Abstract
The yeast Rgt1 repressor is a bifunctional protein that acts as a transcriptional repressor and activator. Under glucose-limited conditions, Rgt1 induces transcriptional repression by forming a repressive complex with its corepressors Mth1 and Std1. Here, we show that Rgt1 is converted from a transcriptional repressor into an activator under high glucose conditions and this occurs through two independent but consecutive events mediated by two glucose signaling pathways: (1) disruption of the repressive complex by the Rgt2/Snf3 pathway; (2) phosphorylation of Rgt1 by the cAMP-PKA (cAMP-dependent protein kinase) pathway. Rgt1 is phosphorylated by PKA at four serine residues within its amino-terminal region, but this does not occur until the repressive complex is disrupted. While phosphorylation of any one of these sites is sufficient to enable Rgt1 to induce transcriptional activation, phosphorylation of all the sites results in the release of Rgt1 from DNA. We discuss how the bifunctional properties of Rgt1 are regulated through differential phosphorylation.
Keywords: Glucose, Phosphorylation of Rgt1, Degradation of Mth1, Rgt2/Snf3 pathway, cAMP-PKA Pathway, Glucose sensing and signaling
Introduction
Glucose sensing and signaling in the budding yeast Saccharomyces cerevisiae represents an important paradigm for understanding how extracellular signals lead to changes in the gene expression program in eukaryotes. Due to their specialized mode of glucose metabolism—aerobic fermentation, yeast cells must pump large amounts of glucose to supply sufficient ATP via glycolysis. To accomplish this, yeast cells enhance expression of the glucose transporter genes (HXT) when glucose is available in their environment. Glucose-induction of HXT expression is largely regulated by Rgt1, a member of the Gal4-family of transcription factors that contains a Zn2Cys6 binuclear cluster DNA-binding domain (Ozcan and Johnston, 1995; Ozcan et al., 1996; Kim et al., 2003; Kim, 2004; Kim, 2009). Under glucose-limited conditions, Rgt1 recruits the general corepressor complex Ssn6-Tup1, which mediates repression of many genes, to the HXT promoters (Kim et al., 2003; Ozcan and Johnston, 1995). Rgt1 does this in conjunction with its corepressors Mth1 and Std1, paralogous proteins that interact with Rgt1 (Tomas-Cobos et al., 2002; Lakshmanan et al., 2003; Polish et al., 2005). Furthermore, chromatin immunoprecipitation analyses show that Mth1 and Std1 are associated with the HXT3 promoter along with Ssn6 and Tup1, suggesting that Rgt1 forms a protein complex with these proteins to inhibit transcription of the HXT genes (Kim et al., 2003).
The presence of extracellular glucose appears to cause a disruption of the protein complex (Flick et al., 2003; Moriya and Johnston, 2004; Kim et al., 2006). Mth1 and Std1 are subject to glucose-dependent degradation through phosphorylation-mediated ubiquitination (Flick et al., 2003; Kim et al., 2006). In this process, Mth1 and Std1 are phosphorylated by the plasma membrane-tethered yeast casein kinases (Yck1/2) (Moriya and Johnston, 2004) and ubiquitinated by the SCFGrr1 ubiquitin ligase (Spielewoy et al., 2004). The resulting ubiquitinated Mth1 and Std1 are then targeted for degradation by the 26S proteasome. It is not clearly understood how Yck1/2 are activated in response to glucose (Pasula et al., 2010), but the kinases have been shown to interact with the two plasma membrane-spanning glucose sensors Rgt2 and Snf3 (Moriya and Johnston, 2004), which are activated upon glucose binding (Ozcan et al., 1996). Therefore, it has been proposed that the glucose sensors trigger a signaling pathway in response to glucose that ultimately alters the function of Rgt1 in the nucleus (Ozcan et al., 1998; Pasula et al., 2007).
It has been also shown that Rgt1 function is regulated by PKA. Induction of HXT1 is prevented in cells with attenuated PKA activity (Kim and Johnston, 2006; Palomino et al., 2006) or cells expressing Rgt1 without the putative PKA phosphorylation sites in Rgt1 (Kim and Johnston, 2006). Genome-wide expression profiling has also shown that the cAMP-PKA pathway is involved in the regulation of HXT expression (Wang et al., 2004; Zaman et al., 2009). These findings lead us to posit that the two glucose signaling pathways—the Rgt2/Snf3 pathway and the cAMP-PKA pathway—collaborate to suppress the repressor function of Rgt1 (Kim and Johnston, 2006).
