 bp of peak summits were averaged to generate a Myc/Flag value for each time point for each target. On average 8 probes contributed to the Myc/Flag signal for Rap1 targets. Peaks summits were used to assign target regions to promoters or coding regions for further analysis.
Telomeric regions were tiled using only uniquely mapping probes, making signal discontinuous in these regions and making peak calling difficult. For this reason, telomeres were defined by annotations from the Saccharomyces genome database (http://www.yeastgenome.org/). We excluded telomeres from any analysis that relied on our turnover metric because they contain many arrayed Rap1 binding sites within their AC-rich repeats. In theory, as the number of Rap1 binding sites detected by an individual microarray probe increases, the probability that either isoform of Rap1 will be detected at that probe increases. This violates some assumptions of our turnover metric, which would theoretically lead to artificially short residence time estimates. Despite this, empirically we see no relation between Rap1 residence times and motif number or density (Supplementary Fig. 10).
Motif discovery
The 439 Rap1 bound target regions (excluding telomeres), were placed into 4 categories based on their turnover properties: Longest (110 targets), Long (110 targets), Short (110 targets) and Shortest (109 targets). The DNA sequences for each Rap1 target region in each group were then used as input for the web-based interface for BioProspector36 (http://ai.stanford.edu/~xsliu/BioProspector/). Default parameters were used except the width of the first motif block was changed to “13” and “S. cerevisiae intergenic” was used as a genome background model. Rap1’s telomeric motif was determined from the full telomeric sequences of the 26 telomeres which were uniquely mappable on our arrays. Weblogo37 (http://weblogo.berkeley.edu/logo.cgi) was used to generate visual representation of the position weight matrix output from Bioprospector. The 439 Rap1 targets were similarly grouped by their occupancy properties to determine Rap1 motifs for Rap1 targets grouped by occupancy. The default settings on the motif scanning program Clover38 were used to detect Rap1 motifs genome wide using a previously published Rap1 PWM10.
External data sets
Values from existing data sets with a one-to-one correspondence to the arrayed elements in our study were used as published. For data sets derived from arrays that did not match our probe set, log2 ratios and z-scores were calculated for each array probe, for each replicate of the external data set. Z-scores were defined as the number of standard deviations a probe’s log2 ratio was from the mean log2 ratio of all probes on the array. In cases with multiple replicates, average z-scores were used to represent each probe. To map the data back to our experiments, the average z-scores of the array probes for the specific data set that were contained within the promoter or coding region assigned to each Rap1 target were used comparison. For histone H3 turnover data, the highest value for a probe that fell within promoters associated with peak summits for target regions was used for our analysis6. For Rap1 nucleosome interaction data we summed all the detected interactions which fell within each Rap1 target region.
Supplementary Material
Acknowledgments
We thank Tommy Kaplan and Oliver Rando for help with their turnover model, Timothy Palpant and Sheera Adar for help with time course experiments, and Roche-Nimblegen for pre-release custom HD4 12-plex microarrays. This work was supported by NIH Grant R01-GM072518 to JDL. FM was supported in part by the Region Ile-de-France in the framework of C’Nano IdF, the nanoscience competence center of Paris Region.
Footnotes
Author contributions C.R.L., S.E.H, and J.D.L. designed the study. C.R.L. and S.E.H. performed the experiments. F.M. developed and implemented the binding dynamics model. C.R.L., F.M., J.G.M. and J.D.L. performed data analysis. C.R.L., F.M., J.G.M, and J.D.L. wrote the paper.
Author information Data is available under the following GEO accession numbers: ChIP-chip data GSE32351; ChIP platform GPL14612; RNA expression array data GSM677030-GSM677033; Expression platform GPL4414. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Mueller F, Wach P, McNally JG. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J. 2008;94:3323–39. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- 3.Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Molecular and Cellular Biology. 2004;24:2682–97. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karpova TS, et al. Concurrent Fast and Slow Cycling of a Transcriptional Activator at an Endogenous Promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- 5.MacArthur S, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome biology. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–8. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 7.van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol. 2009;16:1043–8. doi: 10.1038/nsmb.1674. [DOI] [PubMed] [Google Scholar]
- 8.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–34. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 9.Piña B, Fernández-Larrea J, García-Reyero N, Idrissi FZ. The different (sur)faces of Rap1p. Molecular Genetics and Genomics. 2003;268:791–798. doi: 10.1007/s00438-002-0801-3. [DOI] [PubMed] [Google Scholar]
- 10.Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–28. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 12.Layer JH, Miller SG, Weil PA. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J Biol Chem. 2010;285:15489–99. doi: 10.1074/jbc.M110.104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelechano V, Chavez S, Perez-Ortin JE. A complete set of nascent transcription rates for yeast genes. PLoS One. 2010;5:e15442. doi: 10.1371/journal.pone.0015442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–49. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 15.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–56. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–50. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pokholok DK, et al. Genome -wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Anderson JD, Lowary PT, Widom J. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2001;307:977–85. doi: 10.1006/jmbi.2001.4528. [DOI] [PubMed] [Google Scholar]
- 19.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature genetics. 2011;43:264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc Natl Acad Sci U S A. 2004;101:16843–8. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell. 2009;35:889–902. doi: 10.1016/j.molcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganapathi M, et al. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 2011;39:2032–44. doi: 10.1093/nar/gkq1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee S, et al. Rapid analysis of the DNA -binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–9. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosisio D, et al. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. The EMBO journal. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voss TC, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–54. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–5. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 28.Perlmann T, Eriksson P, Wrange O. Quantitative analysis of the glucocorticoid receptor-DNA interaction at the mouse mammary tumor virus glucocorticoid response element. The Journal of biological chemistry. 1990;265:17222–9. [PubMed] [Google Scholar]
- 29.Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Molecular Cell. 2008;30:486–97. doi: 10.1016/j.molcel.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossetti L, et al. Specific interactions of the telomeric protein Rap1p with nucleosomal binding sites. J Mol Biol. 2001;306:903–13. doi: 10.1006/jmbi.2001.4458. [DOI] [PubMed] [Google Scholar]
Methods references
- 31.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–62. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 33.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Molecular and Cellular Biology. 2001;21:2098–106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman CS. Preparation of yeast DNA, RNA, and proteins. In: Ausubel FM, et al., editors. Current Protocols in Molecular Biology. Vol. 2. John Wiley and Sons, Inc.; New York, NY: 1997. pp. 13.11.1–13.11.4. [DOI] [PubMed] [Google Scholar]
- 35.Cesaroni M, Cittaro D, Brozzi A, Pelicci PG, Luzi L. CARPET: a web-based package for the analysis of ChIP-chip and expression tiling data. Bioinformatics. 2008;24:2918–20. doi: 10.1093/bioinformatics/btn542. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput. 2001:127–38. [PubMed] [Google Scholar]
- 37.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frith MC, et al. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–81. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.