Abstract
The Gene Expression Omnibus (GEO) database is a major repository that stores high-throughput functional genomics data sets that are generated using both microarray-based and sequence-based technologies. Data sets are submitted to GEO primarily by researchers who are publishing their results in journals that require original data to be made freely available for review and analysis. In addition to serving as a public archive for these data, GEO has a suite of tools that allow users to identify, analyze and visualize data relevant to their specific interests. These tools include sample comparison applications, gene expression profile charts, data set clusters, genome browser tracks, and a powerful search engine that enables users to construct complex queries.
Keywords: database, microarray, next-generation sequence, gene expression, epigenomics, functional genomics, data mining
1. Introduction
The Gene Expression Omnibus (GEO) database (1) was launched in 2000 by the National Center for Biotechnology Information (NCBI) to support the storage, use, and dissemination of high-throughput gene expression data (2). High-throughput methodologies have evolved considerably since GEO’s inception to include both array- and sequence-based methodologies that generate a wide variety of functional genomics data types. Due to GEO’s flexible design and ability to store diverse data structures, GEO’s current holdings are much more diverse than implied by its name. Table 1 illustrates the diversity and relative quantities of both array- and sequence-based functional genomics studies that are currently represented in GEO.
Table 1.
Listing of GEO study types and the number of Series records with those types, correct at the time of writing. The types describe both the general application (e.g., expression profiling) as well as the technology (e.g., high-throughput sequencing). Users can retrieve studies of a particular type using the ‘DataSet Type’ field in the GEO DataSets query interface.
| Application | Technology | Number of Series |
|---|---|---|
| expression profiling | by array | 17988 |
| non-coding RNA profiling | by array | 348 |
| genome binding/occupancy profiling | by array | 73 |
| genome variation profiling | by array | 314 |
| methylation profiling | by array | 46 |
| protein profiling | by protein array | 31 |
| SNP genotyping | by SNP array | 151 |
| genome variation profiling | by SNP array | 272 |
| expression profiling | by genome tiling array | 305 |
| non-coding RNA profiling | by genome tiling array | 82 |
| genome binding/occupancy profiling | by genome tiling array | 849 |
| genome variation profiling | by genome tiling array | 410 |
| methylation profiling | by genome tiling array | 118 |
| expression profiling | by high throughput sequencing | 134 |
| non-coding RNA profiling | by high throughput sequencing | 234 |
| genome binding/occupancy profiling | by high throughput sequencing | 250 |
| methylation profiling | by high throughput sequencing | 31 |
| expression profiling | by SAGE | 206 |
| expression profiling | by RT-PCR | 25 |
| expression profiling | By MPSS | 21 |
Most data in GEO represent original research that is submitted by scientists who are publishing their work in a journal that requires its contributors to deposit data in a public repository as a condition of publication. Consequently, GEO now has supporting data for over 10,000 published manuscripts. In total, GEO is currently comprised of data from almost half a million public samples representing over 1300 different organisms submitted by over 8000 laboratories, and the submission rate exceeds 10,000 new sample deposits per month. GEO has been under constant development to keep up with the growing diversity of data and to provide useful tools to help researchers effectively query the database in order to identify data that are relevant to a specific area of interest (3). This chapter addresses the practical aspects of effectively utilizing GEO search mechanisms to find and retrieve data of interest, and explores the use of tools developed for visualizing and interpreting specific data types.
2. Methods
2.1. ‘GEO accession’ query box
This is a simple retrieval mechanism that works with Series (GSExxx), Sample (GSMxxx), Platform (GPLxxx) and DataSet (GDSxxx) accession numbers (see Note 1) to retrieve the queried entry. This feature is used primarily for straightforward retrievals of data that has been quoted in a publication when one has possession of an accession number and wishes to retrieve the corresponding GEO entry. To retrieve an entry using an accession number: (a) go to the GEO home page (1), (b) enter the accession number to be retrieved in the ‘GEO accession’ query box, (c) Click ‘GO’. The ‘GEO accession’ query box is also available at the top of most GEO pages.
2.2. Searching Entrez GEO DataSets and Entrez GEO Profiles
NCBI has a powerful search and retrieval system called Entrez that can be used to search the content of its network of integrated databases (4). This system can be used to query individual databases or all databases from a single interface (5). GEO data are available in two separate Entrez databases referred to as GEO DataSets and GEO Profiles.