Rgt1 also acts as a transcriptional activator. Unlike other HXT genes such as HXT3, glucose-induced expression of HXT1 is ∼ 6-fold decreased in the rgt1 mutant (Ozcan and Johnston, 1995). In addition, a LexA-Rgt1 fusion strongly activates transcription of the CYC1-lacZ reporter gene that contains a lexO site (Ozcan et al., 1996; Polish et al., 2005). Also, Rgt1 is converted from a transcriptional repressor to an activator upon glucose-mediated phosphorylation (Mosley et al., 2003). However, the underlying mechanism of this conversion is still unclear. In this study, we examined how the bifunctional Rgt1 repressor is regulated through an interaction between the Rgt2/Snf3 and cAMP-PKA glucose signaling pathways.
Materials and Methods
Yeast Growth
Except where indicated, yeast strains were grown in YP (2% bacto-peptone, 1% yeast extract) or SYNB (synthetic yeast nitrogen base media; 0.17% yeast nitrogen base with 0.5% ammonium sulfate) supplemented with the appropriate amino acids. The yeast strains used were BY4742 (MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 (Brachmann et al., 1998)), YM6370 (BY4742 rgt2∷kanMX snf3∷kanMX (Kaniak et al., 2004)), DC124 (MATa his3 leu2 ura3 trp1 ade8 (Cameron et al., 1988), and RS-13-58A-1 (MATa his3 leu2 ura3 trp1 ade8 tpk1w1 tpk2∷HIS3 tpk3∷TRP1 bcy1∷LEU2 (Cameron et al., 1988).
Western Blotting
Western blotting was performed as described previously (Kim et al., 2003). Briefly, 5 ml of yeast cells (O.D600=1.2∼1.5) expressing GFP-Mth1 were collected by centrifugation at 3,000 rpm in a table-top centrifuge for 5 min, and the cell pellets were resuspended in 100 μl of SDS-buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% β-mercaptoethanol) and boiled for 5 min. After the lysates were cleared by centrifugation at 12,000 rpm for 10 min., soluble proteins were resolved by SDS-PAGE and transferred to PVDF membrane (Millipore). The membrane was incubated with anti-GFP-antibody in TBST buffer (10 mM Tris-HCl, pH 7.5, 150mM NaCl, 0.05% Tween-20) and proteins were detected by the enhanced chemiluminescence (ECL) system (Pierce).
Chromatin Immunoprecipitation (CHIP)
ChIP was carried out as described previously (Kim et al., 2003). Yeast cells expressing HA-Rgt1 were grown to mid-log phase (OD600 = 1.0∼1.2) and incubated with formaldehyde (1% final concentration) for 15 to 20 min at room temperature. The cross-linking reaction was quenched by adding glycine to a final concentration of 125 mM for 5 min. The cells were disrupted by vortexing with glass beads in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% Na-deoxycholate). The lysate was sonicated (ultrasonic cell disruptor with a microtip) five times with 10-s pulses. The genomic DNA fragments, which averaged 200 to 500 bp in length, were immunoprecipitated with the monoclonal HA antibody (Santa Cruz). The immunoprecipitated DNA and 1/100 of the input DNA were used as templates in a 25-cycle PCR. The sequences of primer pairs used for ChIP of the HXT1 gene promoters were 5′ATATAATTCCCCCCTCCTGAAG-3′ and 5′-TGATTCTACGTTTTTGCAAG C -3′.
Fluorescence Microscopy
Cells expressing GFP-fluorescent proteins were visualized using a Zeiss LSM 510 META confocal laser scanning microscope with a 63× Plan-Apochromat 1.4 NA Oil DIC objective lens. Images were acquired with the Zeiss LSM 510 software version 3.2.
β-Galactosidase Assay
To assay β-galactosidase activity with yeast cells expressing the HXT1-lacZ reporter, the yeast cells were grown to mid-log phase and the assay was performed as described previously (Kaniak et al., 2004). Results were given in Miller Units [(1,000 × OD420)/(T × V × OD600), where OD420 was the optical density at 420 nm, T was the incubation time in minutes, and V is the volume of cells in milliliters]. The reported enzyme activities were averages of results from triplicates of three different transformants.
