Abstract
Ciliated protozoa harbor two different types of nuclei in each cell. The diploid micronucleus is the transcriptionally inactive generative nucleus, while the macronuclous contains a highly amplified transcriptionally active genome of lower complexity. The macronuclear genes encoding the two largest subunits of both RNA polymerases I and II of Euplotes octocarinatus were identified by a novel method of two step PCR walking, employing primer pairs derived from telomeric sequences of the organism and known conserved RNA polymerase polypeptide sequences, respectively. The relative gene dosage was determined. The genes are present in equal copy numbers for the respective matching subunits. Northern hybridizations showed comparable amounts of transcripts, as well, within the matching pairs. Mapping of the 5'-termini of the transcripts of the gene sized chromosomes showed that the upstream nontranscribed regions are very short and contain characteristic sequence motifs which could be the determinants of equal promoter strengths for subunits of a common RNA polymerase.
Full text
PDF

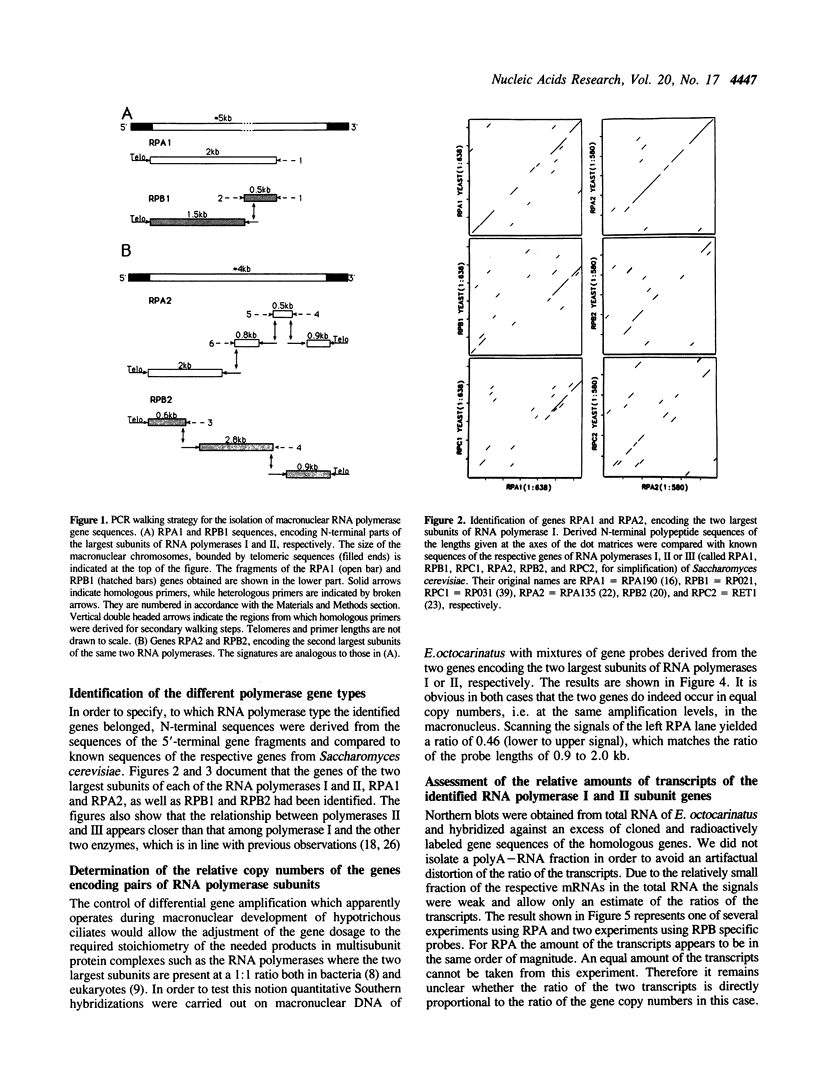



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Ammermann D., Steinbrück G., von Berger L., Hennig W. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma. 1974 May 10;45(4):401–429. doi: 10.1007/BF00283386. [DOI] [PubMed] [Google Scholar]
- Baird S. E., Klobutcher L. A. Characterization of chromosome fragmentation in two protozoans and identification of a candidate fragmentation sequence in Euplotes crassus. Genes Dev. 1989 May;3(5):585–597. doi: 10.1101/gad.3.5.585. [DOI] [PubMed] [Google Scholar]
- Baird S. E., Klobutcher L. A. Differential DNA amplification and copy number control in the hypotrichous ciliate Euplotes crassus. J Protozool. 1991 Mar-Apr;38(2):136–140. doi: 10.1111/j.1550-7408.1991.tb06033.x. [DOI] [PubMed] [Google Scholar]
- Brunk C. F., Sadler L. A. Characterization of the promoter region of Tetrahymena genes. Nucleic Acids Res. 1990 Jan 25;18(2):323–329. doi: 10.1093/nar/18.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J. L., Pannetier C., Jaulin C., Kourilsky P. Optimal conditions for directly sequencing double-stranded PCR products with sequenase. Nucleic Acids Res. 1990 Jul 11;18(13):4028–4028. doi: 10.1093/nar/18.13.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Cornelissen A. W., Evers R., Köck J. Structure and sequence of genes encoding subunits of eukaryotic RNA polymerases. Oxf Surv Eukaryot Genes. 1988;5:91–131. [PubMed] [Google Scholar]
- Delidakis C., Kafatos F. C. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 1989 Mar;8(3):891–901. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Furter-Graves E. M., Hall B. D. DNA sequence elements required for transcription initiation of the Schizosaccharomyces pombe ADH gene in Saccharomyces cerevisiae. Mol Gen Genet. 1990 Sep;223(3):407–416. doi: 10.1007/BF00264447. [DOI] [PubMed] [Google Scholar]
- Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- James P., Whelen S., Hall B. D. The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. Structural analysis of the wild-type and ret1-1 mutant alleles. J Biol Chem. 1991 Mar 25;266(9):5616–5624. [PubMed] [Google Scholar]