2.2.1. Entrez GEO DataSets
The Entrez GEO DataSets search interface is directly accessible at (6). This ‘study-level’ database is where users can search for studies relevant to their interests. The database stores all original submitter-supplied records, as well as curated gene expression DataSets. As explained in Section 2.3, while GEO DataSets can be searched using many different attributes including organism, DataSet type, supplementary file types and authors, it is also possible to retrieve useful data simply by entering relevant keywords. For example, to find studies that examine lung cancer, just type “lung cancer” into the search box. Retrievals include a summary of each study matching the search criteria and a listing of the Samples they include.
2.2.2. Entrez GEO Profiles
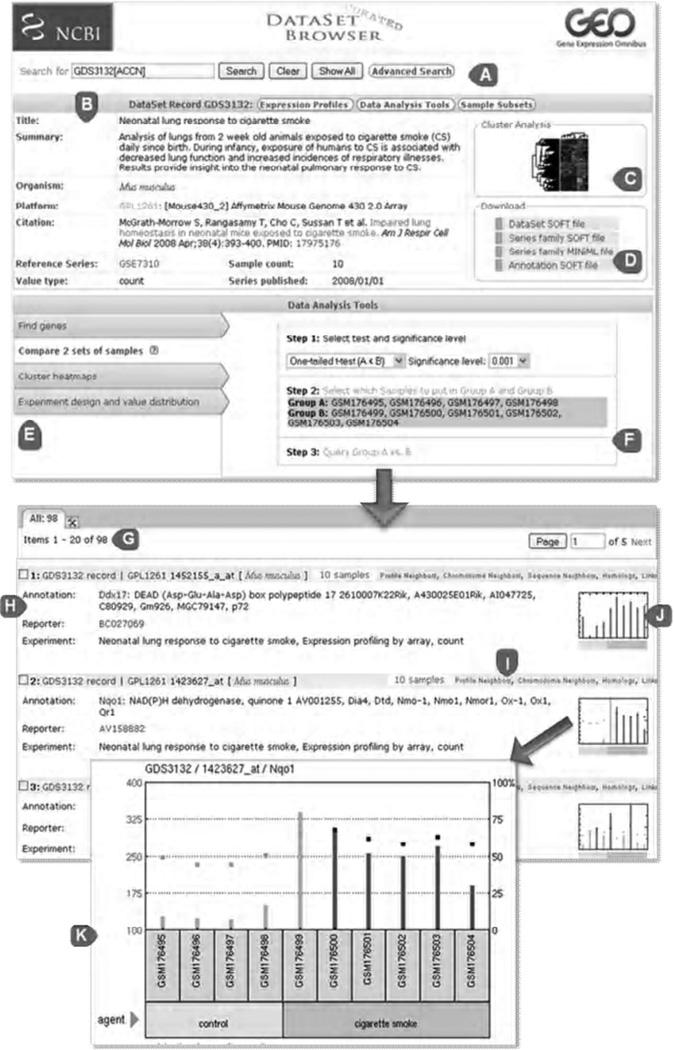
The Entrez GEO Profiles search interface is directly accessible at (7). This ‘gene-level’ database is where users can search for specific genes of interest, either across all DataSet records or within specific DataSets. The database stores individual gene expression profiles from curated DataSets (see Note 1; GEO Profiles are generated only for DataSet entries, so only a subset of GEO data is represented as profiles). As explained in Section 2.3, while GEO Profiles can be searched using many different attributes including gene names, GenBank accession numbers, Gene Ontology (GO) terms, or genes flagged as being differentially expressed, it is also possible to retrieve useful data simply by entering relevant keywords. For example, to find profiles for gene Nqo1, just type “Nqo1” into the search box. Retrievals include gene names and individual thumbnail images that depict the expression values of a particular gene across each Sample in a DataSet (Fig. 1). Experimental context is provided in the bars at the foot of the charts making it possible to see at a glance whether a gene is expressed differentially across experimental conditions. Clicking on the thumbnail image enlarges the chart to reveal the full profile details, expression values, and the DataSet subsets that reflect experimental design.
Figure 1.