Results
Differential Activation of the RGT2/SNF3 and cAMP-PKA Glucose Signaling Pathways by Different Sugars
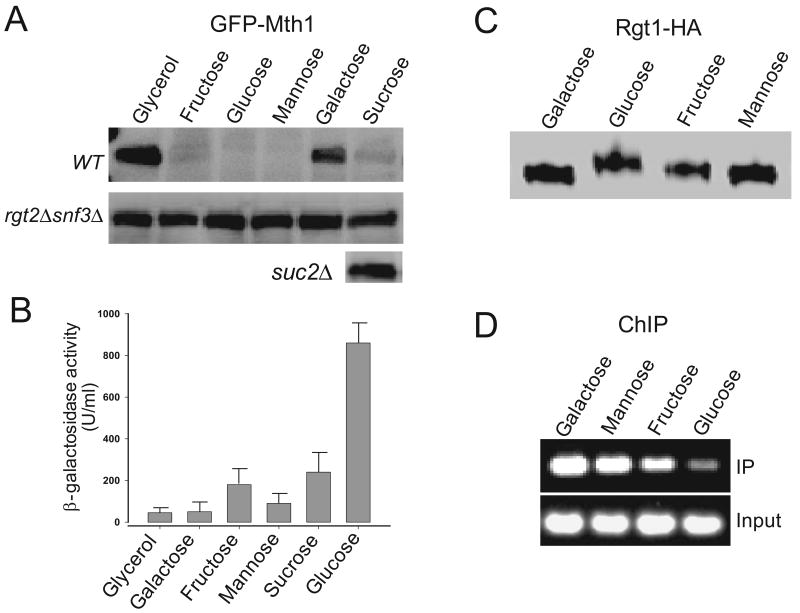
To understand the respective roles of the two pathways in glucose-regulation of Rgt1 function, we first attempted to identify sugars that can differentially activate the two pathways. To this end, we tested 15 sugars for their ability to activate the Rgt2/Snf3 pathway by measuring degradation of Mth1. The sugars tested include glucose, galactose, mannose, fructose, xylose, sucrose, lactose, maltose, cellobiose, raffinose, arabinose, allose, talose, isomaltose, and sorbose. Std1, the Mth1 paralog, is also subject to glucose-dependent degradation, but Std1 degradation is obscured by feedback regulation of glucose-induced STD1 expression (Kaniak et al., 2004; Kim et al., 2006). For this reason, Std1 was not thoroughly examined in this study. Mth1 degradation occurs in a glucose-sensor dependent manner in only cells grown on glucose, fructose, or mannose (Fig. 1A). In sucrose medium, Mth1 is degraded in wild type cells but not in the suc2 mutant cells that lack the invertase activity. Sucrose is a disaccharide that consists of glucose and fructose. Therefore, it is likely that the glucose sensors in sucrose-fermenting cells were activated by the glucose and fructose produced by hydrolysis of sucrose, rather than by sucrose itself. Glucose and galactose only differ with respect to C-4, yet galactose does not activate the glucose sensors, suggesting that the glucose sensors have remarkable substrate specificity.
Fig. 1.
Mth1 degradation does not directly activate expression of HXT1. (A) Yeast cells (wild type, rgt2Δsnf3Δ, and suc2Δ) expressing GFP-Mth1 under the control of the glucose-independent MET25 promoter were grown to mid-log phase (O.D600=1.2∼1.5) in minimal medium (MM) containing 2% galactose. Aliquots were washed with MM without sugar and then transferred to MM containing the various sugars (2%) indicated above in each lane. After incubation at 30°C for 30 min, yeast cell extracts were prepared and resolved by SDS-PAGE, followed by Western blotting with anti-GFP antibody. (B) Yeast cells expressing HXT1-lacZ were grown as described in panel (A). Triplicates of three different transformants were assayed for their β-galactosidase activity in permeabilized cells grown to mid-log phase. Activities are given in Miller Units. (C) Rgt1-HA proteins were analyzed by Western blotting using the HA-antibody. (D) Chromatin was precipitated from cells expressing Rgt1-HA with the anti-HA antibody as described in the Material and Methods section. The immunoprecipitated DNA and 1/100 of the input DNA (input) were used as templates for PCR amplification.