- Kolodziej P. A., Woychik N., Liao S. M., Young R. A. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol Cell Biol. 1990 May;10(5):1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontermann R., Sitzler S., Seifarth W., Petersen G., Bautz E. K. Primary structure and functional aspects of the gene coding for the second-largest subunit of RNA polymerase III of Drosophila. Mol Gen Genet. 1989 Nov;219(3):373–380. doi: 10.1007/BF00259609. [DOI] [PubMed] [Google Scholar]
- Larson D. D., Umthun A. R., Shaiu W. L. Copy number control in the Tetrahymena macronuclear genome. J Protozool. 1991 May-Jun;38(3):258–263. doi: 10.1111/j.1550-7408.1991.tb04439.x. [DOI] [PubMed] [Google Scholar]
- Leffers H., Gropp F., Lottspeich F., Zillig W., Garrett R. A. Sequence, organization, transcription and evolution of RNA polymerase subunit genes from the archaebacterial extreme halophiles Halobacterium halobium and Halococcus morrhuae. J Mol Biol. 1989 Mar 5;206(1):1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- Mahadevan S., Struhl K. Tc, an unusual promoter element required for constitutive transcription of the yeast HIS3 gene. Mol Cell Biol. 1990 Sep;10(9):4447–4455. doi: 10.1128/mcb.10.9.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Mémet S., Saurin W., Sentenac A. RNA polymerases B and C are more closely related to each other than to RNA polymerase A. J Biol Chem. 1988 Jul 25;263(21):10048–10051. [PubMed] [Google Scholar]
- Osheim Y. N., Miller O. L., Jr Novel amplification and transcriptional activity of chorion genes in Drosophila melanogaster follicle cells. Cell. 1983 Jun;33(2):543–553. doi: 10.1016/0092-8674(83)90435-x. [DOI] [PubMed] [Google Scholar]
- Pühler G., Leffers H., Gropp F., Palm P., Klenk H. P., Lottspeich F., Garrett R. A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze Dieckhoff H., Freiburg M., Heckmann K. The isolation of gamones 3 and 4 of Euplotes octocarinatus. Eur J Biochem. 1987 Oct 1;168(1):89–94. doi: 10.1111/j.1432-1033.1987.tb13391.x. [DOI] [PubMed] [Google Scholar]
- Spradling A. C. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell. 1981 Nov;27(1 Pt 2):193–201. doi: 10.1016/0092-8674(81)90373-1. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Swanton M. T., Greslin A. F., Prescott D. M. Arrangement of coding and non-coding sequences in the DNA molecules coding for rRNAs in Oxytricha sp. DNA of ciliated protozoa. VII. Chromosoma. 1980;77(2):203–215. doi: 10.1007/BF00329545. [DOI] [PubMed] [Google Scholar]
- Swanton M. T., Heumann J. M., Prescott D. M. Gene-sized DNA molecules of the macronuclei in three species of hypotrichs: size distributions and absence of nicks. DNA of ciliated protozoa. VIII. Chromosoma. 1980;77(2):217–227. doi: 10.1007/BF00329546. [DOI] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wünning I. U., Lipps H. J. A transformation system for the hypotrichous ciliate Stylonychia mytilus. EMBO J. 1983;2(10):1753–1757. doi: 10.1002/j.1460-2075.1983.tb01653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991 Feb;11(2):754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]