Screenshot of a GEO DataSet record, data analysis tools, and corresponding GEO Profiles. (A) DataSet Browser search box. (B) Area containing descriptive information about that DataSet, including the title, summary, organism and citation ((27) for this example). (C) Thumbnail image of cluster heatmap. Click the image to be directed to the full interactive cluster from where regions may be selected and exported. (D) Download section containing various file format options; mouseover each option for description of content. (E) Data Analysis Tools options. Select from ‘Find genes’, ‘Compare 2 sets of Samples’, ‘Cluster heatmaps’ and Experiment design and value distribution’. (F) ‘Compare 2 sets of Samples’ analysis. In this example, the user has opted to perform a one-tailed t-test in order to find genes more highly expressed in mouse lung Samples exposed to cigarette smoke, compared to controls. (G) Results of the previous t-test; 98 genes were retrieved in this case. (H) Gene annotation area. (I) ‘Neighbors’ links that connect the targeted profile to genes related by expression pattern (Profile neighbors), sequence similarity (Sequence neighbors) or physical proximity (Chromosome neighbors). (J) Thumbnail image of gene expression profile. (K) Full profile image that in this example depicts how gene Nqo1 is more highly expressed in smoke-exposed Samples compared to controls. Each bar in the chart represents the expression level of Nqo1 in a Sample. The bars at the foot of the chart represent the experimental variables, in this case ‘control’ or ‘cigarette smoke’.
2.3. Advanced Entrez queries
As mentioned in the previous section, Entrez searches may be effectively performed by simply entering appropriate keywords and phrases into the search box. However, given the large volumes of data stored in these databases, it is often useful to perform more refined queries in order to filter down to the most relevant data. GEO data are indexed under many different fields. This enables sophisticated queries to be performed by restricting searches to specific fields and combining terms with Boolean operators (AND, OR, NOT) using the following syntax:
term[field] OPERATOR term[field]
A query tutorial page (8) was recently released to explain to users how to build complex, fielded queries in the GEO DataSets and GEO Profiles databases. The tutorial includes an exhaustive listing of the field qualifiers that are available for each database, as well as clickable examples to demonstrate their use (see Note 2). Furthermore, new tools are available on ‘Advanced Search’ and ‘Limits’ pages, which are linked from the Entrez home pages, to assist users to quickly construct multi-part, fielded queries.
Search Builder: This section includes a complete listing of all the fields that can be searched, and the values indexed under each field. To use, the following basic steps are performed: (a) select a search field from the drop-down menu, (b) type a search term -OR- select search term from list after clicking ‘Show Index’, (c) choose desired Boolean operator (AND, OR, NOT) and click ‘Add to Search Box’, (d) repeat steps 1–3 for additional search terms until query has been completed, and (e) execute search by clicking ‘Search’ (alternatively, click ‘Preview’ to see the result count of your query in the Search History section).
Limits: This section presents a specific box for several of the most popular and useful search fields. The user simply enters keywords, or selects search terms from the drop-down menus, hits ‘Add to Search Box’ and the query is automatically constructed.
Search History: This section stores the results of previous searches for up to eight hours (see Note 3). Each search is assigned a number, e.g., “#2”. Users can use these numbers to construct new queries or find the intersection of multiple queries., e.g., (#2 NOT #3) AND human.
Users typically perform multiple searches of both GEO DataSets and GEO Profiles to arrive at the data they are interested in. For example, if a user wants to locate studies that examine the effect of smoke on lung tissue, derived from any organism except human, and having raw Affymetrix .cel files, he could search GEO DataSets with:
(lung[Description] AND smok*[Description]) NOT human[Organism] AND cel[Supplementary Files]
At the time of writing, this search retrieves three independently-generated DataSets, GDS3622, GDS3548 and GDS3132. If the user then wants to search these three DataSets to see how his favorite gene, Nqo1, is expressed under these conditions, he could search GEO Profiles with:
(GDS3622 OR GDS3548 OR GDS3132) AND Nqo1[Gene Symbol]
This returns three profiles, all of which indicate that Nqo1 is upregulated upon smoke exposure in lung. If the user wants to explore any of these DataSets in more depth, he could use the advanced data mining tools described in Sections 2.4 and 2.5 and Fig. 1.
2.4. Advanced data mining features for GEO DataSets
As discussed in Note 1, DataSet records are assembled by GEO staff using the data and information derived from select Series records. In addition to querying the GEO DataSets interface for these records as discussed in the previous section, it is also possible to directly browse and query these entries using the ‘DataSet Browser’ (9) (Fig. 1). The Search bar at the top of the browser can be used to filter the list of DataSets by entering relevant keywords (e.g., heart, mouse, lymphoma, GPL81, etc.). Selecting a row in the browser displays the corresponding DataSet record in the panel below.