Glucose-Dependent Degradation of MTH1 is Required but is Not Sufficient for Induction of HXT1
Next, we explored whether Mth1 degradation directly results in induction of HXT1 by measuring expression of the HXT1-lacZ reporter gene (Fig. 1B). Interestingly, HXT1 expression is activated by glucose, but not significantly by fructose and mannose, even though all of these sugars have equal ability to induce degradation of Mth1 through the Rgt2/Snf3 pathway (Fig. 1A). Rgt1 proteins expressed in cells grown on glucose, fructose or mannose are differently phosphorylated (Fig. 1C) and have different binding affinities for the HXT1 promoter (Fig. 1D). This is perhaps due to different PKA activities in those cells, because the cAMP-PKA pathway is activated by glucose, but not significantly activated by fructose and mannose (Rolland et al., 2002).
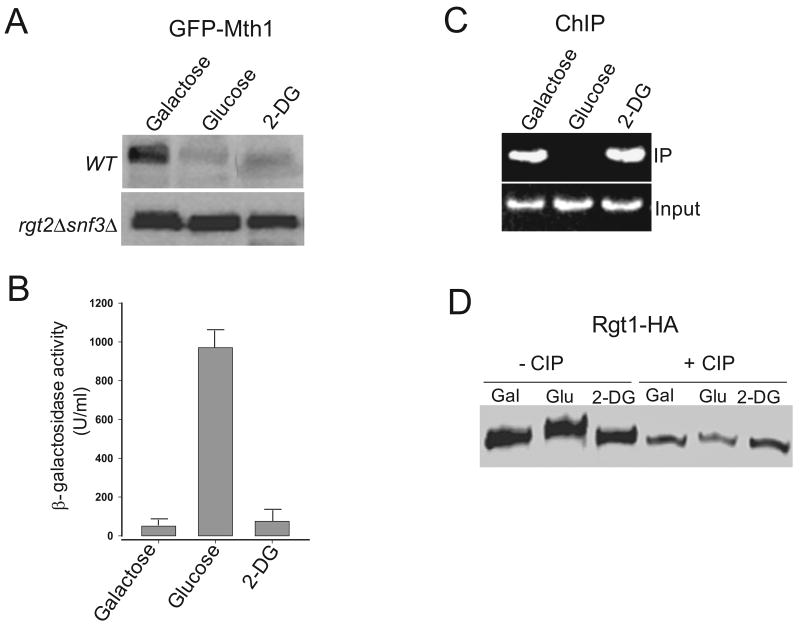
In yeast, glucose, fructose, and mannose are all taken up by the glucose transporters and metabolized (Reifenberger et al., 1995), raising the possibility that the sugar metabolites might affect induction of HXT genes. We examined this possibility by testing whether 2-deoxy glucose (2-DG), a glucose analog that is transported into the cells but not further metabolized, can induce expression of HXT1. Mth1 degradation normally occurs in 2-DG-treated cells in a glucose sensor-dependent manner (Fig. 2A), supporting the idea that glucose metabolism is not required for activation of the glucose sensors (Ozcan, 2002; Dietvorst et al., 2010). In those cells, however, neither significant expression of HXT1 (Fig. 2B) nor removal of Rgt1 from the HXT1 promoter is observed (Fig. 2C). In addition, the characteristic hyperphosphorylation of Rgt1 is not also observed (Fig. 2D). These results suggest that Rgt1 is not phosphorylated by PKA in those cells, because 2-DG does not activate the cAMP-PKA pathway (Lemaire et al., 2004).
Fig. 2.
2-DG activates the Rgt2/Snf3 pathway but not the cAMP-PKA pathway. Yeast cells were grown as described in Fig. 1, and the aliquots of the cells were transferred to MM containing galactose (Gal), glucose (Glu), or 2-DG (final concentration, 2%). Western blotting (A), β-galactosidase activity (B), and ChIP (C) assays were carried out as described in Fig. 1. (D) Immunoprecipitated Rgt1-HA was incubated with 10 U of calf intestinal alkaline phosphatase (CIP) (Roche) at 37°C for 30 min and analyzed by Western blotting using the HA antibody.
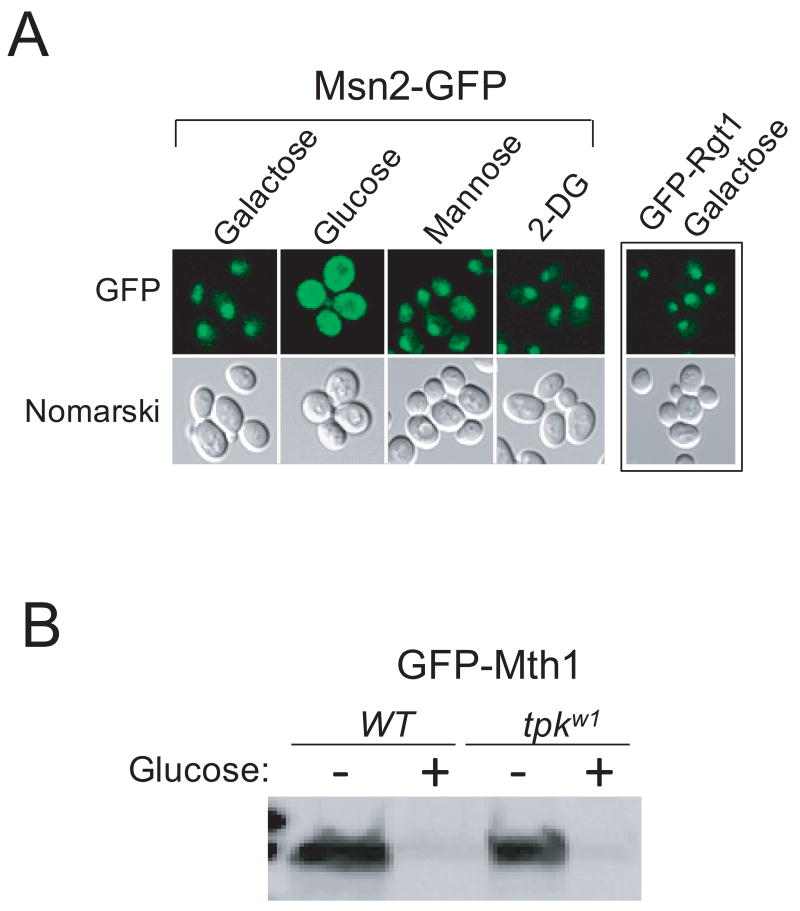
Since subcellular localization of Msn2, a transcription factor that is activated in stress conditions, is regulated by PKA-mediated phosphorylation, we examined the localization of Msn2-GFP in cells grown on the sugars tested above (Fig. 3A). Msn2-GFP (Huh et al., 2003) is found in the cytoplasm in cells grown in high-glucose medium, as reported previously (Smith et al., 1998). On the contrary, Msn2-GFP is localized to the nucleus in cells grown on galactose, mannose, or 2-DG, suggesting that the cAMP-PKA pathway is activated only by glucose but not the others. Taken together, our results indicate that Mth1 degradation is required but not sufficient for induction of HXT1, because Rgt1 is not released from the HXT1 promoter until it is phosphorylated by PKA.
Fig. 3.
PKA is not involved in degradation of Mth1. Yeast cells expressing Msn2-GFP were grown to mid-log phase under glucose-limiting condition (2% galactose). Aliquots of the cells were washed with MM and further incubated in MM containing the various sugars (final concentration, 2%) indicated above each lane. Localization of Msn2-GFP was examined by fluorescent microscopy. Rgt1, a nuclear protein (Kim et al., 2003), was used as a control. (B) GFP-Mth1 was expressed in wild type cells or cells carrying the tpkw1 allele (bcy1tpk1w1tpk2tpk3) that encodes a functionally attenuated PKA catalytic subunit and analyzed by Western blotting using the GFP antibody as described in Fig. 1.
The Target of the cAMP-PKA Pathway is the RGT1 Repressor but not its Corepressor MTH1
Our results above indicate that glucose-induction of HXT1 expression is achieved through the two glucose-dependent events—the degradation of Mth1 and the release of Rgt1 from DNA—that are regulated by an interaction between the two glucose signaling pathways. To gain more insight into these events, we determined whether PKA is also involved in degradation of Mth1. Our Western blot analysis shows that glucose-induced degradation of Mth1 normally occurs in the tpkw1 cells with attenuated PKA activity (Fig. 3B). There are two putative PKA consensus phosphorylation sites ((R-(R/K/S)-X-(S/T), phosphorylatable Ser or Thr are underlined (Kennelly and Krebs, 1991)) at Ser-112 and Ser-358 in Mth1. Changing these serine residues to alanines does not significantly inhibit degradation of Mth1 (Data not shown). Therefore, these results suggest that the cAMP-PKA pathway is not involved in the degradation of Mth1.