DataSet records have integrated ‘Data Analysis Tools’ (Fig. 1) that facilitate examination and interrogation of the data in order to identify potentially interesting genes. These tools include:
Find Genes: Allows users to retrieve specific expression profiles in that DataSet using gene names or symbols, or to retrieve expression profiles that have been flagged as potentially showing differential expression across experimental variables.
Compare 2 sets of Samples: Allows users to retrieve expression profiles based on specified statistical parameters. Users select which Samples to include in their comparison, the type of statistical comparison to be performed, and the significance level or cut-off to apply.
Cluster heatmaps: Allow users to visualize several types of precomputed cluster heatmaps of data and to select regions of interest for further study. GEO cluster heatmap images are interactive; cluster regions of interest may be selected, enlarged, charted as line plots, viewed in GEO Profiles, and the original data downloaded.
Experimental design and value distribution: Provides users with a graphic representation of the study’s experimental design showing experimental subsets, and a box and whiskers plot displaying the distribution of expression values of each Sample within the DataSet.
2.5. Advanced data mining features for GEO Profiles
The GEO Profiles results page (Fig. 1) includes features that enable users to identify additional gene expression profiles based on similarity to a given profile of interest, and to link to related information in other NCBI Entrez databases.
Profile Neighbors: Retrieves profiles with similar patterns of expression within the same DataSet. This feature assists in the identification of genes that may show coordinated regulation.
Chromosome Neighbors: Retrieves profiles for up to 20 of the closest-found chromosome neighbors within the same DataSet. This feature assists in the identification of available data for genes within the same chromosomal region.
Sequence Neighbors: Retrieves profiles based on BLAST nucleotide sequence similarity across all DataSets. This feature assists in the identification of profiles representing sequence homologs and orthologs.
Homologs: Retrieves profiles that belong to the same HomoloGene group across all DataSets. HomoloGene is a NCBI resource for automated detection of homologs among the annotated genes of several completely sequenced eukaryotic genomes.
2.6. Programmatic Access to GEO DataSets and GEO Profiles
The GEO DataSets and GEO Profiles databases can be accessed programmatically using a suite of programs collectively referred to as the Entrez Programming Utilities (E-utilities). GEO has a help page (10) describing some common examples and uses but more advanced users, for example those wishing to perform sophisticated retrievals using Perl scripts, should consult the E-utilities help page (11) for further guidance.
2.7. GEO BLAST query
This feature, linked to from the GEO home page, allows users to retrieve gene expression profiles based on BLAST (12) nucleotide sequence similarity. Entered nucleotide sequences or accession identifiers are queried against nucleotide sequences corresponding to the GenBank identifiers represented on microarray Platforms of DataSet entries. The initial output of a GEO BLAST query is similar to conventional BLAST output showing significant alignments between query and subject sequences. On the BLAST output page, users can click the “E” icon to view GEO Profiles corresponding to a particular subject sequence of interest. This query method can be used to find GEO data representing sequence homologs and orthologs, or for gaining insight into potential roles of uncharacterized nucleotide sequences.
2.8 Specialized resources for next-generation sequence data
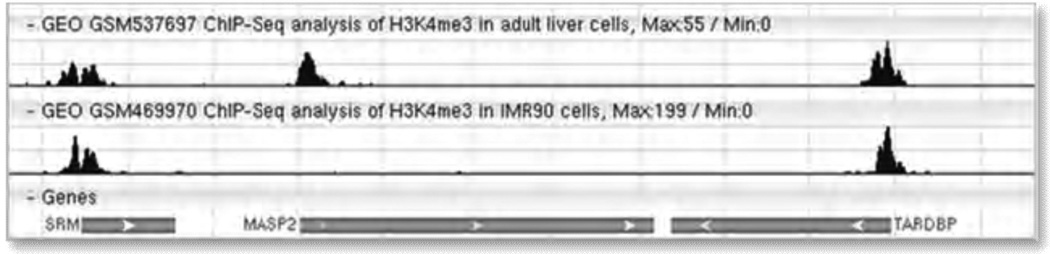
Increasingly, the microarray community is switching to next-generation sequence technologies to perform functional genomics analyses. Table 1 lists the major categories of sequence study types handled by GEO. GEO hosts the processed and analyzed sequence data, together with descriptive information about the Samples, protocols and study; raw data files are brokered to NCBI's Sequence Read Archive (SRA) database. Next-generation sequence studies can be located in GEO DataSets using the same search strategies as described for array-based studies. However, sequence data present new challenges in terms of data analysis and visualization. As a first step, hundreds of GEO Samples have been selected for integration into NCBI’s new Epigenomics resource (13). This resource maps the sequence reads to genomic coordinates to generate data ‘tracks’ that can be viewed using genome browsers. Multiple tracks can be viewed side-by-side, allowing data for specific genes to be visualized and compared across different Samples (Fig. 2). The GEO records selected for this advanced processing can be identified using the following cross-database search in GEO DataSets: "gds epigenomics"[Filter].