Glucose Induces Hyperphosphorylation of RGT1
Mosley et al. (2003) have shown that glucose-mediated phosphorylation converts Rgt1 from a transcriptional repressor to an activator. To identify the domains of Rgt1 that are phosphorylated in response to glucose, three different regions of Rgt1 [1-392 (I), 392-782 (II), and 782-1171 (III)] were isolated from cells grown in medium with or without glucose and treated with calf intestinal phosphatase (CIP). The mobility of LexA-Rgt1 fragments I and II are increased upon treatment with CIP, indicating that the changes in mobility are due to phosphorylation (Fig. 4A). These results are consistent with the previous results showing that Rgt1 lacking the first 392 amino acids, unlike the full-length Rgt1, does not exhibit the characteristic phosphorylation-dependent mobility shift upon SDS-PAGE (Mosley et al., 2003). Therefore, glucose-induced phosphorylation likely occurs in the amino-terminal region of Rgt1.
Fig. 4.
Glucose-induced Rgt1 phosphorylation occurs at its amino-terminal region. (A) Different domains of LexA-Rgt1 [1-392 (I), 392-782 (II), and 782-1171 (III)] were expressed in yeast cells grown in galactose- or glucose-containing medium. The proteins were harvested with the anti-LexA antibody-conjugated beads. The immunoprecipitated proteins were incubated with (+) and without (-) 10 U of calf intestinal alkaline phosphatase (CIP) (Roche) for 30 min and then subjected to Western blotting with the anti-LexA antibody (Santa cruz). (B) Tryptic phosphopeptide analysis of [32P]-labeled LexA-Rgt1. LexA-Rgt1 (1-392) were labeled with [32P] orthophosphate in yeast cells grown on galactose or glucose. Radiolabeled proteins were immunoprecipitated with the LexA antibody-conjugated beads and digested with trypsin. Tryptic phosphopeptides were analyzed on thin layer cellulose plates by electrophoresis followed by chromatography, and phosphopeptides were visualized by autoradiography.
To identify phosphorylated amino acid residues, LexA-Rgt1 (1-392) was metabolically labeled with [32P] in cells grown on galactose or glucose, and harvested using LexA-antibody conjugated beads. After incubation with trypsin, the tryptic phosphopeptides of [32P]-labeled LexA-Rgt1 (1-392) were resolved in 2-dimensional gels (Fig. 4B). The resulting autoradiographs show that there are four major phosphopeptides from galactose samples (No. 1 - 4) and five from glucose samples (No. 1 - 5) and that there are also several minor phosphopeptides present only in the glucose samples. These results suggest that the N-terminal Rgt1 is phosphorylated at multiple sites, and some of which are induced by glucose.
PKA Catalyzes the Phosphorylation of RGT1 that Converts it from a Transcription Repressor to an Activator
PKA was initially identified as an Rgt1 kinase by an in vitro phosphorylation assay (Kim and Johnston, 2006) using 119 known and predicted yeast kinases (Zhu et al., 2000). Rgt1 has 10 putative PKA phosphorylatable Ser residues, 5 of the 10 serines (positions at 146, 202, 283, 284, and 293) are well conserved in yeast. We have previously shown that changing the serines to alanines fully prevents glucose induction of HXT1 expression (Kim and Johnston, 2006). To identify the amino acid residues that are crucial for the activator function of Rgt1, all of these serines were individually mutated to alanines. The resulting mutant LexA-Rgt1 proteins were tested for transcriptional activation of the CYC1-lacZ reporter, which contains a lexO site in place of the CYC1 UAS (Ozcan et al., 1996) (Fig. 5A). Our results show that mutations altering Ser-202, Ser-283, or Ser-284 inhibit the Rgt1- and glucose-dependent transcriptional activation of CYC1-lacZ expression without affecting Rgt1 stability (Fig. 5B); in contrast, mutation at Ser-146 or at Ser-293 causes only about 25% reduction in expression of the reporter gene.
Fig. 5.

PKA-mediated phosphorylation at either Ser-202, Ser-283, or Ser-284 is sufficient for converting Rgt1 from a transcriptional repressor to an activator. (A) The serines in the 10 potential PKA phosphorylation sites in Rgt1 were changed to alanines, and the mutant Rgt1 proteins were tested for their ability to activate transcription of the CYC1-lacZ reporter. Cells were grown in MM-galactose to the mid-log phase and transferred to MM-glucose medium. β-galactosidase activity was assayed as described in Fig. 1. (B) Levels of some of mutant Rgt1 proteins (S202A, S283A, S284A, and 3SA) were analyzed by Western blotting with the anti-LexA antibody. (C) In vitro phosphorylation of LexA-Rgt1 by Tpk1. The wild-type LexA-Rgt1, LexA-Rgt1 (3SA) with the three serines substituted for alanines (S202A, S283A, and S284A), and Gst-Tpk1 were affinity-purified using the LexA-beads and Gst-beads, respectively. Gst-Tpk1 proteins were incubated with (+) or without (-) the LexA-Rgt1 proteins in buffer containing [32P] ATP for 30 min and resolved by SDS-PAGE and detected by autoradiography.
To make certain that PKA catalyzes phosphorylation of the three serines (Ser-202, Ser-283, and Ser-284), we examined the in vitro phosphorylation of the mutant Rgt1 (3SA) using Gst-Tpk1 as described previously (Kim and Johnston, 2006). As shown in Fig. 5C, Gst-Tpk1 phosphorylates wild-type LexA-Rgt but not alanine-substituted LexA-Rgt1 (3SA). These results suggest that PKA-mediated phosphorylation of any one of these residues is sufficient to convert Rgt1 from a transcriptional repressor to an activator.
Discussion
Derepression of Rgt1-repressed genes such as the HXT genes is achieved when Rgt1 is inactivated by disruption of the Rgt1 repressor-corepressor complex through crosstalk between glucose signaling pathways (Johnston and Kim, 2005). Cellular level of Mth1 is crucial for regulating Rgt1 function, and is tightly controlled by two mechanisms operating differently. First, Mth1 is highly regulated at the translational level; it undergoes glucose-dependent proteasomal degradation triggered by the Rgt2/Snf3 pathway. Second, the MTH1 gene is subject to transcriptional control by the Snf1-Mig1 pathway. Mig1, a downstream target of Snf1 kinase, binds to the MTH1 promoter and represses transcription of the gene. These results indicate that the Rgt2/Snf3 and Snf1-Mig1 pathways are intertwined in a regulatory circuit to reinforce Mth1 degradation by glucose repression of MTH1 (Kaniak et al., 2004). The present study describes that glucose-induced inactivation of Mth1 is not sufficient to suppress completely the repressor function of Rgt1. Rgt1 remains bound to the HXT1 promoter until it is phosphorylated by the cAMP-PKA pathway, which was identified as the third glucose signaling pathway that regulates Rgt1 function (Kim and Johnston, 2006; Palomino et al., 2006). It now becomes clear that the process to alter Rgt1 function consists of two consecutive steps: (1) disruption of the Rgt1 repressor-corepressor complex via degradation of Mth1; (2) phosphorylation-induced release of Rgt1 from DNA.
It is not fully understood why Rgt1 is regulated through these two distinctive steps, but this may be due in part to the dual function properties of Rgt1 acting as a transcriptional repressor and activator. The repressor function of Rgt1 is abolished through its intramolecular interaction (Polish et al., 2005) which occurs in a phosphorylation -dependent manner (Kim and Johnston, 2006). This process requires removal of Mth1; otherwise, it sterically hinders the phosphorylation-induced conformational change of Rgt1 by blocking access of PKA to Rgt1. PKA-mediated phosphorylation likely regulates Rgt1 function via two different modes; inhibition of its transcriptional repressor function and stimulation of its activator function. Rgt1 is differentially phosphorylated by PKA at the four putative PKA sites within its N-terminus. Phosphorylation of any one of the three serines—Ser-202, Ser-283, and Ser-284—is sufficient to convert Rgt1 to a transcriptional activator (Fig. 5); phosphorylation at all of these four serines—Ser-146, Ser-202, Ser-283, and Ser-284—results in a significant decrease in the DNA-binding activity of Rgt1 that leads to the suppression of its repressor function (Kim and Johnston, 2006). Therefore, the distinctive bifunctional properties of Rgt1 are conferred by its differential phosphorylation. We surmise that Rgt1 is readily phosphorylated at any one of these three serines and activates transcription, and then acts in this manner until it is removed from DNA upon phosphorylation at all of these four serines. Thus, this model is similar to the ‘hit-and-run’ mechanism as described previously (Cosma et al., 1999; Frolova et al., 1999; Fletcher et al., 2002). TAF12 (TAF61), a subunit of the TFIID and SAGA complexes, is also associated with Rgt1 (Sanders et al., 2002), suggesting that Rgt1 can directly activate transcription.