Figure 2.
Chromatin immunoprecipitation sequence (ChIP-seq) tracks displayed in NCBI’s Sequence Viewer. Histone H3 lysine 4 trimethylation (H3K4me3) peaks are typically observed at the 5′ end of transcriptionally active genes. In this example, there is a clear peak next to MASP2 in the adult liver cells (top track, GEO Sample GSM537697) but not in the IMR90 cells (lower track, GEO Sample GSM469970).
Additionally, GEO has a new centralized page (14) dedicated to the organization and presentation of next-generation sequence data derived from the NIH RoadMap Epigenomics Project. Features available on this page include the ability to link to the original GEO records, filter for records based on keywords, download data, and view selected Samples as tracks on either the NCBI Sequence Viewer or the UCSC Genome Browser (15).
2.9 Data Download
Data are made available for bulk download in several formats from the GEO FTP site (16) (see Note 4). There are currently five DATA/subdirectories:
SeriesMatrix/: This directory contains tab-delimited value-matrices generated from the VALUE column of the Sample tables of each Series entry. Files also include Series and Sample metadata and are ideal for opening in spreadsheet applications such as Microsoft Excel. Most users find SeriesMatrix files the most convenient format for handling data that have not been assembled into a DataSet.
SOFT/: This directory contains files in ‘Simple Omnibus Format in Text’ (SOFT). SOFT files are generated for DataSet entries, as well as for Series and Platform entries (subdirectories are included for each entry type). The Series and Platform files are actually “family files” that include the metadata and complete data tables of all related entries in the family. In contrast, the DataSet SOFT files include the metadata of the DataSet entry only, plus a matrix table containing the extracted gene annotations and Sample values used in GEO Profiles.
MINiML/: This directory includes files in MINiML (MIAME Notation in Markup Language) format. MINiML is essentially an XML rendering of SOFT format, and the files provided here are the XML-equivalents of the Series and Platform family files provided in the SOFT/ directory.
supplementary/: This directory contains supplementary files organized according to entry type (Platforms, Samples, Series). Platform supplementary files are typically related to the array design (e.g., .gal, .bpmap or .cdf), Sample supplementary files are typically native files representing raw (e.g., .cel, .gpr or .txt) (see Note 5) or processed data (e.g., .chp, .bed, .bar, .wig or .gff), and Series files would typically include results of upper-lever analyses such as ANOVA tables or significant genes lists. In addition, there is a compressed archive for each Series entry (GSExxxx_RAW.tar) that is comprised of the supplementary files gathered from all related Samples and Platforms. The ‘RAW’ part of the name is a misnomer since these files often include more than just raw data, but they enable users to download all supplementary files associated with a given Series entry in one step.
annotation/: This directory includes gene annotations for Platforms that participate in DataSet entries and, consequently, GEO Profiles. The annotations are derived by extracting stable sequence tracking identifiers directly from GEO Platform tables (e.g., GenBank accession numbers, clone identifiers, etc.) and using them to retrieve up-to-date gene annotations from the Entrez Gene and UniGene databases. This helps to ensure that the gene annotations associated with GEO Profiles are as up-to-date as possible.
3 Conclusions
Functional genomics assays employing microarrays and next-generation sequencing have become standard tools in biological research. Deposition of such data sets in public repositories is mandated by many journals for the purpose of allowing the research community to access and critically evaluate the data discussed in manuscripts. This requirement has resulted in astonishing growth in the numbers of studies and data types that are now available in the GEO database.