Acknowledgments
We thank Dr. Sabire Özcan (University of Kentucky, Lexington, KY) for providing plasmids. This work was supported by Grant GM087470 (JK) from the National Institute of General Medical Sciences, the National Institute of Health.
References
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cameron S, Levin L, Zoller M, Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell. 1988;53:555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Dietvorst J, Karhumaa K, Kielland-Brandt MC, Brandt A. Amino acid residues involved in ligand preference of the Snf3 transporter-like sensor in Saccharomyces cerevisiae. Yeast. 2010;27:131–138. doi: 10.1002/yea.1737. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford R, Warren BS, Hager GL. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol. 2002;22:3255–3263. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, Wittenberg C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell. 2003;14:3230–3241. doi: 10.1091/mbc.E03-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova E, Johnston M, Majors J. Binding of the glucose-dependent Mig1p repressor to the GAL1 and GAL4 promoters in vivo: regulationby glucose and chromatin structure. Nucleic Acids Res. 1999;27:1350–1358. doi: 10.1093/nar/27.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Johnston M, Kim JH. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:247–252. doi: 10.1042/BST0330247. [DOI] [PubMed] [Google Scholar]
- Kaniak A, Xue Z, Macool D, Kim JH, Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- Kim JH. Immobilized DNA-binding assay, an approach for in vitro DNA-binding assay. Anal Biochem. 2004;334:401–402. doi: 10.1016/j.ab.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Kim JH. DNA-binding properties of the yeast Rgt1 repressor. Biochimie. 2009;91:300–303. doi: 10.1016/j.biochi.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Brachet V, Moriya H, Johnston M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:167–173. doi: 10.1128/EC.5.1.167-173.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Johnston M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem. 2006;281:26144–26149. doi: 10.1074/jbc.M603636200. [DOI] [PubMed] [Google Scholar]
- Kim JH, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan J, Mosley AL, Ozcan S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet. 2003;44:19–25. doi: 10.1007/s00294-003-0423-2. [DOI] [PubMed] [Google Scholar]
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 2004;16:293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Lakshmanan J, Aryal BK, Ozcan S. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J Biol Chem. 2003;278:10322–10327. doi: 10.1074/jbc.M212802200. [DOI] [PubMed] [Google Scholar]
- Ozcan S. Two different signals regulate repression and induction of gene expression by glucose. J Biol Chem. 2002;277:46993–46997. doi: 10.1074/jbc.M208726200. [DOI] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino A, Herrero P, Moreno F. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 2006;34:1427–1438. doi: 10.1093/nar/gkl028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasula S, Chakraborty S, Choi JH, Kim JH. Role of casein kinase 1 in the glucose sensor-mediated signaling pathway in yeast. BMC Cell Biol. 2010;11:17–26. doi: 10.1186/1471-2121-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasula S, Jouandot D, 2nd, Kim JH. Biochemical evidence for glucose-independent induction of HXT expression in Saccharomyces cerevisiae. FEBS Lett. 2007;581:3230–3234. doi: 10.1016/j.febslet.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polish JA, Kim JH, Johnston M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics. 2005;169:583–594. doi: 10.1534/genetics.104.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol. 1995;16:157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x. [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielewoy N, Flick K, Kalashnikova TI, Walker JR, Wittenberg C. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol Cell Biol. 2004;24:8994–9005. doi: 10.1128/MCB.24.20.8994-9005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas-Cobos L, Sanz P. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem J. 2002;368:657–663. doi: 10.1042/BJ20020984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pierce M, Schneper L, Guldal CG, Zhang X, Tavazoie S, Broach JR. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:610–622. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol. 2009;5:245–258. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, Klemic KG, Smith D, Gerstein M, Reed MA, Snyder M. Analysis of yeast protein kinases using protein chips. Nat Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]