This chapter provides an overview of strategies for navigating the data in GEO and locating information relevant to the users’ particular interests. Approaches include simple and complex text-based searches, tools that identify genes with specific patterns of expression, as well as various easily interpretable graphical renderings of select data. GEO is a well-used resource, typically receiving over 40,000 web hits and 10,000 bulk downloads per day. A review of the literature reveals that the community is applying GEO data to their own studies in diverse ways; see (17) for a listing of over 1000 papers that cite usage of GEO data. It is clear that researchers use these data to address questions far beyond those for which the original studies were designed to address. Examples include using GEO data to test new algorithms (18), functionally characterize genes (19), create new added-value targeted databases (20), perform massive meta-analyses across thousands of independently-generated assays (21), and identify diagnostic protein biomarkers for disease (22).
GEO will continue to support these endeavors by improving the utility of the data in several ways, including enhancing data annotation standards, expanding integration with related resources, and by developing new analysis tools that can be used by as many users as possible.
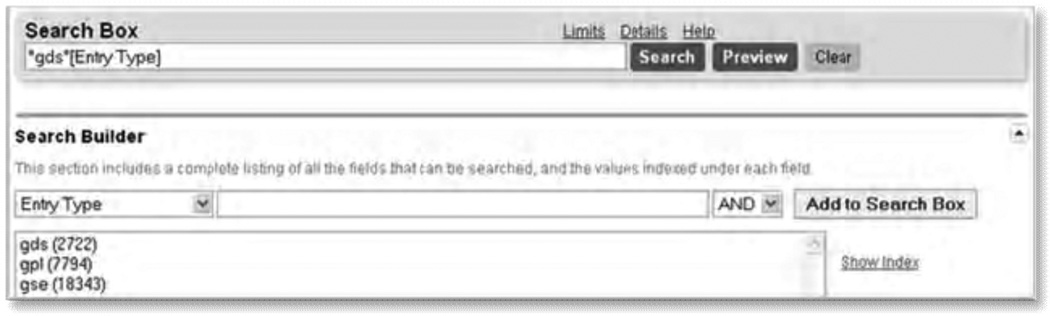
Figure 3.
Screenshot of Search Builder results, demonstrating fixed list terms for the ‘Entry type’ field.
Acknowledgments
This chapter is an official contribution of the National Institutes of Health; not subject to copyright in the United States. The authors unreservedly acknowledge the expertise of the whole GEO curation and development team—Pierre Ledoux, Carlos Evangelista, Irene Kim, Kimberly Marshall, Katherine Phillippy, Patti Sherman, Michelle Holko, Dennis Troup, Maxim Tomashevsky, Rolf Muertter, Oluwabukunmi Ayanbule, Andrey Yefanov and Alexandra Soboleva.
Funding
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
Entry types, accession codes and their relationships to each other are described in detail at (23). There are three primary entry types, referred to as Platform (GPLxxx), Sample (GSMxxx), and Series (GSExxx) entries. Platform entries are used to list the elements being detected by the experiment, e.g., oligonucleotide sequences, gene symbols or representative GenBank accession numbers. Sample entries are used to describe the biomaterials under investigation and the treatments to which they were subjected, and to provide access to the associated hybridization protocols and measurements. Series entries are used to group experimentally-related Samples and provide summary and design details. A fourth entry type, referred to as DataSets (GDSxxx), are assembled by the GEO curation staff from the three primary entries. DataSet entries contain essentially the same data and information as in the three primary entries, but the format has been arranged such that the submitter-supplied normalized data can be visualized and interrogated using downstream analysis tools. Only array-based expression data are currently considered for DataSet creation, and not all expression data qualify (for instance, due to having experimental designs or data processing methods that are incompatible with GEO tools). Furthermore, many expression studies have not yet been reviewed by the curation staff for DataSet creation. The net result is that only about 20% of the expression data in GEO are currently represented as DataSets and analyzable using GEO’s analysis tools.
- go to the GEO DataSets advanced search page (24)
- select the ‘Entry Type’ field from the drop-down list in the Search Builder section, and
- click ‘Show Index’
The results are shown as shown in Fig. 3. This result indicates that the GEO DataSets ‘Entry Type’ field can be queried only for “gds”, “gpl”, and “gse” terms. The numbers in parentheses are the total number of each entry type. For example, all DataSet entries can be retrieved by searching GEO DataSets with “gds[Entry Type]”. ‘Show Index’ can be used to see a listing of the indexed terms for any field listed in the drop-down list, but is mostly useful for identifying searchable terms for fixed list fields.
To save Entrez searches indefinitely, create a My NCBI account (25).When logged in, after performing your query you should see a 'Save Search' option next to the search box. Additionally, you will be presented with the option to receive e-mail alerts when new data matching your search criteria have been added to the database.
FTP directory content and file formats are described in detail in the README file (26). In many cases, direct links to the FTP site are provided on records. For instance, Series and Platform entries contain a direct link to their corresponding SOFT and MINiML family files, and SeriesMatrix files. Supplementary files are directly accessible using the links provided at the foot of Series, Sample and Platform entries, and DataSet entries contain links to the DataSet SOFT file, Series family SOFT and MINiML files, and the annotation SOFT file. SOFT and MINiML formats can also be exported using the toolbar located at the top of Series, Sample and Platform records. Furthermore, document summaries can be exported from the GEO DataSets and GEO Profiles results pages by setting the tool bar at the head of the page to ‘Send to: File’.
Studies that have supplementary files of specific types may be identified by constructing a query using the [Supplementary Files] field in GEO DataSets. This is useful for users who want to identify, download and reanalyze, for example, all .cel files for a specific Affymetrix platform.
References
- 1. http://www.ncbi.nlm.nih.gov/geo/
- 2.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. http://www.ncbi.nlm.nih.gov/gquery/
- 6. http://www.ncbi.nlm.nih.gov/gds/
- 7. http://www.ncbi.nlm.nih.gov/geoprofiles/
- 8. http://www.ncbi.nlm.nih.gov/geo/info/qqtutorial.html.
- 9. http://www.ncbi.nlm.nih.gov/sites/GDSbrowser/
- 10. http://www.ncbi.nlm.nih.gov/geo/info/geo_paccess.html.
- 11. http://www.ncbi.nlm.nih.gov/books/NBK25501/
- 12.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Fingerman IM, McDaniel L, Zhang X, et al. NCBI Epigenomics: A new public resource for exploring epigenomic datasets. Nucleic Acids Res. 2011;39:D908–D912. doi: 10.1093/nar/gkq1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. http://www.ncbi.nlm.nih.gov/geo/roadmap/epigenomics/
- 15.Rhead B, Karolchik D, Kuhn RM, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. ftp://ftp.ncbi.nih.gov/pub/geo/DATA/
- 17. http://www.ncbi.nlm.nih.gov/geo/info/ucitations.html.
- 18.Bhattacharya A, De RK. Divisive Correlation Clustering Algorithm (DCCA) for grouping of genes: detecting varying patterns in expression profiles. Bioinformatics. 2008;24:1359–1366. doi: 10.1093/bioinformatics/btn133. [DOI] [PubMed] [Google Scholar]
- 19.Pierre M, DeHertogh B, Gaigneaux A, et al. Meta-analysis of archived DNA microarrays identifies genes regulated by hypoxia and involved in a metastatic phenotype in cancer cells. BMC Cancer. 2010;10:176. doi: 10.1186/1471-2407-10-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogata Y, Suzuki H, Sakurai N, et al. CoP: a database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics. 2010;26:1267–1268. doi: 10.1093/bioinformatics/btq121. [DOI] [PubMed] [Google Scholar]
- 21.Liu S. Increasing alternative promoter repertories is positively associated with differential expression and disease susceptibility. PLoS One. 2010;5:e9482. doi: 10.1371/journal.pone.0009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen R, Sigdel TK, Li L, et al. Differentially Expressed RNA from Public Microarray Data Identifies Serum Protein Biomarkers for Cross-Organ Transplant Rejection and Other Conditions. PLoS Comput Biol. 2010;6(9) doi: 10.1371/journal.pcbi.1000940. e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. http://www.ncbi.nlm.nih.gov/geo/info/overview.html.
- 24. http://www.ncbi.nlm.nih.gov/gds/advanced/
- 25. http://www.nlm.nih.gov/pubs/techbull/jf05/jf05_myncbi.html#register.
- 26. ftp://ftp.ncbi.nih.gov/pub/geo/README.TXT.
- 27.McGrath-Morrow S, Rangasamy T, Cho C, et al. Impaired lung homeostasis in neonatal mice exposed to cigarette smoke. Am J Respir Cell Mol Biol. 2008;38:393–400. doi: 10.1165/rcmb.2007-0104OC. [DOI] [PMC free article] [PubMed] [Google Scholar]